Abstract
The implementation of fiber tracking or tractography modules in commercial navigation systems resulted in a broad availability of visualization possibilities for major white matter tracts in the neurosurgical community. Unfortunately the implemented algorithms and tracking approaches do not represent the state of the art of tractography strategies and may lead to false tracking results. The application of advanced tractography techniques for neurosurgical procedures poses even additional challenges that relate to effects of the individual anatomy that might be altered by edema and tumor, to stereotactic inaccuracies due to image distortion, as well as to registration inaccuracies and brain shift.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- Diffusion modeling
- Diffusion-weighted magnetic resonance imaging
- Fiber tracking
- Functional navigation
- Major white matter tracts
- Tractography
Introduction
Diffusion-weighted magnetic resonance imaging (DW-MRI) is a noninvasive imaging technique providing information about the microstructure of the brain in vivo. It is based on measuring the direction-dependent diffusion of water molecules, i.e., it depicts differences in tissue anisotropy. Diffusion is anisotropic, i.e., orientation-dependent, in areas with a strong aligned microstructure, like major white matter tracts.
DW-MRI has been increasingly used in imaging neuroscience over the last decade. An early form of this technique, diffusion tensor imaging (DTI) was rapidly implemented by major MRI scanner companies as a scanner selling point. Due to the ease of use of such implementations, and the plausibility of some of their results, DTI was leapt on by imaging neuroscientists who saw it as a powerful and unique new tool for exploring the structural connectivity of the human brain. However, DTI is a rather approximate technique, and its results have frequently been given implausible interpretations that have escaped proper critique and have appeared misleadingly in journals of high reputation [31].
Despite the wealth of publications, performing clinical research using DW-MRI is absolutely not straightforward. There is a plethora of available processing and analysis methods for DW-MRI, including multiple software platforms, data models, algorithms, and philosophies. Complicating the picture further, changes in the most commonly measured quantities are not specific to particular brain pathology. It is clear we can measure statistically significant brain changes with DW-MRI, but what do they mean? Today, the chief pitfall in applying DW-MRI to clinical research may well be the challenge of understanding and interpreting its meaning [53].
Tractography is the name given to any computational method that attempts to reconstruct white matter fiber tracts or “trace brain connections” based on DW-MRI data. Accurately estimating the course of the brain’s connections from DW-MRI is a difficult technical problem; therefore many methods for performing tractography have been developed [29, 53].
Most neurosurgical publications applying tractography techniques to real-world surgical procedures are dealing with DTI-based tractography approaches, despite that the DTI-based approach has several methodological limitations and pitfalls that may explain the failure of tractography in a clinical setting. Visualization of major white matter tracts has become routine in many neurosurgical centers due to the broad availability of free tractography software packages and integration of tractography algorithms in commercial navigation systems [14, 47, 51]. There are various reports on the beneficial effects of tractography techniques in several different neurosurgical procedures ranging from glioma surgery, surgery for vascular lesions, epilepsy surgery, and deep brain stimulation procedures [57]. However, there are also several reports and comments that emphasize that these techniques should not be used in the surgical environment uncritically [20, 21].
It is beyond this review to give a comprehensive overview on DW-MRI and tractography methods; it is rather the aim to show some examples for potential pitfalls of tractography techniques and to nevertheless provide some advice and how these techniques can be applied in a neurosurgical setting as well as to encourage to apply advanced DW-MRI and tractography methods.
DTI-Based Tractography
In 1994 the tensor model was introduced to describe the diffusion properties of water in white matter [6]. With DTI it was possible to noninvasively measure the organization and integrity of white matter fibers by quantifying the movement of water molecules inside the tissue. The tensor model was the simplest and probably most elegant way to characterize the diffusion, requiring only six parameters to be estimated. Understandably, the tensor model oversimplifies the underlying neuroanatomy. Thus it is important to interpret results derived from the tensor model with care [63].
Visualization
For visualization of these tensors, glyphs, generally defined as small visual representations of multivariate information, in the shape of ellipsoids were used. Isotropic diffusion can be represented as a sphere, whereas anisotropic diffusion is expressed as an ellipsoid, with the water molecules moving along the long axis of a fiber bundle and less movement perpendicularly. The ellipsoid can capture directionality and magnitude of all three eigenvectors. Tensors of rank two would be sufficient to describe the directionality of a voxel if the contents were all aligned in the individual voxel. However, mapping a single tensor at each voxel is not sufficient to describe more complex fiber configurations and is in fact misleading [42].
Tractography
Soon after the introduction of the tensor model describing the diffusion behavior, tractography algorithms were proposed to reconstruct 3-D trajectories of major white matter tracts. The basic aim of tractography is to compute paths through the directional information that is visualized using glyphs [7, 17, 32, 44].
Tractography is probably the most clinically appealing and understandable technique for representing major white matter tracts in the neurosurgical context. Various tracking algorithms which compare local tensor field orientations measured by DTI from voxel to voxel have been developed, allowing a noninvasive tracing of large fiber tract bundles in the human brain.
DTI gained a wide clinical application in brain tumors, spinal cord diseases, epilepsy, diffuse axonal injury, multiple sclerosis, Alzheimer disease, and ischemic stroke; for an overview, see [38]. DTI tractography was established in the clinical routine in neurosurgery in the last decade. This was facilitated due to multiple free software packages, as well as the integration of tractography modules in the major commercial navigation software systems, so that DTI-based tractography has gained a broad application in neurosurgery [23]. DTI tractography provides information about the course, the displacement, or interruption of white matter tracts around a tumor, and a widening of fiber bundles due to edema or tumor infiltration can be detected.
Deterministic/Probabilistic Tractography
There are several principally different approaches to reconstruct major white matter tracts. Most tractography algorithms in common use rely on line propagation techniques to delineate white matter pathways. This general class of methods is also often referred to as deterministic streamline fiber tractography. These rely on the identification of a suitable position from which to initiate the algorithm (the seed point), the propagation of the track along the estimated fiber orientation, and the termination of the track when appropriate termination criteria are met.
Noise in the DW-MRI measurements will inevitably introduce uncertainty in the estimated fiber orientations, which may in turn introduce errors in the delineated pathway. These errors can lead to completely different connections being identified, as a small error at one point in the track can cause the algorithm to enter and follow a different white matter pathway. Unfortunately, deterministic tractography algorithms only provide a single estimate of the path of white matter fibers from each supplied seed point, without any indication of the confidence interval that can be placed around this estimate. Probabilistic tractography algorithms attempt to address this limitation by providing their results in the form of a probability distribution, rather than a single “best fit” estimate. It should be emphasized that probabilistic methods are not more “accurate” than their deterministic counterparts, as they rely on the same underlying model. Many probabilistic tractography methods are based on deterministic techniques and hence suffer from the same limitations. As with deterministic approaches, manual guidance such as region of interest (ROI)-based editing may be needed to ensure the validity of the probabilistic results. The main benefit of probabilistic approaches, however, is that they can provide an estimate of the “precision” with which a tract pathway has been reconstructed. It is also critical to emphasize that the probability values produced by these algorithms are in no way related to the “connectivity” (e.g., number of axons) of the corresponding white matter pathways; they merely reflect the confidence that the particular connection of interest exists [63].
