Abstract
The use of medical images by medical practitioners has increased to an extent that computers have become a necessity in the image processing and analysis. This research investigates if the Edge density and Local Directional Pattern can be used to characterize medical images. The performance of the Edge density and Local Directional Pattern features is assessed by finding their accuracy to retrieve images of the same group from a database. The combination of the Edge density and Local Directional Pattern features has shown to produce good results in both, classification of medical images and image retrieval. For the classification using the nearest neighbor and 5-nearest neighbor techniques yielded 98.2 % and 99.6 % classification success rates respectively and 99.4 % for image retrieval. The results achieved in this research work are comparable to other approaches used in literature.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
1 Introduction
There exist various medical imaging devices which have been used for many years in medicine. Magnetic resonance imaging (MRI), computerized tomography (CT), digital mammography, X-rays and ultrasound images provide effective means for creating images of the human body for the purpose of medical diagnostics. These allow medical professionals to clearly isolate different parts of the human body and determine if disease or injury is present and also improves the decisions made in treatment planning. With the increasing size and number of medical images being produced by various imaging modalities, the use of computers and image processing techniques to facilitate their processing and analysis has become necessary. These medical images can be characterized by the use of image processing techniques to assist medical practitioners with medical diagnostics. Medical images can be characterized by color, texture, shape and region-based descriptors.
In this paper, the Edge density and Local Directional Pattern features are investigated to determine if they can be used to classify medical images. This paper provides an overview of the system used and explains the details of the image enhancement and feature extraction techniques used. And finally assesses the results obtained from using the edge density feature to classify the medical images and to retrieve images from a database.
2 Background and Related Work
Prior to 2005, automatic classification of medical images was often restricted to a small number of classes and has evolved from a task of 57 classes to a task of almost 200 classes in 2009 [1]. Maria et al. [2] proposed a method to characterize mammograms into two categories; normal and abnormal. In their research four features where extracted; mean, variance, skewness and kurtosis, along with these features additional features were added to the training of the classification system, these features are; the type of tissue and the position of the breast. The results obtained where 81.2 % and 69.1 % using neural networks and association rule mining respectively. Yanxi et al. [3] developed a semantic-based image retrieval system centered classification using features that related to size, shape and texture obtained 80 % accuracy on classification and retrieval. Vanitha et al. [4] used the support vector machine (SVM) to characterize medical images and used three approaches for feature extraction, namely, structural, statistical and spectral approaches, with results of 97.5 % during training and 93.33 % during testing.
Edge information can provide essential information of an image, in the universal model for content-based image retrieval done by Nandagopalan et al. [5], edge histogram descriptors were used and compared with other statistical features such as colour and texture. The colour outperformed all other statistical features but the edge histogram descriptor was shown to be successful in precision and recall. In the survey done by Surya et al. [6], the feature proposed by Phung et al. [7], namely edge density, which differentiates objects from non-objects in an image using edge characteristics, was shown to have a good discriminating capability when compared to other features such as Haar-Like features. Phung et al. [7] have also used the concept of edge density to detect people in images and in this work edge density was also found to have stronger discriminating capabilities and can be easily implemented. The work done on texture classification and defect detection by Propescu et al. [8] had used many statistical features including edge density. It was found that edge densities once again had good discriminating capabilities but can achieve better results in texture classification by combining second order type statistical features.
3 Methods and Techniques
The system uses part of the content based image retrieval model proposed by [5]. The system is trained with a set of known images and tested with an unseen image. The feature vectors of both training and unseen images are constructed in exactly the same way. Firstly, the feature vector of every training image is taken and stored in a feature database and, secondly, the Euclidean distance is taken from the feature vector of the unseen image to every image in the feature vector database. The images are ranked and only the images corresponding to the feature vectors that produce the smallest Euclidean distance are retrieved. All images, before undergoing feature extraction, have to be preprocessed.
The medical images characterization system is divided into five processes; image preprocessing, feature extraction, building the feature vector, similarity comparison and image retrieval and classification. The image preprocessing is divided into three steps, finding the edges within the image and creating a new image consisting of these edges, then sharpening of the edges and the image is finally converted to a binary image. The image is then passed to feature extraction where the image is windowed and the edge densities of these windows are computed including the global edge density of the image, as well as computing the Local Directional Pattern features. After feature extraction the feature vector is constructed using these values and used for similarity comparison to finally retrieve images from the database and classifies the query image. The similarity comparison is done using the nearest neighbor method.
3.1 Preprocessing
Image preprocessing involves methods which enhance the quality of the images and prepare the image for feature extraction. Before feature extraction takes place the image is segmented by detecting the edges within the image followed by an enhancement to sharpen the edges. Finally the image is converted to a binary image. In this paper, the feature extraction is performed on the binary image produced by the preprocessing techniques. These preprocessing techniques used ensures that all images provided to the feature extraction mechanism contains pixels that are either black or white and that the edge density is only influenced by pixels that form the main edges of the image. This ensures that noisy edge pixels that have a negative influence on the results are removed.
