Abstract
Genome structure in higher eukaryotes is highly dependent on the type and abundance of transposable elements, particularly retrotransposons , in their non-coding DNA. Retrotransposons are generally viewed as genomic parasites that must be suppressed in order to ensure genome integrity. This perception is based on the instances of retrotransposons having caused deleterious structural variation in genomes. Recent data are beginning to provide a more positive view of the impact of retrotransposons , particularly in mammals, where the evolution of the placenta has depended on the exaptation of a type of retrotransposon , endogenous retroviruses. Finally, exosome trafficking of retrotransposons between cells has been shown to induce the innate immune system gene expression, possibly indicative of a role for retrotransposons in the regulation of the innate immune system. It may be time for us to review the status of retrotransposons and reclassify them as symbionts rather than parasites.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
1 Evolutionary Origin and Structure of Retrotransposons
Genome structure and function are two sides of the same coin, and retrotransposons (AKA retrotransposable elements, retroelements and retroposons ), self-replicating DNA sequences that are found in all eukaryotic taxa, have the capacity to make larger changes to genome structure than other sources of variation—such as DNA polymerase errors that lead to single nucleotide variation (SNV). Because retrotransposons can account for the majority of the genome sequence in eukaryotes, their accumulation and clade specificity have been implicated in speciation, regulation of gene expression, exaptation and structural variation. Understanding the mechanisms that govern retrotransposon distribution and replication is thus of fundamental importance.
The evolutionary origin of retrotransposons is a matter of debate, but sequence similarity of their reverse transcriptases with the catalytic subunit of telomerase (Eickbush 1997; Lingner et al. 1997) and phylogenetic studies of reverse transcriptase sequences can be interpreted to indicate that reverse transcriptase may have evolved from telomerase, or telomerase is the result of co-opting reverse transcriptase. However, there are also good arguments for the ancient, prokaryotic origin of reverse transcriptase as a descendant of group II introns , which are mobile, self-splicing introns (Boeke 2003).
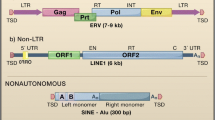
Retrotransposons can be divided into four major classes (Eickbush and Jamburuthugoda 2008). This classification is based on the reverse transcriptase enzyme required for replication and encoded by these elements. In vertebrates, retrotransposons can account for half of the genome sequence, and in plants, up to 70 % of the genome. This chapter is focused on the mammalian/vertebrate retrotransposons and these are commonly described as falling into two broad categories: those containing long terminal repeats (LTR) and those not containing LTR (non-LTR) (Jurka et al. 2007).
Non-LTR retrotransposons encode their own internal promoter and one or two open reading frames (ORFs) with reverse transcriptase and endonuclease activities that are used for replication (Fig. 4.1). LTR containing retrotransposons resemble (endogenous) retroviruses (ERVs) in that they can contain additional ORFs similar to those found in retroviruses , and these are referred to as endogenous retrovirus -like elements (ERVL). ERVL LTR retrotransposons are believed to have evolved from DNA transposons (Bao et al. 2010) and then acquired additional genes from viruses such as env, allowing them to become retrovirus -like and to produce infectious particles.
Retrotransposon life cycle: A TEs are transcribed by RNA Pol II and exported to the cytoplasm (Swergold 1990). B In the cytoplasm, ORF1 and ORF2 are both translated. The ORF1 protein (ORF1p) is an RNA-binding protein believed to aid the entry of LINE L1 RNA into the nucleus (Martin 2006). The ORF2 protein (ORF2p) has both endonuclease and reverse transcriptase activities (Feng et al. 1996; Moran et al. 1996). C To enter the nucleus, ORF1p and ORF2p form a complex with the L1 RNA known as a ribonuclear protein (RNP) (Martin 2006). D The endonuclease activity of ORF2p creates double-stranded breaks without insertion of TEs (Gasior et al. 2006). E The endonuclease activity is essential for the process of target-primed reverse transcription (TPRT). TPRT requires that ORF2p creates a nick in each strand at the integration site. The LINE L1 RNA is then used as a template for the reverse transcriptase activity of ORF2p (Cost et al. 2002). F L1 RNA is able to insert into and aid in repairing double-stranded breaks independent of the endonuclease activity of ORF2p (Morrish et al. 2002)
2 The Retrotransposon Life cycle
Retrotransposons replicate via an RNA intermediate that is reverse transcribed and reinserted into the genome (Fig. 4.1) at short target motifs (Fig. 4.2) (Cost and Boeke 1998). For non-LTR retrotransposons , also called long interspersed elements (LINE ), transcription is initiated by an internal Pol II promoter and the resulting transcript is then translated to produce two proteins, one of which, ORF2p has both reverse transcriptase and endonuclease activities (Feng et al. 1996; Moran et al. 1996). ORF2p has the ability to recognise short target sequences and initiate nicks at those locations which subsequently serve to prime the reverse transcription of the retrotransposon RNA directly into the genome (Eickbush and Jamburuthugoda 2008; Morrish et al. 2002).
Target-primed Reverse Transcription (TPRT) is how retrotransposons are inserted into the genome. ORF2p endonuclease activity creates a nick in the DNA at the AA/TTTT target site (Cost and Boeke, 1998). ORF2p reverse transcriptase activity then uses the cDNA copy as a template for DNA synthesis. Next ORF2p endonuclease activity creates a second nick in the DNA. The second DNA strand is then synthesised via double-strand break (DSB) repair and results in the formation of short target site duplications (TSD)
Some retrotransposons do not contain ORFs (non-autonomous) and are dependent on retrotransposons that do (autonomous) (Jurka et al. 2007). Autonomous retrotransposons are longer (LINE s), whereas the shorter, non-autonomous elements are called short interspersed elements (SINE s). While LINE s are usually ubiquitously distributed across taxa, SINE s are usually clade specific, as they result from the fusion of an internal promoter containing transcript with the 3’ end of a LINE .