Compared to deterministic approaches in which the estimated fiber orientation (e.g., direction of maximum diffusivity for the tensor model) is assumed to represent the best estimate to propagate streamlines, probabilistic methods generate multiple solutions to reflect also the variability or “uncertainty” of the estimated fiber orientation. These methods, therefore, provide additional information on the reproducibility of each tractography reconstruction by mapping the intrinsic uncertainty of individual diffusion data sets. The magnetic resonance noise, partial volume effects, and inaccuracy of the chosen diffusion model mainly drive the uncertainty quantified by probabilistic tractography. Therefore, the probability of individual maps should not be considered as a direct measure of the anatomical probability of the tract. Indeed, in some cases trajectories based on artifacts can have high probability similar to true anatomical pathways. Ultimately, in datasets without noise, both deterministic and probabilistic approaches based on the same diffusion model would generate identical tractography maps [18].
A common misconception in the clinical setting is that the problems experienced using DTI-based tractography methods can be addressed by the application of more complex fiber tracking algorithms to fiber orientations estimated using the tensor model. The direct comparison of tensor-based data analyzed using a deterministic algorithm versus a probabilistic algorithm emphasizes that, while there remain some advantages to using probabilistic algorithms, the application of such an algorithm cannot compensate for fundamental limitations of the fiber orientation estimates obtained using the tensor model. The probabilistic tractography remains limited by the poor-quality fiber orientation information provided by the diffusion tensor model and does not alone provide an acceptable clinical solution [22].
Initial Neurosurgical Application
Bringing tractography techniques to neurosurgical applications the primary attempt was the visualization of the pyramidal tract, since the pyramidal tract represents the largest tract system, that should be traceable most easily and that should be most robust against tracking errors. DTI tractography was increasingly used in the resection of both high- and low-grade gliomas [1, 19]. More complex situations include the visualization of the optic radiation and the complex language tract system [12].
Challenges in Tractography
To highlight the aspects of challenges of tractography strategies that apply also for the clinical setting, some aspects of the reconstruction of the pyramidal tract are detailed as most practitioners feel that it is the easiest traceable structure. The placement of seed regions becomes crucial for reconstruction of complex tract systems, even for the most prominent tract system, the pyramidal tract. This is of importance as emphasized by Kamali et al.: Many DTI studies seed the ROIs for tractography of the corticospinal tract at the pons and higher levels including the midbrain, internal capsule, or motor cortex. These studies mix the corticospinal and corticopontocerebellar tracts specifically the frontopontocerebellar tract which runs side by side with the corticospinal tract and inserts into the motor cortex. By adding a ROI below the level of the pons, for example, at the pontomedullary junction or medulla, the corticopontocerebellar pathways will be excluded as they have already crossed to the contralateral cerebellum at the level of the pons. In vivo depiction of three-dimensional anatomy of the major white matter tracts by fiber tracking is becoming more commonly used in preoperative and intraoperative planning of lesions located close to these eloquent brain structures to avoid postoperative deficits. It is very important to realize that even small misplacement of the ROI for the tracking algorithm may result in significantly different reconstructed fiber tracts. Better understanding of technical limitations and accurate placement of ROIs to distinguish the complex anatomical relationships between fiber tracts are essential to avoid confusing the neighboring fiber bundles with variant physiologic significance [33].
The challenges facing DTI tractography of the corticobulbar tract have been the crossing fibers at the white matter of the superior corona radiata at the centrum semiovale just lateral to the lateral ventricles. At the centrum semiovale, there is a heavy load of fiber bundles directed in the anterior–posterior orientation such as the superior longitudinal fasciculus intersecting with the vertically oriented ascending and descending fiber bundles. These crossing fibers result in intra-voxel orientation heterogeneity, which lowers the sensitivity and specificity of both probabilistic and deterministic tractography algorithms. Complex fiber architecture within the voxel may result in abrupt abortion of the tractography algorithm (false-negative result) or creation of incorrect fiber bundles as a result of switching to adjacent fiber tracts (false-positive result) due to the crossing and kissing fiber phenomenon. This phenomenon has been a major obstacle for tractography of the corticobulbar tracts in DTI studies using the single-tensor model. Since the corticobulbar tract projects to the lateral aspect of the motor cortex, the neurons have to bend laterally at the centrum semiovale; hence crossing fibers become the major issue for fiber tracking of the corticobulbar tract. The same issue has been a major obstacle in the way of study of the lateral projections of the corticospinal tract corresponding to the somatotopic distribution of the motor cortex related to the arms and face using single-tensor tractography model [33].
Further Limitations
Despite of its fundamental limitations, DTI-based tractography is still the most widely applied tractography method in neurosurgical settings to delineate major white matter tracts. Correct identification of areas of fiber crossings is not possible by standard DTI because of its inability to resolve more than a single axon direction within each imaging voxel. Techniques, that can resolve multiple axon directions within a single voxel, try to solve the problem of white matter fiber crossings, as well as the problem to reconstruct the correct white matter insertions into the cortex.
The limitations of DTI tractography explain to some extent the heterogeneous and quite controversial evaluation of DTI tractography as a useful clinical tool for neurosurgeons ranging from positive statements like: “The concept of visualizing white matter tract anatomic relationships to guide surgical resection and clinical management certainly offers the potential of a paradigm shift in surgical practice. Just as neuronavigation has become the standard of practice across neurosurgery for discrete anatomic localization, the functional interplay of a given pathology and the eloquent cortical and subcortical structures, as revealed through multimodality imaging technologies, can only serve to improve the safety and efficacy of neurosurgical practice” [9] to very skeptical, even warning statements: “In summary, there is a double risk of DTI, (1) to not select a patient for surgery while the tumor was actually operable, or (2) to stop the resection prematurely, with a lower impact on the natural history of the disease. Last but not least, the risk for young neurosurgeons who use DTI regularly in the operating theater is becoming dependent on neuroimaging. The danger is for them to not learn optimally the functional anatomy of the brain by combining anatomic dissection, intraoperative electrical mapping, and models of cognitive neuroscience and thus to not be able to operate in the central nervous system without any intrasurgical neuroimaging, on the sole basis of their own mental imaging validated by online feedback provided by electrophysiology” [20].
Tractography in the Operating Room
Independent of the basic diffusion modeling and tractography method applied, there are additional challenges due to the specific neurosurgical setting compared to a pure neuroscientific approach that might have its sole focus in identifying and analyzing connectivity patterns.
Special problems in patients compared to healthy volunteers relate to the disease that might directly or indirectly affect the results of tractography. In patients, the time for imaging, i.e., the raw data acquisition, is restricted compared to volunteers, since patients might not be able to lie in a scanner for a longer time, without the risk of movement artifacts. In lesional cases the lesion itself, e.g., a tumor, and edema surrounding the lesion impede imaging and tractography. If the tractography results are to be integrated in a stereotactic/navigational setup, then the spatial accuracy of the raw data becomes a major concern.
The pyramidal tract as the most prominent white matter structure was the first target for intraoperative visualization and integration in modern navigation systems. This led to an extension of the concept of functional navigation [50], which was initially based on integrating functional data from functional magnetic resonance imaging (fMRI) or magnetoencephalography (MEG) for delineation of eloquent cortical brain areas. With the help of DTI tractography, also eloquent subcortical structures could be visualized in the surgical field.