Image Enhancement.
The edges within an image are of primary interest in this process. Image sharpening is performed using the Laplace filter given below.
Image Segmentation.
The segmentation process involves two steps, edge detection and converting to a binary image. The Sobel operator is used to detect the edges and produce a new image with clearly visible edges. The two filters below make up the Sobel operator.
As with the Prewitt operator the Sobel operator also performs smoothing before computing the gradient with an additional part of assigning a higher weight to the current line and column. H S x Performs smoothing over three lines and H S y performs smoothing over three columns before computing the x and y gradients respectively.
An adaptive threshold value t is determined from the processed images histogram. The value is used to convert the image to a binary image which will be used for feature extraction.
Where I(x, y) is the current intensity of pixel (x, y) and I t (x, y) is the resultant intensity after the threshold is performed. The new intensity value is either 0 or 1 where 0 is black and 1 is white.
3.2 Feature Extraction
The global and local edge densities are extracted from the binary image produced by the preprocessing techniques described. These include one global edge density value and seven local edge density values. The image is subdivided into seven smaller regions to obtain these local edge densities. The global edge density alone is not sufficient to distinguish between two images of different classes hence the use of local edge density to improve results.
Image Windowing.
The image is subdivided into seven smaller regions which overlap each other and the entire image region. These regions are given by \( \left( {x_{1} , y_{1} } \right) \) and \( \left( {x_{2} , y_{2} } \right) \), these are the two dimensional co-ordinates of the top-left corner and bottom-right corner of the image respectively.
The regions are described by Eqs. (4)-(11). Where TL i and BR i are the top-left and bottom-right corners of each region \( i = 0 \ldots n, n = 7 \). All co-ordinates are taken with the top-left corner of the image being (0, 0) and W, H are the width and height of the image respectively.
Edge Density.
For any region r with the top-left and bottom-right corners given by \( \left( {x_{1} , y_{1} } \right) \) and \( \left( {x_{2} , y_{2} } \right) \) respectively, and the edge magnitude of the pixels within r given by e (u, v), the edge magnitude of the region r is given by Eq. (12).
Where A r is the area of region r.
Edge Density Feature Vector.
The feature vector is composed of eight components, one component is the edge density of the whole image and the other seven components are the edge densities of the seven sub regions described by Eqs. (5)-(11). Because there is more than one region being used to compute the edge densities for the feature vector a more efficient approach is adopted from [9]. Let \( I\left( {x, y} \right) \) be the input image with height H and width W. The edge magnitude \( E\left( {x, y} \right) \) is computed using the filters given in Eq. (2) and is given by,
The edge magnitude is a combination of the horizontal and vertical edge strengths \( E\left( {x, y} \right)_{H} \) and \( E\left( {x, y} \right)_{V} \) respectively.
From the edge magnitude an integral image \( S\left( {x, y} \right) \) is computed by,
Where \( S\left( {x, y} \right) \) is the sum of edge magnitudes in a rectangular region {(1, 1), (x, y)}.
With just a single pass over the image to compute the edge magnitudes, given a sub region \( r = \left\{ {\left( {x_{1} , y_{1} } \right), \left( {x_{2} , y_{2} } \right)} \right\} \) and the computed edge magnitude integral image, the edge density of region r can be easily computed by Eq. (16).
The feature vector given below will be the final representation after all edge densities of each region within an image is computed.
Where v 0 is the global edge density and v 1 to v 7 are the local edge densities.
Local Directional Pattern (LDP).
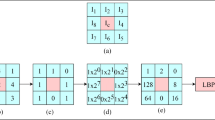
The Local Directional Pattern (LDP) is an eight bit binary code assigned to each pixel of an input [10, 11]. This pattern is computed by comparing the relative edge response value of a pixel in different directions. The eight directional edge response values of a pixel are calculated using Kirsch masks in eight different orientations (M 0-M 7).
Applying eight masks, eight edge response values are obtained \( m_{0} , m_{1} , \ldots q_{7} , \) each representing the edge significance in its respective direction. The presence of edge show high response values in particular directions. The k most prominent directions are used to generate the LDP. The computed top k values |m i | are set to 1 (in this case k = 3). The other (8-k) bit of 8-bit LDP is set to 0 [11].
Local Direction Pattern Feature Vector.
The medical image is represented using the LDP operator, I LDP . Then the histogram of LDP, H LDP is computed from the I LDP . Since k = 3 (from empirical experiments), the histogram labeled image, H LDP is a 56 bin histogram. H LDP histogram contains detail information of an image, such as edges, corner, spot, and other local texture features. In order to incorporate some degree of location information, the medical images are divided into n number of small regions, R 0; R 1; …; R n , and extracted the \( H_{{LDP_{{R_{i} }} }} \) from each region R n . These n H LDP histograms are concatenated to get a spatially combined global histogram for the global image.