The mechanism of SINE creation is still an open question, but most likely is a function of aspects of the LINE life cycle. SINE s have a composite structure: a 5’ end similar to 5’ tRNA, 7SL RNA or 5S rRNA promoters, a unique region and a 3’ end similar to the 3’ tail of LINE s (Piskurek and Jackson 2012). The most accepted hypothesis on SINE origins is based on the proposed template-switching mechanism of Buzdin et al. (Buzdin et al. 2002; Gilbert and Labuda 2000; Gogvadze and Buzdin 2009, Kramerov and Vassetzky 2005; Ohshima and Okada 2005). This template-switching mechanism is based on the study of pseudogenes, where the LINE (L1) reverse transcriptase switches from its own L1 mRNA to other nearby mRNA sequences through an RNA–RNA recombination process, thus creating new recombinant pseudogenes (and possibly SINE s) during L1 insertion (Buzdin et al. 2002; Gogvadze et al. 2007; Ichiyanagi et al. 2007; Piskurek and Jackson 2012). However, other investigators have suggested direct transposon into transposon (TnT) insertion as an alternative mechanism for the creation of novel transposable elements (Giordano et al. 2007; Ichiyanagi et al. 2007; Kriegs et al. 2007). The TnT mode of retrotransposon generation is what has led to the formation of SVA (SINE/VNTR/Alu) elements in humans, which are chimeric elements that can be mobilised by L1 elements and contain Alu-like sequence, Variable Number of Tandem Repeats (VNTR) sequence and SINE-R sequence resulting from a series of TnT events (Ostertag et al. 2003). The template-switching and TnT mechanisms are not mutually exclusive, and it is clear that both operate to create new SINEs, but at present we do not know which mechanism dominates.
Because retrotransposons can control their own expression through internal promoters [Pol II for LINE s and Pol III for SINE s and ERVs (Belancio et al. 2010a; Dieci et al. 2013)], expression is inextricably linked to the retrotransposon replication and to the evolution of new SINE s. As a result of this ability to autonomously insert new copies from expressed sequences into the genome, eukaryotes have evolved mechanisms to keep retrotransposon expression in check in order to avoid large-scale deleterious structural variation.
2.1 Retrotransposon Suppression
There appear to be two main mechanisms for retrotransposon suppression: transcriptional repression and post-transcriptional degradation (Fig. 4.3). Transcriptional repression can be caused by methylation of retrotransposon promoters or alteration of chromatin state to make retrotransposons transcriptionally inaccessible. Proof for the importance of methylation is evident from the phenotype of dnmt3l (DNA (cytosine-5)-methyltransferase 3-like) knockout mice (Bourc’his and Bestor 2004; Webster et al. 2005), which undergo meiotic catastrophe associated with the rampant expression of retrotransposons in male germ cells. The dnmt3l locus encodes a protein that regulates methyl transferase activity required to methylate and suppress the activity of CpG islands in retrotransposon promoters (Vlachogiannis et al. 2015). In addition to CpG island methylation , transcription can be repressed by the alteration of chromatin status (Fadloun et al. 2013), and this may be mediated by piRNA transported to the nucleus (Kuramochi-Miyagawa et al. 2008).
A schematic overview of retrotransposon suppression. Retrotransposons can be suppressed by different mechanisms throughout their life cycle (Crichton et al. 2014). Transcriptional suppression: In most cell types, retrotransposons are in a repressed state due to high levels of DNA methylation or histone modifications (Fadloun et al. 2013; Meissner et al. 2008). In some specific developmental stages and cell types, some retrotransposon RNAs can be transcribed bidirectionally and transported from the nucleus to the cytoplasm (Fadloun et al. 2013). Post-transcriptional suppression: Retrotransposon RNAs can be silenced through the piRNA pathway (mostly in the male germ line) or siRNA pathway (mostly in the female germ line). The ping-pong cycle is a well-characterised model for piRNA synthesis. In the mouse, sense retrotransposon RNAs are processed into primary piRNAs . MILI (or MIWI2) is recruited to cleave antisense retrotransposon RNAs into secondary piRNAs with the guidance of primary piRNAs , and mHEN1 is used to subsequently methylate their 3’ termini. Secondary piRNAs then bind with MIWI2 (or MILI) to cleave sense retrotransposon RNAs into primary piRNAs and close the loop of the ping-pong cycle (Aravin et al. 2008). piRNAs can also be transported to the nucleus to repress the transcription of retrotransposon by directing DNA methylation (Kuramochi-Miyagawa et al. 2008). For the siRNA pathway, sense and antisense retrotransposon transcripts can form double-strand RNAs, which are cleaved into double-strand siRNAs by DICER. Then, double-stranded siRNAs are unwound and loaded into the RISC to guide the degradation of retrotransposons (Ciaudo et al. 2013; Watanabe et al. 2008). Translational suppression: The Tudor domain-containing protein TDRD7 and MILI might be involved in the suppression of retrotransposon activity during translation (Grivna et al. 2006; Tanaka et al. 2011). Other repression mechanisms may also exist at later stages, such as the assembly stage of retrotransposon RNA and retrotransposon-encoded proteins (Goodier et al. 2012)
Post-transcriptional degradation of retrotransposon RNA in the male germ line is mediated by piRNAs derived from retrotransposon sequences and amplified by the ping-pong reaction (Aravin et al. 2008). In the female germ line, the situation appears to be different, with siRNAs shown to mediate retrotransposon transcript destruction via the RNA-induced silencing complex (RISC) pathway (Ciaudo et al. 2013; Watanabe et al. 2008).
There may also be additional mechanisms that can suppress retrotransposons at the translational level (Grivna et al. 2006; Tanaka et al. 2011) or even at the post-translational level to interfere with ORF proteins binding to retrotransposon transcripts (Fig. 4.3) (Goodier et al. 2012). In spite of all of these mechanisms to suppress retrotransposons at various steps in their life cycle, they are still transcribed at some developmental stages and in many somatic tissues (Belancio et al. 2010b). Perhaps suppression is a loaded term in this context and perhaps what we are observing is actually the regulation of retrotransposon expression.