Intraoperative Visualization
So what are the special needs of the neurosurgeon? For the visualization in the operating room and especially in the surgical field applying heads-up technology of modern navigation microscopes, most approaches provided by basic neuroscience did not satisfy the needs for neurosurgical use. Tensor glyphs visualizing DW-MRI data, as well as advanced renderings of anatomical tracts with graph-based representations of functional connectivity data visualizing the human connectome [42], all illustrate different aspects of major white matter tracts and connectivity; however these may not be suitable or the ideal solution for the representation in the clinical routine or even directly in the surgical field.
There is the risk of an information overflow when applying the heads-up display technology in operating microscopes distracting the neurosurgeon from the task of removing a tumor if a plethora of colored streamlines is superimposed on the surgical field. A flexible and user-driven intraoperative visualization is mandatory. For example, the interactive visualization of fiber tracts in the close proximity of a lesion allows an intuitive handling of the tractography data in the surgical context [25].
For clinical intraoperative use, the actual border of a major white matter tract is of main interest. A line or tube representation like the visualization in standard tractography lacks the ability to provide a border, so the user has to interpret the visualization as a model for the whole tract. The generation of hulls is a possibility to overcome this drawback. A surface wrapping a particular subset of previously computed streamlines represents a certain fiber tract bundle [47]. This results in an intuitive visualization of the tract system of interest as a 3-D object. The combination with volume rendering of anatomical MRI data provides a good spatial orientation. Alternatively to a wrapping approach of the streamlines from tractography, volume-growing techniques are also a possibility to generate 3-D objects representing major white tracts.
Spatial Accuracy
When tractography results are incorporated into a 3-D navigational setting, image distortion is an important factor influencing the spatial accuracy, independent of the tractography method and quality. Echo planar imaging (EPI) distortions are caused by magnetic susceptibility differences and concomitant fields and result in displaced tractography results. These distortions mainly manifest themselves as displacements along the phase encoding direction. The correction of EPI distortion using an image-based registration approach showed a significant improvement in tract consistency and accuracy [27, 43]. Propeller EPI is an alternative approach for distortion correction demonstrated in a Q-ball imaging study [15].
All challenges relating to navigation accuracy like registration errors and intraoperative events like brain shift [26, 45, 46] further diminish the spatial accuracy and apply also to tractography integrated in navigational settings [47–49, 51].
Image distortion, registration inaccuracies among different imaging modalities, patient registration inaccuracies, and inaccuracies due to positional shift, and brain shift, have to be taken into account, when intraoperative electrophysiological mapping methods are directly compared in a spatial fashion to tractography – this is independent to all the problems and challenges of DW-MRI, modeling the diffusion behavior and tractography itself.
Validation
To select an appropriate approach to delineate major white matter tracts for neurosurgical needs, the validation of tractography results would be a crucial factor. Tractography relies on complex mathematical models that provide anatomical information indirectly, which cannot be validated easily. In humans, up to now, tractography has mainly been validated by a qualitative comparison with data obtained from dissection. No quantitative comparison was possible because MRI and dissection data are obtained in different reference spaces and because fiber tracts are progressively destroyed by dissection. A recent paper proposes a method of fiber tract dissection in an ex vivo reference space [69], which might be able to overcome this shortcoming. There is no gold standard to which human tractography can be compared. Our knowledge of human neuroanatomy comes primarily from postmortem investigation using techniques such as the Klingler dissection technique and from invasive tracer injection studies in non-human primates. Particularly for white matter pathways involved in language, there could be significant interspecies variation that impacts how easily tracer studies translate to humans. Hence, validation of diffusion MRI consists of scanning animals in which we have a de facto ground truth connectivity profile from other means or scanning phantoms in which the connectivity is known [11].
Up to now in the clinical setting, there was only the possibility of indirect clinical validation by evaluating the postoperative motor or language function, visual field deficits correlation, etc. That is, the clinician analyzes whether the patients have a better neurological outcome and then concludes that the applied method seems to be beneficial. The major question is whether the tractography results actually reflect reality. First attempts correlating the DTI tractography findings to intraoperative electrophysiological measurements showed quite some discrepancies [34] which were probably mainly attributable to a distinct shifting of major white matter tracts during a neurosurgical procedure, which could be demonstrated by comparing pre- and intraoperative fiber tracking, acquired by high-field MRI applied during surgery [39, 49].
Maximal safety may require combining electrophysiological brain mapping with functional navigation that integrates fMRI/MEG data and DTI-based tractography acquired before or during surgery. Like intraoperative electrophysiological mapping can identify cortical eloquent brain areas, subcortical electrical stimulation helps to identify major white matter tracts intraoperatively. Recent studies emphasize that functional navigation and subcortical stimulation are complementary methods that may facilitate the preservation of pyramidal tracts [8, 54, 59].
The intraoperative visualization of the course of the pyramidal tract by microscope-based navigation during the resection of supratentorial gliomas has resulted in reduced neurological deficits, which may serve as a proof of concept per se. This is also supported by studies comparing pre- and postoperative reconstructions of major white matter structures in the brain stem well correlating to clinical deficits. Visual field deficits in temporal lobe surgery for pharmaco-resistant epilepsy provide an ideal model to analyze the clinical validity of changes in tractography by correlating the extent of visual field defects with the changes in pre- and intraoperative DTI tractography-based reconstruction of the optic radiation. The significant correlation between postoperative visual field deficits and the extent of alterations of the optic radiation also proved that reconstruction of major white matter tracts can be reliably used in a clinical setting [13, 56, 68].
All these data clearly support the concept of functional neuronavigation, i.e., adding functional information to 3-D anatomical datasets to reduce postoperative morbidity when operating lesions close to eloquent brain structures. Of course the intraoperative knowledge of the exact position of the pyramidal tract does not prevent neurological deficits per se; intraoperative events such as the necessity to coagulate small vessels close to the pyramidal tract may result in an injury of the pyramidal tract leading to neurological deficits. The distance of how close a reconstructed major white matter tract can be approached is not yet clearly defined. Analyzing DTI tractography-based navigation data in regard to the distance between tumor and pyramidal tract revealed that a distance of 5 mm seems to be a critical distance, which should be taken into account as safety margin [52]. This corresponds well to an identical critical distance of about 5 mm when approaching functionally eloquent cortical brain areas delineated by fMRI or MEG data.
Additional hulls around the reconstructed 3-D objects representing major white matter tracts are a possibility to visualize these safety margins. These encompassing hulls ideally would vary in thickness respective to the quality and reliability of the reconstructed fiber bundle. In case of noisy unreliable data, a thick hull would be added, while in highly reliable data, the hull would be thinner. The technical, as well as clinical, definition of the extent of these safety margins has still to be established.
Multimodal Navigation
The integration of tractography data into a navigation environment offers the possibility to correlate tractography findings in a multimodal setup, e.g., correlations with MR spectroscopy, and PET become possible. Besides preservation of neurological deficits, tractography-based navigation also allows to directly correlate histological findings with diffusion imaging parameters. Thus tumor invasion of major white matter tracts can be detected and quantified [60, 61].