3.3 Feature Normalization
The scales of Edge density features and Local Directional Pattern features are completely different. Firstly, this disparity can be due to the fact that each feature is computed using a formula that can produce various ranges of values. Secondly, the features may have the same approximate scale, but the distribution of their values has different means and standard deviations. A statistical normalization technique is used to transform each feature in such a way that each transformed feature distribution has means equal to 0 and variance of 1.
The similarity between the query image and a training image is given by the Euclidean distance between the two feature vectors. The distances from the query image to all training images are then ordered and images resulting in the smallest distance to the query image are regarded as the relevant images.
Where the i th entry of the test images feature vector is q i , t i is the i th entry of the query images feature vector and n is the dimension of the feature vector.
The classification of the query images uses the nearest neighbor technique. The query image will be classified as an image of the class of the closest image retrieved. The nearest neighbor technique is split into the first nearest neighbor (NN) and the 5-nearest neighbor (5-NN).
4 Results and Discussions
The experiments were carried out using a dataset of 2500 medical images obtained from the medical research centre. The dataset consisted of 125 images of each the researched body region images (hand, pelvis, breast, skull and chest), and other human body images. The researched model is evaluated in two ways, using content based image retrieval and using the nearest neighbor technique for classification. The researched model is used to assess the performance of the edge density feature, LDP features, and the combination of both features. The classification of the query image is based on the first nearest neighbor technique and using the 5-nearest neighbor technique. With the nearest neighbor technique, the class of the closest image to the query image is assigned to the query image, whereas with the 5-nearest neighbor, the class which has the most number of entities within the five closest images is assigned to the query image.
Table 1 shows the results of the classification accuracy along with the image retrieval precision achieved from the characterization of the images. It shows that the Edge density and LDP features complement each other in extraction local distinctive features for global characterization of images. Hence, the combination of the two techniques improved the overall accuracy rate. The 5-NN technique achieved better results than the NN method on all the techniques used. The results obtained show that the edge density and LDP can classify medical images and also be used in content based image retrieval systems for medical applications.
5 Conclusions
An effective approach based on Edge density and LDP for characterizing medical images has been presented. The results obtained were 98.2 % and 99.6 % classification success using the first nearest neighbor and the 5-nearest neighbor respectively. The LDP has also shown to work well in the retrieval of medical images. The results obtained are comparable to the results in the literature. The overall experimental results show that the proposed approach has a high true positive rate. Further investigation of the effect of the proposed approach with image processing in general is envisioned.
References
Tommasi, T., Deselaers, T: The medical image classification task, Experimental Evaluation in Visual Information Retrieval Series, vol. 32, 1st Edition (2010)
Antonie, M., Osmar, R.Z., Coman, A.: Application of data mining techniques for medical image classification. In: Proceedings of the Second International Workshop on Multimedia Data Mining (2001)
Yanxi, L., Deselaers, F., Rotthfus, W.E.: Classification driven semantic based medical image indexing and retrieval, The Robotics Institute, Curnegie Mellon University, Pittsburg
Vanitha, L., Venmathie, A.R.: Classification of medical images using support vector machine. Int. Conf. Inform. Netw. Technol. Singapore 4, 63–67 (2011)
Nandagopalan, S., Adigu, B.S., Deepak, N.: A universal model for content based image retrieval. World Academy of Science. Eng. Technol. 46, 644–647 (2008)
Surya, S.R., Sasikala, G.: Survey on content based image retrieval. Indian J. Comput. Sci. Eng. 2, 691–696 (2011)
Phung, S.L., Bouzerdoum, A.: A new image feature for fast detection of people in images. Int. J. Inform. Syst. Sci. 3(3), 383–391 (2007)
Popescu, D., Dobrescu, R., Nicolae, M.: Texture classification and defect detection by statistical features. Int. J. Circ. Syst. Signal Processing 1, 79–84 (2007)
Phung, S.L., Bouzerdoum, A., Detecting people in images: an edge density approach (2007)
Jabid, T., Kabir, H., Chae, O.: Gender classification using local directional pattern. In: International Conference on Pattern Recognition (2010)
Jabid, T., Kabir, H., Chae, O.: Robust facial expression recognition based on local directional pattern. ETRI J. 32(5), 784–794 (2010)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2015 Springer International Publishing Switzerland
About this paper
Cite this paper
Viriri, S. (2015). Characterization of Medical Images Using Edge Density and Local Directional Pattern (LDP). In: Kamel, M., Campilho, A. (eds) Image Analysis and Recognition. ICIAR 2015. Lecture Notes in Computer Science(), vol 9164. Springer, Cham. https://doi.org/10.1007/978-3-319-20801-5_43
Download citation
DOI: https://doi.org/10.1007/978-3-319-20801-5_43
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-20800-8
Online ISBN: 978-3-319-20801-5
eBook Packages: Computer ScienceComputer Science (R0)