2.2 Retrotransposon Expression
At certain phases of the mammalian life cycle, retrotransposons are negatively regulated to a lesser degree and are therefore transcribed and able to retrotranspose. Because methylation of cytosine to 5-methyl-cytosine (5mC) is critical to retrotransposon silencing, retrotransposons are potentially most active at times of low genomic 5mC content, which occurs in mouse embryos at around 3.5 days of embryonic development and also in primordial germ cells (Hackett and Surani 2013). However, it is primarily in early embryos that L1 retrotransposons are transcribed and retrotranspose (Kano et al. 2009). Presumably, other suppression mechanisms keep retrotransposons in check in primary germ cells. In spite of significant levels of global 5mC in the genome at other stages of development, retrotransposons are also activated in specific somatic tissues, indicating that retrotransposon suppression is more complex than just ensuring high levels of 5mC, and it may be less stringent in some tissues/cell types. Faulkner et al. (2009) showed that up to 30 % of mouse or human transcripts from all tissues are of retrotransposon origin and that retrotransposons were transcribed in all tissues surveyed. Retrotransposon expression per se does not always mean that retrotransposition is occurring, as some retrotransposons have inserted into UTRs and are therefore transcribed as part of a mRNA. However, it has been shown in both neural progenitor cells and in the human brain that retrotransposition does occur at a detectable level, altering the genomic landscape of that tissue (Baillie et al. 2011; Coufal et al. 2009).
Retrotransposon expression and subsequent retrotransposition have significant impacts on the genomes of both germ line (via germ line insertions and early embryonic insertions) and soma. Germ line insertions can then be transmitted through vertical inheritance, while somatic insertions are not currently believed to contribute to the vertical inheritance of novel insertions. However, there is another mode of retrotransposon transmission: horizontal transfer, where retrotransposon sequences jump to another cell or species, and this type of transfer may be the result of a more general mechanism of intercellular retrotransposon transfer.
3 Horizontal Transfer
Horizontal transfer of transposons has been demonstrated in plants, insects and vertebrates. In the context of retroviruses (including ERVs that have maintained ORFs to support an infectious life cycle), horizontal transfer is a relatively commonplace event. For example, in plants, horizontal transfer of transposable elements is both widespread and frequent (El Baidouri et al. 2014). In animals, horizontal transfer of DNA transposons is also widespread (Ivancevic et al. 2013). A good example is in Drosophila melanogaster where P-elements swept through the population starting in the 1950s via horizontal transfer (Daniels et al. 1990). Mariner elements are also horizontally transmitted between species, including both insects and mammals (Lampe et al. 2003; Lohe et al. 1995; Maruyama and Hartl 1991). Furthermore, Space Invader (SPIN) elements have been horizontally transferred in mammals and other tetrapods, as have OC1 elements (Gilbert et al. 2010; Pace et al. 2008). It was not until the 1990s that the first evidence for horizontal transfer of retrotransposons was published, when the patchy phylogenetic distribution and likely horizontal transfer of BovB retrotransposons was first reported (Kordis and Gubensek 1998, 1999a).
3.1 BovB : An Example of Widespread Horizontal Transfer
The BovB retrotransposon (also known as LINE-RTE) is a 3.2 kb LINE with at least one large ORF encoding a reverse transcriptase and a possible small ORF1 overlapping with the large ORF (Malik and Eickbush 1998). In cattle and sheep, over a thousand full length BovB, hundreds of thousands of 5’ truncated BovB fragments and derived SINEs (Bov-tA and Bov-tA2 (Lenstra et al. 1993; Okada and Hamada 1997) account for ~25 % of the genome sequence (Adelson et al. 2009; Jiang et al. 2014). The high degree of sequence conservation of BovB with sequences detected from the venom gland of Vipera ammodytes gave the first support to the idea of horizontal transfer of this retrotransposon (Kordis and Gubensek 1998, 1999b). BovB is now known to have a widespread, but patchy phylogenetic distribution, coupled to a high degree of sequence conservation, two of the hallmarks of horizontally transferred DNA (Fig. 4.4).
BovB phylogeny Maximum likelihood tree of aligned BovB sequences based on Walsh et al. (2013), showing the sporadic distribution, sequence similarity and abundance of BovB elements across taxa. Local support values are only shown if <0.9. The labels at each branch tip give the species common name and (in brackets) the percentage of genome sequence identified as BovB elements for that species. Reptile Tick 1 is Bothriocroton hydrosauri, Reptile Tick 2 is Amblyomma limbatum; and the BovB genome coverage for these ticks is unknown
Even though BovB has horizontally transferred across a wide range of species, it has not always colonised the genome to the same extent in different species. Some lineages such as ruminants and afrotheria have a high percentage of their genomes derived from BovB, whereas in other species BovB has not retrotransposed as prolifically (Fig. 4.4). This difference may be indicative of either variability in how different species suppress retrotransposons or it may simply reflect stochasticity in the population dynamics of retrotransposon expansion in different genomes. Presumably, the initial horizontal transfer event that results in retrotransposition and replication needs only a single germ line incorporation which can either replicate exponentially or “fizzle out” within the “genomic ecosystem” (Brookfield 2005; Le Rouzic et al. 2007). It is clear based on the currently available small and biased (towards mammals) sample of available genome sequences that retrotransposons as exemplified by BovB are capable of widespread and near ubiquitous horizontal transfer , and that this transfer might be enabled by parasites, such as ticks , that feed on blood. However, what is currently lacking is/are the molecular mechanism(s) for these transfers.
3.2 Possible Mechanisms/Modes of Transfer
A number of vectors , including arthropods , viruses, snails and DNA transposons , have been proposed for horizontal transfer, and the current state of knowledge was recently summarised by Ivancevic et al. (2013). It is relatively easy to see how a virus or transposon might act as a vector to package or transpose retrotransposons , but at the molecular level, it is not as obvious how eukaryotic vectors might effect the transfer of retrotransposon sequences between species, let alone into the germ line of another species.
3.2.1 Viruses as Vectors
For retrotransposons , the only example at present of a molecular virus vector is the taterapox virus (a dsDNA virus) which may have mediated transfer of Sauria SINE between reptiles and West African rodents (Piskurek and Okada 2007). This can be viewed as a highly unusual transfer, as a non-autonomous retrotransposon should not be as likely to colonise a new genome after transfer as an autonomous retrotransposon , such as a LINE . However, if cognate autonomous LINE s are present in both source and recipient species, a non-autonomous SINE could replicate effectively in the recipient species. RNA viruses have also been proposed as vectors of horizontal transfer for retrotransposons as they might package non-LTR retrotransposon transcripts inside infectious virus particles, but a tangible example for this type of transfer has yet to be demonstrated. Interestingly, Mariner-like DNA transposons are the plausible vectors for transfer of the CR1 retrotransposon in butterflies and moths (Sormacheva et al. 2012).