Tumor Effects
Further important aspects influencing the quality of the reconstructed systems, especially when tractography is applied in the vicinities of a tumor, are the disruption of the fibers by tumor invasion and a widening of the fibers by the effects of edema surrounding a tumor. The standard diffusion modeling and tractography algorithms do not account enough for the effects of the edema and the tumor itself impedes the correct tracking so that either existing fibers are not visualized at all or even an erroneous tracking may result. An approach with generalized q-sampling imaging (GQI) seems to provide better results for visualization of tracts in edema than DTI-based tractography [70].
The special challenges for the neurosurgical application of tractography relate to the problem that different tract systems have different degrees of complexity, so that different reconstruction strategies might be necessary. The biggest and potentially most robust tract system in respect to visualization is the pyramidal tract. It might be sufficient to use a more simple approach to delineate the pyramidal tract than the complex language tract systems. However, the algorithms easily available for the neurosurgeons, due to the integration into the commercial navigation systems, might not be suitable for reconstruction of complex systems, despite the fact that they might produce trustworthy results for a large and more robust system.
Function and Tractography
It is important that the neurosurgeon is aware of the fact that the ability to reconstruct a tract system prior to surgery not automatically implies that the system the tract belongs to has normal function. On the other hand, alterations of reconstructed tract systems in pre-/postoperative comparison might well correspond to neurological deficits due to surgery. A conceptual issue is that DTI tractography should not be considered as a tool of functional mapping, but only as a tool allowing an indirect study of fiber anatomy [20]. Intraoperative monitoring with direct cortical stimulation and subcortical stimulation enables preserving essential tissue during surgery, while preoperative functional imaging including fMRI and DTI tractography allows to assess the surgical risk [55].
Combining the various methods from functional navigation, integrating fMRI and tractography information, in parallel to intraoperative electrophysiological measurements increases the safety for the patient. The different methods are not rivaling against each other but should be used in a complementary fashion.
In contrast to tractography in cohorts of healthy volunteers like it is applied in basic neuroscience, single-subject tractography like it is used and needed for surgical planning and guidance falls in the category of exploring unknown connectivity, because an individual variation even of known pathways, as well as a variation with disease, has to be expected. This is especially important when dealing with complex tract systems, e.g., like the language tracts [11].
Advanced Tractography Techniques
Diffusion-weighted imaging is inherently a noise-sensitive and artifact-prone MRI technique. To obtain a reliable representation of major white matter tracts, the following three steps are required in the process of DW-MRI tractography: the acquisition of appropriate diffusion-weighted image data; the correct estimations of fiber orientations, i.e., choosing the appropriate model; and finally the appropriate tracking algorithm.
Limitations of the DTI Approach
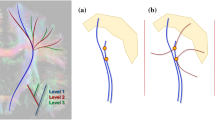
The DTI approach to model the complex anatomical diffusion information has some distinct limitations. It is far beyond the possibility of this chapter to provide a comprehensive overview on all different advanced techniques that are available and published. The review “Diffusion tensor imaging and beyond” by Tournier et al. [63] is recommended for further reading: Although attractive in its simplicity, the diffusion tensor model has been shown to be inadequate in the many regions of the brain that contain so-called “crossing fibers,” whereby two or more differently oriented fiber bundles are colocated within the same voxel. The term “crossing fibers” is itself somewhat misleading, as it includes any situation where multiple fiber orientations contribute to the signal measured for the same imaging voxel. Therefore, this also applies to configurations that may not initially have been thought of as “crossing fibers,” e.g., fiber bundles that “brush” past each other within the same imaging voxel, or even curving or “fanning” fibers (Fig. 1). Crossing fibers are endemic to DW-MRI, due to its coarse resolution (2–3 mm) compared with the white matter structures of interest; even the pyramidal tracts are only 3 mm thick in subcortical regions. Indeed, recent studies have shown that a significant proportion of the white matter contains crossing fibers with the most recent estimating that multiple fiber orientations can be detected in over 90 % of white matter voxels. Crossing fibers are even more problematic for tensor-based tractography methods: if one corrupt orientation estimate is encountered, the tracking algorithm may venture off course into an adjacent white matter structure, leading to both false-positive and false-negative connections. Moreover, the problem is far greater than might initially be expected: any given white matter tract of interest will traverse a large number of voxels, any of which might contain crossing fibers. It can readily be appreciated that the proportion of tracts traversing at least one affected voxel must be much greater than the proportion of affected voxels. If as much as 90 % of white matter voxels are affected, it is unlikely that any tracts will remain unaffected throughout their entire course.
A recent literature research by Abhinav et al. [3] identified 1838 papers dealing with fiber tracking or tractography among them the majority on DTI-based tractography approaches. Among the 735 papers applying tractography techniques beyond DTI, 84 papers with clinical applications were identified; among these 57 % applied a ball and stick (B&S) model and 15 % diffusion spectrum imaging (DSI), followed by 11 % for constrained spherical deconvolution (CSD) techniques. Sixty-four percent of the studies used probabilistic, and 36 % deterministic fiber tracking.
Advanced Diffusion Models
Among the advanced diffusion models replacing the DTI model as basis for tractography are sophisticated approaches including multitensor models, high angular resolution diffusion imaging (HARDI), hybrid diffusion imaging (HYDI), diffusion spectrum imaging (DSI), Q-ball imaging (QBI), Q-space imaging (QSI), the constraint spherical deconvolution model (CSD), and persistent angular structure MRI (PAS-MRI) [4, 5, 28, 37, 63–67]. These methods have gained increasing popularity, replacing the traditional tensor model for tractography. For instance, DSI and QBI use probability density functions instead of single tensors, which can describe the diffusion process in many different directions at each voxel. This however comes with the limitation of requiring longer acquisition times as it needs more encoding directions [58].
The major improvement for both probabilistic and deterministic tractography approaches is the introduction of these advanced diffusion models for the estimation of multiple fiber orientations. These models may be grouped in (overview taken from: [18]):
-
1.
Multiparametric methods (e.g., multitensor or “ball and stick” (B&S) models) are model-dependent approaches in which the diffusion data are fitted with a chosen model that assumes a discrete number of fiber orientations (e.g., two or more).
-
2.
Nonparametric, model-independent methods such as DSI, QBI, or diffusion orientation transform have been developed to better characterize the water molecular displacement by using a spherical function or the diffusion orientation distribution function (dODF). The multilobe shape of the dODF provides information on the number of fiber orientations, their orientation, and the weight of each fiber component.
-
3.
Methods that try to take advantage of both approaches by extracting directly the underlying fiber orientation (i.e., fiber ODF) using a specific diffusion model for white matter fibers. The latter approaches are usually described as spherical deconvolution methods and they generally show higher angular resolution (i.e., the ability to resolve crossing fibers at smaller angles) compared with methods based on dODFs. Spherical deconvolution methods are becoming the methods of choice in an increasing number of studies, as they require acquisition protocols that are close to clinical DTI protocols (e.g., low number of diffusion gradient directions and b-values that are accessible in most clinical scanners).
Advanced Tractography Algorithms
In parallel to all these advanced diffusion models, there are advanced tractography algorithms taking into account that with the new diffusion models describing multiple fiber directions in a single voxel, there is the increased risk of false-positive reconstructions. However, most of the current tractography algorithms are still based on the same tracking strategies originally introduced by the first tractography approaches. These strategies apply rules to avoid, for example, unrealistic fiber bending (i.e., angular thresholds) or tracking outside white matter regions (i.e., anisotropy thresholds) and are effective in reducing some of the reconstructions based on artifacts. Different approaches have been recently proposed to guide the propagation of the tractography algorithm across regions with multiple fiber orientations and try to discriminate between crossing, kissing, and bending configurations. Some of these approaches use “directional consistency” or similarity between fiber orientations across neighboring voxels; others use tract-specific properties or microstructural characteristics (e.g., axonal diameter) to propagate and differentiate tracts [18].
Global tractography is an alternative method in which the entire tract is generated simultaneously without a direct propagation of streamlines. By piecing together smaller tracts, the entire pathway is globally fitted to a chosen model that maximizes the consistency of the whole tract with the corresponding diffusion data. Because of small local errors, the final pathway can be formed by different anatomical tracts; for this reason anatomical constraints (or priors) are applied to distinguish between true tracts and artifacts.
In a recent paper giving an overview on global tracking techniques, Mangin et al. emphasize: “In the early days of MR diffusion-based tractography, the potential impact of the technique was so uplifting that the neuroscience community was comfortably blind to the “ill-posed” nature of the problem: the step-by-step reconstruction of a fiber bundle trajectory cannot afford any serious mistake in the evaluation of the local fiber orientations. This major risk was difficult to deal with because it does not exist in the well-known invasive techniques used with animals: a marker injected in a neuron is trapped inside the axon except when it can be transmitted into another neuron via synaptic connections. Hence invasive methods are not at risk of losing a bundle during tracking. Unfortunately, apart from the large bundles of deep white matter where axons are parallel, the evaluation of local fiber orientations in diffusion data is difficult. Indeed the myriad axons passing through a given MRI voxel usually have different orientations. Numerous ambiguities arise when one gets close to grey matter because of crossing, kissing and more exotic configurations. Considering the over-simplistic tensor models used at the beginning of the field, it is easy to understand why the first tractograms were full of spurious forks leading to barely exploitable connectivity maps” [41].
There are various technical attempts to approach the limitations of tractography; an agreed standard, or ideal solution, is not yet defined. It will be important to compare the different approaches especially in respect to their reliability and also clinical applicability.
So for the clinician, it is more or less impossible at the moment to find the right algorithm. Up to now, there is no definite solution. It seems to be obvious that advanced white matter imaging techniques have advantages over the simplified DTI-based tractography approach also in a clinical setting [2, 3, 10, 22, 35, 36, 70]. Figures 2 and 3 illustrate some typical advantages of advanced tractography techniques.
The DTI tractography reconstruction of the pyramidal tract (a) misses a lot of fibers that are part of the pyramidal tract connecting to the lateral aspects of the precentral gyrus; this is much better represented by advanced tractography techniques (b, multidirectional tractography with an HARDI/ODF approach based on spherical ridgelets)
DTI tractography of the arcuate fasciculus (a) often shows a false continuation that can be resolved correctly applying advanced tractography techniques (b), where the middle longitudinal fasciculus (red) is separated from the arcuate fasciculus (green) (tractography results rendered as 3-D objects: (a) DTI and (b) multidirectional HARDI/ODF approach based on spherical ridgelets)
The most widely used clinical tractography method (i.e., DTI-based tractography) results in systematically unreliable and clinically misleading information. Higher-order tractography models, using the same diffusion-weighted data clearly demonstrate fiber tracts more accurately, providing improved estimates of safety margins that may be useful in neurosurgical procedures. We therefore need to move beyond the diffusion tensor framework [22]. However, even highly sophisticated recently published methods like multi-tissue constrained spherical deconvolution approaches for the improved analysis of multi-shell diffusion MRI data might be compromised by the mass effect of brain pathologies, like a tumor or edema [30].
A comparison of several clinically available tractography programs [23] demonstrated significant anatomical differences among nine different tractography programs (NeuroQLab (modified tensor deflection [TEND] algorithm), Sörensen DTI task card (modified streamline tracking technique algorithm), Siemens DTI module (modified fourth-order Runge-Kutta algorithm), six different software packages from Trackvis (interpolated streamline algorithm, modified FACT (fiber assignment continuous tracking) algorithm, second-order Runge-Kutta algorithm, Q-ball [FACT algorithm], tensorline algorithm, Q-ball [second-order Runge-Kutta algorithm]), DTI Query (modified streamline tracking technique algorithm), Medinria (modified TEND algorithm), Brainvoyager (modified TEND algorithm), DTI Studio modified FACT algorithm, and the BrainLab DTI module based on the modified Runge-Kutta algorithm). As a main pitfall of clinical tractography, the fact is identified that results can easily be manipulated by “cleaning up” the image with exclusion regions of interest. The final image shows fibers only where the user wants them to be shown, potentially hiding relevant tracts; this can present a serious problem when planning a surgical procedure where displaced fibers could unintentionally be hidden.
The Medical Image Computing and Computer Assisted Intervention Society (MICCAI) performs so-called MICCAI challenges where different research groups compare their tractography approaches in relation to identical data (for details, see the website dti-challenge.org). A quantitative evaluation of ten different algorithms applying a diffusion phantom demonstrated, as expected, that single-tensor-based methods performed worse than others in crossing regions for the obvious reason that a single tensor is unable to correctly characterize the two-fiber compartment specific of those regions. However, the single DTI model is still able to correctly characterize numerous fiber bundles. Notably, the DTI model with only few degrees of freedom is by essence less sensitive to noise than more complex models, which often makes it the unique alternative in clinical applications. Second, in case of good-quality datasets, the best option seems to use a fiber orientation distribution function in conjunction with a streamline tractography algorithm where the next direction of propagation is directly inferred from the fiber orientation distribution (FOD) maxima. Indeed, with reasonable SNR (signal to noise ratio) datasets, FODs seem successful in modeling the fiber directions within a voxel and can be trusted. Finally, for datasets of medium and low quality as it is often encountered in real situations, several options are possible but all of them are using a spatial prior to make the model estimation more robust to noise. Conversely, without spatial prior, not any diffusion model was shown to correctly estimate the different fiber contributions within a voxel and consequently should be used with extreme caution [24].
Practical Guidelines and Proposal for the Future
Despite all complex possibilities and sophisticated developments in basic research, how can an individual neurosurgeon try to use “simple” tractography tools for the benefit of the patients? Most important seem to be: (1) to stay critical to the results provided by a software tool, (2) to compare the results of several tools, and (3) to have a good knowledge of anatomy, to be able to judge whether tractography results may be possible at all. Furthermore it is advisable not to rely exclusively on tractography to guide surgery, as well as, to combine several methods, like additional integration of functional MRI for identification of eloquent brain areas and like applying intraoperative electrophysiological methods, since tractography provides information on structure but not actual reliable information about function.