3.2.2 Endogenous Retroviruses /LTR Retrotransposons
As mentioned in Sect. 4.1, LTR retrotransposons are believed to have arisen from retrotransposons that acquired viral genes allowing them to become infectious, possibly leading to the evolution of retroviruses (Shimotohno and Temin 1981). In addition, waves of retroviral invasions into eukaryotic genomes have resulted in the formation of ERVs. While some ERVs have remained endogenous, occasionally they are able to become infectious and transfer to other genomes, where they can cause disease and eventually become domesticated. This is currently the case for a rodent ERV that has infected Koalas and is causing leukaemia in its new host while colonising the germ line as a new ERV (Tarlinton et al. 2006). Over time, domesticated retroviruses (ERVs) have contributed significantly to the genomic landscape of eukaryotes and have been co-opted into various aspects of eukaryotic biology (Feschotte and Gilbert 2012). In addition to this evolution of the capacity for horizontal transfer via infection, it is possible that retroviruses could package non-infectious non-LTR retrotransposons as a part of their viral payload. While there is no solid evidence for such transfer, exosomes /microvesicles are able to incorporate virus particles and transfer them to adjacent cells. This raises the question of whether exosomes can also transfer retrotransposon sequences directly.
3.2.3 Exosomes /Vesicles as Vectors
Exosomes are a class of membrane vesicle that has recently been shown to contain protein and RNA including miRNAs, piRNA s and retrotransposon sequences that they can transport from cell to cell (Batagov and Kurochkin 2013, Li et al. 2013; Skog et al. 2008; Valadi et al. 2007; Villarroya-Beltri et al. 2013; Yuan et al. 2009). Furthermore, exosome transport of Pol III -produced retrotransposon sequences has been specifically shown to regulate cancer therapy resistance pathways, including interferon -stimulated genes by direct activation of retinoid acid-inducible gene 1 (RIG-I) (Boelens et al. 2014). One of the hallmarks of Pol III transcripts is their 5’ triphosphate group, which is recognised specifically by RIG-I as a trigger for activation. Pol III is responsible for the transcription of primarily housekeeping-type genes such as tRNAs and rRNAs, but it also transcribes many other loci, including SINE s that have originated from a fusion of Pol III promoter containing transcripts with LINE 3’ sequences (Belancio et al. 2010b; Dieci et al. 2013). Because retrotransposons are known to be somatically expressed (see Sect. 4.2.2) in many tissues and cell types, they are likely to be present in exosomes exported by those cell types.
In the context of horizontal transfer , one can envision a number of potential scenarios for intercellular transport of retrotransposon sequences by exosomes (Fig. 4.5). Exosome-mediated transfer could allow transfer of retrotransposon sequences from a mammal or reptile to somatic cells of a parasite such as a tick through blood-borne exosomes . Within the tick , exosome -mediated transfer could then allow transmission to the germ line from the soma and eventual transmission back to other species used as food sources by that species of tick .
Possible scenarios of intercellular transfer of transposable elements via exosomes . TEs packaged in exosomes can be transferred between both somatic and germline cells. Within an organism, a TE can travel from a somatic, exosome-generating cell directly (e.g. through the blood) into a somatic, exosome-target cell by fusing with the plasma membrane and undergoing endocytosis. Similarly, TEs can be horizontally transferred between the somatic cells of different organisms or species, via some kind of vector (e.g. a parasite). Exosomes can also carry TEs from the soma to the germ line, making them a permanent change in the genome that is eventually passed down to the offspring. Note that for simplicity only entry to the male germ line is shown above. In addition to the transfer of TEs, once inside the target cell, this “foreign RNA” from the TE can trigger an interferon pathway response by inducing the interferon signal transduction pathway via RIG-I. For example, in ruminants , exosomes loaded with ERV/TE RNAs trigger pattern recognition receptors, stimulating the innate immune system and production of interferon -tau, which plays a role in pregnancy recognition and placentation (see Sect. 4.4.4)
While one might envision that the existing piRNA -based suppression system might degrade these retrotransposon sequences rapidly, it also appears that retrotransposon sequences (as exosome cargo) have been co-opted into a signalling role for the innate immune system in vertebrates and used to activate interferon-stimulated genes in the absence of interferon (Dreux et al. 2012; Li et al. 2013). This would not be the first time that retrotransposon sequences have been co-opted for gene regulation (Feschotte 2008; Feschotte and Gilbert 2012), but it introduces a new dimension of intercellular regulation of gene expression in the context of the evolutionary impact of retrotransposons .
4 Evolutionary Impacts
Retrotransposons are known to affect genome structure and hence function. The specific types of structural changes they introduce upon retrotransposition can have a wide-ranging set of subsequent effects in terms of genome structure, gene expression and gene function. More recently, it has become clear that retrotransposons have had a profound impact on the evolution of placentation in mammals.
4.1 Genome Structure
Retrotransposon insertion can directly perturb gene structure, but it can also have significant effects on a larger scale (Fig. 4.6). In particular, if retrotransposons form an array of elements with the same orientation on a chromosome, they can serve as a substrate for non-allelic homologous recombination (NAHR) leading to segmental duplication (Fig. 4.6a) (Stankiewicz and Lupski 2002; Startek et al. 2015). However, statistical analysis of repeats in flanking regions of segmental duplications found that only ~10 % of segmental duplications could be attributed to flanking repetitive elements (Zhou and Mishra 2005). Other types of rearrangements have been shown to result from arrays of repeats such as inversions (Fig. 4.6b) and gene conversion (Fig. 4.6c).
Retrotransposons can lead to changes in genome structure. a Changes in CNVs result from non-allelic homologous recombination (NAHR) caused by the insertion of many TEs from the same family (Stankiewicz and Lupski 2002; Startek et al. 2015). b Chromosomal inversion is also the result of NAHR (Stankiewicz and Lupski 2002). c SINE elements have potential to drive change through gene conversion (Roy et al. 2000)
While it is clear that retrotransposons can have indirect effects on genome structure as mentioned above, given the limitations inherent in identifying small segmental duplications and copy number variants the precise magnitude of these effects is unknown.
4.2 Gene Expression
As shown in Fig. 4.7, transposable elements can insert into and next to genes, affecting gene expression through multiple mechanisms, including epigenetic silencing of transcription, shortening a transcript via premature poly-Adenylation, driving piRNA expression or altering 3’ UTR structure to affect mRNA stability. Analysis of retrotransposon insertions into or near genes has shown that many genes have been altered in ways that are likely to alter expression (Jjingo et al. 2011; Jordan et al. 2003) and analysis of enhancers has shown that retrotransposons drive the evolution of eukaryotic enhancers (McDonald et al. 1997). All of these effects on gene expression are subject to selection and are therefore part of the evolutionary process. Not all insertions into genes will affect regulation of gene expression, some can directly affect the coding sequence or coding potential of genes through exaptation .