Guidelines toward a clinical application of tractography proposed by Chung et al. [16] are: always control the original image quality, choose the algorithm you understand, perform reproducibility tests on healthy subjects, and bear limitations in mind, when applying it to clinical practice.
Qualitative data from tractography-based studies with DTI or advanced white matter imaging techniques should be interpreted with a sound knowledge of the perilesional or loco-regional anatomy. In the setting of mass lesions, this problem is accentuated further as an abnormal trajectory of a fiber may be either technical in origin or be real. Good neuroanatomical knowledge therefore may help with an accurate interpretation of the data. In order to reduce the subjectivity of the interpretation of qualitative data, particularly in the setting of mass lesions, the use of a combined qualitative and quantitative approach may be helpful [2, 3].
The combined use of DW-MRI and cortical and subcortical stimulations could offer better information and a higher predictive value in preserving motor functions. For the validation and improvement of fiber tracking algorithms for neurosurgical planning, the use of electrophysiological data and functional image guidance can be very useful, especially under difficult pathological conditions. Using the most appropriate diffusion model combined with an adequate choice of tractography algorithm will increase the clinical relevance of the use of DW-MRI in neurosurgical planning. This reliability can be further improved by the combination with intraoperative mapping, which should be assessed systematically [10, 40].
Tractography for connectivity analysis in neuroscience has the problems of finding the exact termination of connections, detecting collaterals, tracking the very dense network of horizontal intracortical connections, discriminating between afferents and efferents, detecting synapses, etc. [29], which may be of lower relevance when applying tractography in neurosurgery, where these methods are mainly used to delineate a tract system close to a lesion, to decide on the extent of a resection or choosing a low-risk approach.
Future improvements in the accuracy of diffusion tractography will require innovations in MRI hardware, sequence design, data acquisition strategies, diffusion modeling, and tractography algorithms. Although such advances will lead to incremental improvements in the overall accuracy, they may not overcome the inherent ambiguities in inferring long-range axonal connectivity based on local diffusion displacement profiles. One suggestion, therefore, is to select the tractography method, or combination of methods, most appropriate for a specific objective. For example, if the objective is to reduce the possibility of identifying spurious pathways, a tractography method with better specificity, such as DTI, QBI, or B&S probabilistic tractography (using a conservative threshold), should be used. Alternatively, if it were the objective to reduce the likelihood of missing salient pathways, a tractography technique with relatively high sensitivity, such as CSD and B&S probabilistic tractography (using a liberal visualization threshold), would be more appropriate. For example, to avoid inadvertent transection of critical fibers of passage, as in the case of surgery for brain tumors, a tractography technique with low specificity but high sensitivity would be appropriate [62].
It is important to keep in mind that DW-MRI tractography alone is unlikely to provide an anatomically accurate map of the brain connectome. It is crucial to complement tractography results with a combination of histological or neurophysiological methods to map structural connectivity accurately [62].
The neurosurgical community has to find a consensus for adequate tractography strategies for different clinical situations and demands. It should be evident whether a rough estimation of the course of a well-defined tract system, which might be adequately described by a simple DTI-based tractography approach accompanied with some additional safety hulls, is sufficient for an intraoperative situation or whether the neurosurgeon has the demand to apply more reliable techniques; then advanced, sophisticated diffusion modeling and tractography approaches beyond the DTI approach have to be used.
Commercial navigation systems have to be open for the integration of research platforms, so that these sophisticated tractography approaches can be used in the clinical routine, and these techniques should also be implemented in the commercial systems directly.
A practical solution in the clinical setting includes besides using good clinical judgment the parallel application of different tractography approaches in complicated cases and comparing the results of these in each individual case. The clinician has to learn the pros and cons of the available software tools in the clinical setting like any surgical technique. Looking at the raw data should not be forgotten, using a cookbook strategy with an identical evaluation sequence to minimize errors is essential, and the additional use of complementary techniques like intraoperative mapping and monitoring to gain additional safety is recommended.
Conclusion
DW-MRI provides a unique way of probing tissue microstructure in vivo and noninvasively and is by far the most promising tool for studying white matter and its organization in living humans. It is, however, a difficult technique to apply correctly due to its unique imaging artifacts, the often very intricate interactions between microstructure and signal, the sophistication of the reconstruction algorithms used, and the shear complexity of white matter itself [63].
There is a distinct delay of application of modern methods to reconstruct white matter tracts in neurosurgical procedures compared to the state of the art of basic neurosciences. Progress was made in recent years in all three major steps for the reconstruction of white matter tracts: raw data acquisition, modeling the diffusion behavior, and tractography approaches. Each of these steps influences the others and is not independent. Despite of all these developments, still most neurosurgeons use the DTI tractography method, because it is easily available, e.g., as software package part of commercial navigation systems. It is mandatory that either these commercial systems become more open to facilitate integration of better solutions that exist outside the operating room or the technical advantages are directly implemented into these commercial systems, so that they are available for the whole neurosurgical community.
References
Abdullah KG, Lubelski D, Nucifora PG, Brem S (2013) Use of diffusion tensor imaging in glioma resection. Neurosurg Focus 34, E1
Abhinav K, Pathak S, Richardson RM, Engh J, Gardner P, Yeh FC, Friedlander RM, Fernandez-Miranda JC (2014) Application of high-definition fiber tractography in the management of supratentorial cavernous malformations: a combined qualitative and quantitative approach. Neurosurgery 74:668–680; discussion 680–681