Retrotransposons can alter gene expression. a 5’ insertion of a retrotransposon with respect to a gene. a TEs are able to act as alternative promoters to adjacent genes (Faulkner et al. 2009; Speek 2001). b TEs are able to act as transcription factor binding sites (TFBS) and are thereby able to modulate gene expression (Bourque et al. 2008). c In plants, epigenetic silencing of TEs silences nearby genes; this is also likely to occur in animals (Buckley and Adelson 2014; Hollister and Gaut 2009). b 3’ insertion of a retrotransposon a polyA signal/tail of the retrotransposon can result in shortened transcripts (Lee et al. 2008; Perepelitsa-Belancio and Deininger 2003). b Retrotransposon insertion in the 3’ UTR of a gene can provide a target site for piRNAs which down-regulate gene expression (Watanabe et al. 2014). c Intergenic insertion of TEs. a Insertion of TEs into a piRNA cluster results in piRNAs that can target genes carrying TE-derived sequences (Yamamoto et al. 2013). b TEs involved in the origin and evolution of lncRNA (Kapusta et al. 2013)
4.3 Exaptation
When retrotransposons contribute to non-coding or protein coding exon sequences, they are referred to as exaptations . These exaptations may or may not be subject to immediate purifying selection, depending on the type of change they cause. Some exaptations that prove beneficial are selected for, but these are rare. Many examples of exaptation come from non-coding transcripts, where retrotransposon insertions have led to novel piRNA and miRNA transcripts (Jurka et al. 2007; Yamamoto et al. 2013). In fact, only ~50 instances of coding sequences derived from LTR retrotransposons syntenic between human and mouse have been identified (Jurka et al. 2007). One of these encodes the PEG10 (paternally expressed gene 10) locus, which is required for placentation . Occasionally, insertion of a retrotransposon sequence into an intron can lead to exonisation of part of the retrotransposon sequence as an alternative transcript through the presence of splice donor/acceptor sites in the sequence (Fig. 4.8). When this happens, sometimes the alternative transcripts are deleterious because of impaired function, and the regulation of alternative splicing may then become an additional regulatory mechanism for the affected gene (Lorenz et al. 2007).
Retrotransposon exaptation influences mRNA processing and can cause multiple splice variants. At the top, the UCSC browser (Kent et al. 2002) track for the human NOS3 gene is shown, including repeat element annotation. Below, a schematic of the 3’ end of the human NOS3 gene illustrating an Alu element (black bar) inserted into intron 13. This retrotransposon provides exon 14 alternative splicing version 1. An adjacent L1 insertion can result in exon 14 alternative splicing version 2 (Lorenz et al. 2007). Dashed lines indicate a splicing event
4.4 Innate Immunity/Pregnancy Recognition
Some exaptations of retrotransposon sequences have been well-characterised, particularly in terms of the evolution of placentation . There is strong evidence for exaptation of ERV genes in both mouse and hominoid primates required for placental function (Chuong 2013; Haig 2012; Mallet et al. 2004). One of the most striking such exaptations is the role of endogenous jaagsiekte retrovirus (enJSRV) in ruminant pregnancy recognition and placentation . The domestic ruminant conceptus expresses interferon -tau (IFNT) from days 10 to 12, which dramatically alters gene expression in the uterine epithelium and stroma (Bazer et al. 2008; Dunlap et al. 2006; Gray et al. 2006; Spencer and Bazer 1995). At the same time, enJSRVs are released into the ruminant reproductive tract and they are known to regulate key peri-implantation development in the embryo and placenta (Dunlap et al. 2005, 2006). enJSRVs therefore have been exapted to regulate key aspects of development associated with implantation and placentation by virtue of their ability to trigger expression of IFNT expression in the conceptus. Recently, exosomes have been shown to be part of the specific mechanism used to trigger IFNT expression in this system, but without specifically testing for retrotransposon RNA content (Ruiz-Gonz ez et al. 2014, 2015). We speculate that exosomes loaded with retrotransposon sequences may also be involved in pregnancy recognition more generally in order to activate the STAT1 pathway in an interferon -free fashion.
SINE/ERV transcripts packaged into exosomes can trigger RIG-I in target cells leading to IFN independent activation of the IFN pathway, leading us to speculate that the role of retrotransposons is broader than previously thought, and that they may be involved in global regulation of the innate immune system.
5 Conclusion
Retrotransposons are abundant, found in a broad phylogenetic distribution and yet in spite of clade specific non-autonomous variants, exhibit a significant degree of commonality. Furthermore, their transcription is highly regulated, rather than suppressed at all times. These facts, along with the evidence of pervasive and widespread horizontal transfer and an exosome -based mechanism for transfer that has likely co-evolved with the innate immune system and placentation , suggest to us that retrotransposons are not genomic parasites but rather genomic symbionts. We hypothesise that mammals and other vertebrates depend on these symbionts for cell-to-cell signalling in innate immunity and reproduction.
References
Adelson DL, Raison JM, Edgar RC (2009) Characterization and distribution of retrotransposons and simple sequence repeats in the bovine genome. Proc Natl Acad Sci USA 106:12855
Aravin AA, Sachidanandam R, Bourc’his D, Schaefer C, Pezic D, Toth KF, Bestor T, Hannon GJ (2008) A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol Cell 31:785
Baillie JK, Barnett MW, Upton KR, Gerhardt DJ, Richmond TA, De Sapio F, Brennan PM, Rizzu P, Smith S, Fell M, Talbot RT, Gustincich S, Freeman TC, Mattick JS, Hume DA, Heutink P, Carninci P, Jeddeloh JA, Faulkner GJ (2011) Somatic retrotransposition alters the genetic landscape of the human brain. Nat Cell Biol 479:534
Bao W, Kapitonov VV, Jurka J (2010) Ginger dna transposons in eukaryotes and their evolutionary relationships with long terminal repeat retrotransposons. Mob DNA 1(1):3. doi:10.1186/1759-8753-1-3
Batagov AO, Kurochkin IV (2013) Exosomes secreted by human cells transport largely mrna fragments that are enriched in the 3’-untranslated regions. Biol Direct 8:12. doi:10.1186/1745-6150-8-12
Bazer FW, Burghardt RC, Johnson GA, Spencer TE, Wu G (2008) Interferons and progesterone for establishment and maintenance of pregnancy: interactions among novel cell signaling pathways. Reprod Biol 8(3):179–211
Belancio VP, Roy-Engel AM, Deininger PL (2010a) All y’all need to know ‘bout retroelements in cancer. Semin Cancer Biol 20(4):200–210. doi:10.1016/j.semcancer.2010.06.001
Belancio VP, Roy-Engel AM, Pochampally RR, Deininger P (2010 b) Somatic expression of LINE-1 elements in human tissues. Nucleic Acids Res 38:3909
Boeke JD (2003) The unusual phylogenetic distribution of retrotransposons: a hypothesis. Genome Res 13(9):1975–1983. doi:10.1101/gr.1392003
Boelens MC, Wu TJ, Nabet BY, Xu B, Qiu Y, Yoon T, Azzam DJ, Twyman-Saint Victor C, Wiemann BZ, Ishwaran H, Ter Brugge PJ, Jonkers J, Slingerland J, Minn AJ (2014) Exosome transfer from stromal to breast cancer cells regulates therapy resistance pathways. Cell 159(3):499–513. doi:10.1016/j.cell.2014.09.051
Bourc’his D, Bestor TH (2004) Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking dnmt3 l. Nature 431(7004):96–99. doi:10.1038/nature02886
Bourque G, Leong B, Vega VB, Chen X, Lee YL, Srinivasan KG, Chew JL, Ruan Y, Wei CL, Ng HH, Liu ET (2008) Evolution of the mammalian transcription factor binding repertoire via transposable elements. Genome Res 18(11):1752–1762. doi:10.1101/gr.080663.108
Brookfield JFY (2005) The ecology of the genome—mobile dna elements and their hosts. Nat Rev Genet 6(2):128–136. doi:10.1038/nrg1524
Buckley RM, Adelson DL (2014) Mammalian genome evolution as a result of epigenetic regulation of transposable elements. Biomol Concepts 5(3):183–194. doi:10.1515/bmc-2014-0013
Buzdin A, Ustyugova S, Gogvadze E, Vinogradova T, Lebedev Y, Sverdlov E (2002) A new family of chimeric retrotranscripts formed by a full copy of u6 small nuclear rna fused to the 3’ terminus of l1. Genomics 80(4):402–406
Chuong EB (2013) Retroviruses facilitate the rapid evolution of the mammalian placenta. Bioessays 35:853
Ciaudo C, Jay F, Okamoto I, Chen CJ, Sarazin A, Servant N, Barillot E, Heard E, Voinnet O (2013) Rnai-dependent and independent control of line1 accumulation and mobility in mouse embryonic stem cells. PLoS Genet 9(11):e1003,791. doi:10.1371/journal.pgen.1003791
Cost GJ, Boeke JD (1998) Targeting of human retrotransposon integration is directed by the specificity of the l1 endonuclease for regions of unusual dna structure. Biochemistry 37(51):18081–18093
Cost GJ, Feng Q, Jacquier A, Boeke JD (2002) Human l1 element target-primed reverse transcription in vitro. EMBO J 21(21):5899–5910
Coufal NG, Garcia-Perez JL, Peng GE, Yeo GW, Mu Y, Lovci MT, Morell M, O’Shea KS, Moran JV, Gage FH (2009) L1 retrotransposition in human neural progenitor cells. Nat Cell Biol 460:1127
Crichton JH, Dunican DS, Maclennan M, Meehan RR, Adams IR (2014) Defending the genome from the enemy within: mechanisms of retrotransposon suppression in the mouse germline. Cell Mol Life Sci 71(9):1581–1605. doi:10.1007/s00018-013-1468-0
Daniels SB, Peterson KR, Strausbaugh LD, Kidwell MG, Chovnick A (1990) Evidence for horizontal transmission of the p-transposable element between drosophila species. Genetics 124:339
Dieci G, Conti A, Pagano A, Carnevali D (2013) Identification of rna polymerase iii-transcribed genes in eukaryotic genomes. Biochim Biophys Acta 1829(3–4):296–305. doi:10.1016/j.bbagrm.2012.09.010
Dreux M, Garaigorta U, Boyd B, Décembre E, Chung J, Whitten-Bauer C, Wieland S, Chisari FV (2012) Short-range exosomal transfer of viral rna from infected cells to plasmacytoid dendritic cells triggers innate immunity. Cell Host Microbe 12(4):558–570. doi:10.1016/j.chom.2012.08.010
Dunlap KA, Palmarini M, Adelson DL, Spencer TE (2005) Sheep endogenous betaretroviruses (enjsrvs) and the hyaluronidase 2 (hyal2) receptor in the ovine uterus and conceptus. Biol Reprod 73(2):271–279. doi:10.1095/biolreprod.105.039776
Dunlap KA, Palmarini M, Varela M, Burghardt RC, Hayashi K, Farmer JL, Spencer TE (2006) Endogenous retroviruses regulate periimplantation placental growth and differentiation. Proc Natl Acad Sci USA 103(39):14390–14395. doi:10.1073/pnas.0603836103
Eickbush TH (1997) Telomerase and retrotransposons: which came first? Science (New York, NY) 277(5328):911–912
Eickbush TH, Jamburuthugoda VK (2008) The diversity of retrotransposons and the properties of their reverse transcriptases. Virus Res 134:221