Abhinav K, Yeh FC, Pathak S, Suski V, Lacomis D, Friedlander RM, Fernandez-Miranda JC (2014) Advanced diffusion MRI fiber tracking in neurosurgical and neurodegenerative disorders and neuroanatomical studies: a review. Biochim Biophys Acta 1842:2286–2297
Alexander AL, Wu YC, Venkat PC (2006) Hybrid diffusion imaging (HYDI). Conf Proc IEEE Eng Med Biol Soc 1:2245–2248
Assemlal HE, Tschumperle D, Brun L, Siddiqi K (2011) Recent advances in diffusion MRI modeling: angular and radial reconstruction. Med Image Anal 15:369–396
Basser PJ, Mattiello J, LeBihan D (1994) Estimation of the effective self-diffusion tensor from the NMR spin echo. J Magn Reson B 103:247–254
Basser PJ, Pajevic S, Pierpaoli C, Duda J, Aldroubi A (2000) In vivo fiber tractography using DT-MRI data. Magn Reson Med 44:625–632
Berman JI, Berger MS, Chung SW, Nagarajan SS, Henry RG (2007) Accuracy of diffusion tensor magnetic resonance imaging tractography assessed using intraoperative subcortical stimulation mapping and magnetic source imaging. J Neurosurg 107:488–494
Bi WL, Chiocca EA (2014) From localization to pathways: the continuing evolution of diffusion tensor imaging. World Neurosurg 82:e47–e48
Bucci M, Mandelli ML, Berman JI, Amirbekian B, Nguyen C, Berger MS, Henry RG (2013) Quantifying diffusion MRI tractography of the corticospinal tract in brain tumors with deterministic and probabilistic methods. Neuroimage Clin 3:361–368
Campbell JS, Pike GB (2014) Potential and limitations of diffusion MRI tractography for the study of language. Brain Lang 131:65–73
Chang EF, Raygor KP, Berger MS (2015) Contemporary model of language organization: an overview for neurosurgeons. J Neurosurg 122:250–261
Chen X, Weigel D, Ganslandt O, Buchfelder M, Nimsky C (2009) Prediction of visual field deficits by diffusion tensor imaging in temporal lobe epilepsy surgery. Neuroimage 45:286–297
Cho JM, Kim EH, Kim J, Lee SK, Kim SH, Lee KS, Chang JH (2014) Clinical use of diffusion tensor image-merged functional neuronavigation for brain tumor surgeries: review of preoperative, intraoperative, and postoperative data for 123 cases. Yonsei Med J 55:1303–1309
Chou MC, Huang TY, Chung HW, Hsieh TJ, Chang HC, Chen CY (2013) Q-ball imaging with PROPELLER EPI acquisition. NMR Biomed 26:1723–1732
Chung HW, Chou MC, Chen CY (2011) Principles and limitations of computational algorithms in clinical diffusion tensor MR tractography. AJNR Am J Neuroradiol 32:3–13
Conturo TE, Lori NF, Cull TS, Akbudak E, Snyder AZ, Shimony JS, McKinstry RC, Burton H, Raichle ME (1999) Tracking neuronal fiber pathways in the living human brain. Proc Natl Acad Sci U S A 96:10422–10427
Dell’Acqua F, Catani M (2012) Structural human brain networks: hot topics in diffusion tractography. Curr Opin Neurol 25:375–383
Dimou S, Battisti RA, Hermens DF, Lagopoulos J (2013) A systematic review of functional magnetic resonance imaging and diffusion tensor imaging modalities used in presurgical planning of brain tumour resection. Neurosurg Rev 36:205–214; discussion 214
Duffau H (2014) The dangers of magnetic resonance imaging diffusion tensor tractography in brain surgery. World Neurosurg 81:56–58
Duffau H (2014) Diffusion tensor imaging is a research and educational tool, but not yet a clinical tool. World Neurosurg 82:e43–e45
Farquharson S, Tournier JD, Calamante F, Fabinyi G, Schneider-Kolsky M, Jackson GD, Connelly A (2013) White matter fiber tractography: why we need to move beyond DTI. J Neurosurg 118:1367–1377
Feigl GC, Hiergeist W, Fellner C, Schebesch KM, Doenitz C, Finkenzeller T, Brawanski A, Schlaier J (2014) Magnetic resonance imaging diffusion tensor tractography: evaluation of anatomic accuracy of different fiber tracking software packages. World Neurosurg 81:144–150
Fillard P, Descoteaux M, Goh A, Gouttard S, Jeurissen B, Malcolm J, Ramirez-Manzanares A, Reisert M, Sakaie K, Tensaouti F, Yo T, Mangin JF, Poupon C (2011) Quantitative evaluation of 10 tractography algorithms on a realistic diffusion MR phantom. Neuroimage 56:220–234
Golby AJ, Kindlmann G, Norton I, Yarmarkovich A, Pieper S, Kikinis R (2011) Interactive diffusion tensor tractography visualization for neurosurgical planning. Neurosurgery 68:496–505
Hastreiter P, Rezk-Salama C, Soza G, Bauer M, Greiner G, Fahlbusch R, Ganslandt O, Nimsky C (2004) Strategies for brain shift evaluation. Med Image Anal 8:447–464
Irfanoglu MO, Walker L, Sarlls J, Marenco S, Pierpaoli C (2012) Effects of image distortions originating from susceptibility variations and concomitant fields on diffusion MRI tractography results. Neuroimage 61:275–288
Jansons KM, Alexander DC (2003) Persistent Angular Structure: new insights from diffusion MRI data. Dummy version. Inf Process Med Imaging 18:672–683
Jbabdi S, Johansen-Berg H (2011) Tractography: where do we go from here? Brain Connect 1:169–183
Jeurissen B, Tournier JD, Dhollander T, Connelly A, Sijbers J (2014) Multi-tissue constrained spherical deconvolution for improved analysis of multi-shell diffusion MRI data. Neuroimage 103:411–426
Jones DK, Knosche TR, Turner R (2013) White matter integrity, fiber count, and other fallacies: the do’s and don’ts of diffusion MRI. Neuroimage 73:239–254
Jones DK, Simmons A, Williams SC, Horsfield MA (1999) Non-invasive assessment of axonal fiber connectivity in the human brain via diffusion tensor MRI. Magn Reson Med 42:37–41
Kamali A, Hasan KM (2014) The importance of using a proper technique and accurate seeding of regions-of-interest in diffusion tensor tractography. J Neurol Sci 339:235–236
Kinoshita M, Yamada K, Hashimoto N, Kato A, Izumoto S, Baba T, Maruno M, Nishimura T, Yoshimine T (2005) Fiber-tracking does not accurately estimate size of fiber bundle in pathological condition: initial neurosurgical experience using neuronavigation and subcortical white matter stimulation. Neuroimage 25:424–429
Kuhnt D, Bauer MH, Egger J, Richter M, Kapur T, Sommer J, Merhof D, Nimsky C (2013) Fiber tractography based on diffusion tensor imaging compared with high-angular-resolution diffusion imaging with compressed sensing: initial experience. Neurosurgery 72(Suppl 1):165–175
Kuhnt D, Bauer MH, Sommer J, Merhof D, Nimsky C (2013) Optic radiation fiber tractography in glioma patients based on high angular resolution diffusion imaging with compressed sensing compared with diffusion tensor imaging – initial experience. PLoS One 8, e70973
Landman BA, Bogovic JA, Wan H, El Zahraa ElShahaby F, Bazin PL, Prince JL (2012) Resolution of crossing fibers with constrained compressed sensing using diffusion tensor MRI. Neuroimage 59:2175–2186
Lerner A, Mogensen MA, Kim PE, Shiroishi MS, Hwang DH, Law M (2014) Clinical applications of diffusion tensor imaging. World Neurosurg 82:96–109
Maesawa S, Fujii M, Nakahara N, Watanabe T, Wakabayashi T, Yoshida J (2010) Intraoperative tractography and motor evoked potential (MEP) monitoring in surgery for gliomas around the corticospinal tract. World Neurosurg 74:153–161
Mandelli ML, Berger MS, Bucci M, Berman JI, Amirbekian B, Henry RG (2014) Quantifying accuracy and precision of diffusion MR tractography of the corticospinal tract in brain tumors. J Neurosurg 121:349–358
Mangin JF, Fillard P, Cointepas Y, Le Bihan D, Frouin V, Poupon C (2013) Toward global tractography. Neuroimage 80:290–296
Margulies DS, Bottger J, Watanabe A, Gorgolewski KJ (2013) Visualizing the human connectome. Neuroimage 80:445–461
Merhof D, Soza G, Stadlbauer A, Greiner G, Nimsky C (2007) Correction of susceptibility artifacts in diffusion tensor data using non-linear registration. Med Image Anal 11:588–603
Mori S, Crain BJ, Chacko VP, van Zijl PC (1999) Three-dimensional tracking of axonal projections in the brain by magnetic resonance imaging. Ann Neurol 45:265–269
Nabavi A, Black PM, Gering DT, Westin CF, Mehta V, Pergolizzi RS Jr, Ferrant M, Warfield SK, Hata N, Schwartz RB, Wells WM 3rd, Kikinis R, Jolesz FA (2001) Serial intraoperative magnetic resonance imaging of brain shift. Neurosurgery 48:787–797; discussion 797–798
Nimsky C, Ganslandt O, Cerny S, Hastreiter P, Greiner G, Fahlbusch R (2000) Quantification of, visualization of, and compensation for brain shift using intraoperative magnetic resonance imaging. Neurosurgery 47:1070–1079; discussion 1079–1080
Nimsky C, Ganslandt O, Fahlbusch R (2006) Implementation of fiber tract navigation. Neurosurgery 58:ONS-292–ONS-303; discussion ONS-303–4
Nimsky C, Ganslandt O, Hastreiter P, Wang R, Benner T, Sorensen AG, Fahlbusch R (2005) Intraoperative diffusion-tensor MR imaging: shifting of white matter tracts during neurosurgical procedures–initial experience. Radiology 234:218–225
Nimsky C, Ganslandt O, Hastreiter P, Wang R, Benner T, Sorensen AG, Fahlbusch R (2005) Preoperative and intraoperative diffusion tensor imaging-based fiber tracking in glioma surgery. Neurosurgery 56:130–137; discussion 138
Nimsky C, Ganslandt O, Kober H, Moller M, Ulmer S, Tomandl B, Fahlbusch R (1999) Integration of functional magnetic resonance imaging supported by magnetoencephalography in functional neuronavigation. Neurosurgery 44:1249–1255; discussion 1255–1256
Nimsky C, Ganslandt O, Merhof D, Sorensen AG, Fahlbusch R (2006) Intraoperative visualization of the pyramidal tract by diffusion-tensor-imaging-based fiber tracking. Neuroimage 30:1219–1229
Nimsky C, Ganslandt O, Weigel D, Keller B, Stadbauer A, Akutsu H, Hammen T, Buchfelder M (2008) Intraoperative tractography and neuronavigation of the pyramidal tract. Jpn J Neurosurg 17:21–26
O’Donnell LJ, Pasternak O (2015) Does diffusion MRI tell us anything about the white matter? An overview of methods and pitfalls. Schizophr Res 161:133–141
Ohue S, Kohno S, Inoue A, Yamashita D, Harada H, Kumon Y, Kikuchi K, Miki H, Ohnishi T (2012) Accuracy of diffusion tensor magnetic resonance imaging-based tractography for surgery of gliomas near the pyramidal tract: a significant correlation between subcortical electrical stimulation and postoperative tractography. Neurosurgery 70:283–293; discussion 294
Ottenhausen M, Krieg SM, Meyer B, Ringel F (2015) Functional preoperative and intraoperative mapping and monitoring: increasing safety and efficacy in glioma surgery. Neurosurg Focus 38, E3
Piper RJ, Yoong MM, Kandasamy J, Chin RF (2014) Application of diffusion tensor imaging and tractography of the optic radiation in anterior temporal lobe resection for epilepsy: a systematic review. Clin Neurol Neurosurg 124:59–65
Potgieser AR, Wagemakers M, van Hulzen AL, de Jong BM, Hoving EW, Groen RJ (2014) The role of diffusion tensor imaging in brain tumor surgery: a review of the literature. Clin Neurol Neurosurg 124:51–58
Soares JM, Marques P, Alves V, Sousa N (2013) A hitchhiker’s guide to diffusion tensor imaging. Front Neurosci 7:31
Spena G, Panciani PP, Fontanella MM (2015) Resection of supratentorial gliomas: the need to merge microsurgical technical cornerstones with modern functional mapping concepts. An overview. Neurosurg Rev 38:59–70
Stadlbauer A, Hammen T, Buchfelder M, Bachmair J, Dorfler A, Nimsky C, Ganslandt O (2012) Differences in metabolism of fiber tract alterations in gliomas: a combined fiber density mapping and magnetic resonance spectroscopic imaging study. Neurosurgery 71:454–463
Stadlbauer A, Hammen T, Grummich P, Buchfelder M, Kuwert T, Dorfler A, Nimsky C, Ganslandt O (2011) Classification of peritumoral fiber tract alterations in gliomas using metabolic and structural neuroimaging. J Nucl Med 52:1227–1234
Thomas C, Ye FQ, Irfanoglu MO, Modi P, Saleem KS, Leopold DA, Pierpaoli C (2014) Anatomical accuracy of brain connections derived from diffusion MRI tractography is inherently limited. Proc Natl Acad Sci U S A 111:16574–16579
Tournier JD, Mori S, Leemans A (2011) Diffusion tensor imaging and beyond. Magn Reson Med 65:1532–1556
Tuch DS (2004) Q-ball imaging. Magn Reson Med 52:1358–1372
Tuch DS, Reese TG, Wiegell MR, Makris N, Belliveau JW, Wedeen VJ (2002) High angular resolution diffusion imaging reveals intravoxel white matter fiber heterogeneity. Magn Reson Med 48:577–582
Vos SB, Jones DK, Jeurissen B, Viergever MA, Leemans A (2012) The influence of complex white matter architecture on the mean diffusivity in diffusion tensor MRI of the human brain. Neuroimage 59:2208–2216
Wedeen VJ, Wang RP, Schmahmann JD, Benner T, Tseng WY, Dai G, Pandya DN, Hagmann P, D’Arceuil H, de Crespigny AJ (2008) Diffusion spectrum magnetic resonance imaging (DSI) tractography of crossing fibers. Neuroimage 41:1267–1277
Winston GP, Daga P, White MJ, Micallef C, Miserocchi A, Mancini L, Modat M, Stretton J, Sidhu MK, Symms MR, Lythgoe DJ, Thornton J, Yousry TA, Ourselin S, Duncan JS, McEvoy AW (2014) Preventing visual field deficits from neurosurgery. Neurology 83:604–611
Zemmoura I, Serres B, Andersson F, Barantin L, Tauber C, Filipiak I, Cottier JP, Venturini G, Destrieux C (2014) FIBRASCAN: a novel method for 3D white matter tract reconstruction in MR space from cadaveric dissection. Neuroimage 103:106–118
Zhang H, Wang Y, Lu T, Qiu B, Tang Y, Ou S, Tie X, Sun C, Xu K, Wang Y (2013) Differences between generalized q-sampling imaging and diffusion tensor imaging in the preoperative visualization of the nerve fiber tracts within peritumoral edema in brain. Neurosurgery 73:1044–1053; discussion 1053
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2016 Springer International Publishing Switzerland
About this chapter
Cite this chapter
Nimsky, C., Bauer, M., Carl, B. (2016). Merits and Limits of Tractography Techniques for the Uninitiated. In: Schramm, J. (eds) Advances and Technical Standards in Neurosurgery. Advances and Technical Standards in Neurosurgery, vol 43. Springer, Cham. https://doi.org/10.1007/978-3-319-21359-0_2
Download citation
DOI: https://doi.org/10.1007/978-3-319-21359-0_2
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-21358-3
Online ISBN: 978-3-319-21359-0
eBook Packages: MedicineMedicine (R0)