El Baidouri M, Carpentier MC, Cooke R, Gao D, Lasserre E, Llauro C, Mirouze M, Picault N, Jackson SA, Panaud O (2014) Widespread and frequent horizontal transfers of transposable elements in plants. Genome Res 24(5):831–838. doi:10.1101/gr.164400.113
Fadloun A, Le Gras S, Jost B, Ziegler-Birling C, Takahashi H, Gorab E, Carninci P, Torres-Padilla ME (2013) Chromatin signatures and retrotransposon profiling in mouse embryos reveal regulation of line-1 by rna. Nat Struct Mol Biol 20(3):332–338. doi:10.1038/nsmb.2495
Faulkner GJ, Kimura Y, Daub CO, Wani S, Plessy C, Irvine KM, Schroder K, Cloonan N, Steptoe AL, Lassmann T, Waki K, Hornig N, Arakawa T, Takahashi H, Kawai J, Forrest ARR, Suzuki H, Hayashizaki Y, Hume DA, Orlando V, Grimmond SM, Carninci P (2009) The regulated retrotransposon transcriptome of mammalian cells. Nat Genet 41:563
Feng Q, Moran J, Kazazian H, Boeke J (1996) Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell 87:905
Feschotte C (2008) Opinion—transposable elements and the evolution of regulatory networks. Nat Rev Genet 9:397
Feschotte C, Gilbert C (2012) Endogenous viruses: insights into viral evolution and impact on host biology. Nat Rev Genet 13(4):283–296. doi:10.1038/nrg3199
Gasior SL, Wakeman TP, Xu B, Deininger PL (2006) The human LINE-1 retrotransposon creates DNA double-strand breaks. Journal of Molecular Biology
Gilbert C, Schaack S, Pace JK, Brindley PJ, Feschotte C (2010) A role for host-parasite interactions in the horizontal transfer of transposons across phyla. Nature 464:1347–1352
Gilbert N, Labuda D (2000) Evolutionary inventions and continuity of core-sines in mammals. J Mol Biol 298(3):365–377. doi:10.1006/jmbi.2000.3695
Giordano J, Ge Y, Gelfand Y, Abrusan G, Benson G, Warburton P (2007) Evolutionary history of mammalian transposons determined by genome-wide defragmentation. PLoS Comput Biol 3:e137
Gogvadze E, Buzdin A (2009) Retroelements and their impact on genome evolution and functioning. Cell Mol Life Sci 66(23):3727–3742. doi:10.1007/s00018-009-0107-2
Gogvadze E, Barbisan C, Lebrun MH, Buzdin A (2007) Tripartite chimeric pseudogene from the genome of rice blast fungus magnaporthe grisea suggests double template jumps during long interspersed nuclear element (line) reverse transcription. BMC Genomics 8:360. doi:10.1186/1471-2164-8-360
Goodier JL, Cheung LE, Kazazian HH Jr (2012) Mov10 rna helicase is a potent inhibitor of retrotransposition in cells. PLoS Genet 8(10):e1002941. doi:10.1371/journal.pgen.1002941
Gray CA, Abbey CA, Beremand PD, Choi Y, Farmer JL, Adelson DL, Thomas TL, Bazer FW, Spencer TE (2006) Identification of endometrial genes regulated by early pregnancy, progesterone, and interferon tau in the ovine uterus. Biol Reprod 74(2):383–394. doi:10.1095/biolreprod.105.046656
Grivna ST, Pyhtila B, Lin H (2006) Miwi associates with translational machinery and piwi-interacting rnas (pirnas) in regulating spermatogenesis. Proc Natl Acad Sci USA 103(36):13415–13420. doi:10.1073/pnas.0605506103
Hackett JA, Surani MA (2013) Dna methylation dynamics during the mammalian life cycle. Philos Trans R Soc Lond B Biol Sci 368(1609):20110328. doi:10.1098/rstb.2011.0328
Haig D (2012) Retroviruses and the placenta. Current biology: CB
Hollister JD, Gaut BS (2009) Epigenetic silencing of transposable elements: a trade-off between reduced transposition and deleterious effects on neighboring gene expression. Genome Res 19(8):1419–1428. doi:10.1101/gr.091678.109
Ichiyanagi K, Nakajima R, Kajikawa M, Okada N (2007) Novel retrotransposon analysis reveals multiple mobility pathways dictated by hosts. Genome Res 17:33
Ivancevic AM, Walsh AM, Kortschak RD, Adelson DL (2013) Jumping the fine LINE between species: horizontal transfer of transposable elements in animals catalyses genome evolution. Bioessays 35:12
Jiang Y, Xie M, Chen W, Talbot R, Maddox JF, Faraut T, Wu C, Muzny DM, Li Y, Zhang W, Stanton JA, Brauning R, Barris WC, Hourlier T, Aken BL, Searle SMJ, Adelson DL, Bian C, Cam GR, Chen Y, Cheng S, DeSilva U, Dixen K, Dong Y, Fan G, Franklin IR, Fu S, Fuentes-Utrilla P, Guan R, Highland MA, Holder ME, Huang G, Ingham AB, Jhangiani SN, Kalra D, Kovar CL, Lee SL, Liu W, Liu X, Lu C, Lv T, Mathew T, McWilliam S, Menzies M, Pan S, Robelin D, Servin B, Townley D, Wang W, Wei B, White SN, Yang X, Ye C, Yue Y, Zeng P, Zhou Q, Hansen JB, Kristiansen K, Gibbs RA, Flicek P, Warkup CC, Jones HE, Oddy VH, Nicholas FW, McEwan JC, Kijas JW, Wang J, Worley KC, Archibald AL, Cockett N, Xu X, Wang W, Dalrymple BP (2014) The sheep genome illuminates biology of the rumen and lipid metabolism. Science 344(6188):1168–1173. doi:10.1126/science.1252806
Jjingo D, Huda A, Gundapuneni M, Mariño-Ramrez L, Jordan IK (2011) Effect of the transposable element environment of human genes on gene length and expression. Genome Biol Evol 3:259–271. doi:10.1093/gbe/evr015
Jordan IK, Rogozin IB, Glazko GV, Koonin EV (2003) Origin of a substantial fraction of human regulatory sequences from transposable elements. Trends Genet 19:68
Jurka J, Kapitonov VV, Kohany O, Jurka MV (2007) Repetitive sequences in complex genomes: structure and evolution. Ann Rev Genomics Hum Genet
Kano H, Godoy I, Courtney C, Vetter MR, Gerton GL, Ostertag EM, Kazazian HH (2009) L1 retrotransposition occurs mainly in embryogenesis and creates somatic mosaicism. Genes Dev 23:1303
Kapusta A, Kronenberg Z, Lynch VJ, Zhuo X, Ramsay L, Bourque G, Yandell M, Feschotte C (2013) Transposable elements are major contributors to the origin, diversification, and regulation of vertebrate long noncoding RNAs. PLoS Genet 9:e1003470
Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D (2002) The human genome browser at ucsc. Genome Res 12(6):996–1006. doi:10.1101/gr.229102. Article published online before print in May
Kordis D, Gubensek F (1998) Unusual horizontal transfer of a long interspersed nuclear element between distant vertebrate classes. Proc Natl Acad Sci USA 95(18):10704–10709
Kordis D, Gubensek F (1999a) Horizontal transfer of non-LTR retrotransposons in vertebrates. Genetica 107:121
Kordis D, Gubensek F (1999b) Molecular evolution of bov-b lines in vertebrates. Gene 238(1):171–178
Kramerov DA, Vassetzky NS (2005) Short retroposons in eukaryotic genomes. Int Rev Cytol 247:165–221. doi:10.1016/S0074-7696(05)47004-7
Kriegs JO, Matzke A, Churakov G, Kuritzin A, Mayr G, Brosius J, Schmitz J (2007) Waves of genomic hitchhikers shed light on the evolution of gamebirds (aves: Galliformes). BMC Evol Biol 7:190. doi:10.1186/1471-2148-7-190
Kuramochi-Miyagawa S, Watanabe T, Gotoh K, Totoki Y, Toyoda A, Ikawa M, Asada N, Kojima K, Yamaguchi Y, Ijiri TW, Hata K, Li E, Matsuda Y, Kimura T, Okabe M, Sakaki Y, Sasaki H, Nakano T (2008) Dna methylation of retrotransposon genes is regulated by piwi family members mili and miwi2 in murine fetal testes. Genes Dev 22(7):908–917. doi:10.1101/gad.1640708
Lampe DJ, Witherspoon DJ, Soto-Adames FN, Robertson HM (2003) Recent horizontal transfer of mellifera subfamily mariner transposons into insect lineages representing four different orders shows that selection acts only during horizontal transfer. Mol Biol Evol 20(4):554–562. doi:10.1093/molbev/msg069
Le Rouzic A, Boutin TS, Capy P (2007) Long-term evolution of transposable elements. Proc Natl Acad Sci USA 104(49):19375–19380. doi:10.1073/pnas.0705238104
Lee JY, Ji Z, Tian B (2008) Phylogenetic analysis of mrna polyadenylation sites reveals a role of transposable elements in evolution of the 3’-end of genes. Nucleic Acids Res 36(17):5581–5590. doi:10.1093/nar/gkn540
Lenstra JA, van Boxtel JA, Zwaagstra KA, Schwerin M (1993) Short interspersed nuclear element (sine) sequences of the bovidae. Anim Genet 24(1):33–39
Li CCY, Eaton SA, Young PE, Lee M, Shuttleworth R, Humphreys DT, Grau GE, Combes V, Bebawy M, Gong J, Brammah S, Buckland ME, Suter CM (2013) Glioma microvesicles carry selectively packaged coding and non-coding rnas which alter gene expression in recipient cells. RNA Biol 10(8):1333–1344. doi:10.4161/rna.25281
Lingner J, Hughes TR, Shevchenko A, Mann M, Lundblad V, Cech TR (1997) Reverse transcriptase motifs in the catalytic subunit of telomerase. Science (New York, NY) 276:561
Lohe AR, Moriyama EN, Lidholm DA, Hartl DL (1995) Horizontal transmission, vertical inactivation, and stochastic loss of mariner-like transposable elements. Mol Biol Evol 12(1):62–72
Lorenz M, Hewing B, Hui J, Zepp A, Baumann G, Bindereif A, Stangl V, Stangl K (2007) Alternative splicing in intron 13 of the human enos gene: a potential mechanism for regulating enos activity. FASEB J 21(7):1556–1564. doi:10.1096/fj.06-7434com
Malik H, Eickbush T (1998) The RTE class of non-LTR retrotransposons is widely distributed in animals and is the origin of many SINEs. Mol Biol Evol 15:1123
Mallet F, Bouton O, Prudhomme S, Cheynet V, Oriol G, Bonnaud B, Lucotte G, Duret L, Mandrand B (2004) The endogenous retroviral locus ERVWE1 is a bona fide gene involved in hominoid placental physiology. Proc Natl Acad Sci USA 101:1731
Martin SL (2006) The orf1 protein encoded by line-1: structure and function during l1 retrotransposition. J Biomed Biotechnol 2006(1):45621. doi:10.1155/JBB/2006/45621
Maruyama K, Hartl DL (1991) Evidence for interspecific transfer of the transposable element mariner between Drosophila and Zaprionus. J Mol Evol 33:514
McDonald JF, Matyunina LV, Wilson S, Jordan IK, Bowen NJ, Miller WJ (1997) Ltr retrotransposons and the evolution of eukaryotic enhancers. Genetica 100(1–3):3–13
Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, Gnirke A, Jaenisch R, Lander ES (2008) Genome-scale dna methylation maps of pluripotent and differentiated cells. Nature 454(7205):766–770. doi:10.1038/nature07107
Moran JV, Holmes SE, Naas TP, DeBerardinis RJ, Boeke JD, Kazazian HH Jr (1996) High frequency retrotransposition in cultured mammalian cells. Cell 87(5):917–927
Morrish TA, Gilbert N, Myers JS, Vincent BJ, Stamato TD, Taccioli GE, Batzer MA, Moran JV (2002) Dna repair mediated by endonuclease-independent line-1 retrotransposition. Nat Genet 31(2):159–165. doi:10.1038/ng898
Ohshima K, Okada N (2005) Sines and lines: symbionts of eukaryotic genomes with a common tail. Cytogenet Genome Res 110(1–4):475–490. doi:10.1159/000084981
Okada N, Hamada M (1997) The 3’ ends of trna-derived sines originated from the 3’ ends of lines: a new example from the bovine genome. J Mol Evol 44(1):52–56
Ostertag E, Goodier J, Zhang Y, Kazazian H (2003) SVA elements are nonautonomous retrotransposons that cause disease in humans. Am J Hum Genet 73:1444
Pace JK, Gilbert C, Clark MS, Feschotte C (2008) Repeated horizontal transfer of a DNA transposon in mammals and other tetrapods. Proc Natl Acad Sci USA 105:17023
Perepelitsa-Belancio V, Deininger P (2003) Rna truncation by premature polyadenylation attenuates human mobile element activity. Nat Genet 35(4):363–366. doi:10.1038/ng1269
Piskurek O, Jackson DJ (2012) Transposable elements: from dna parasites to architects of metazoan evolution. Genes (Basel) 3(3):409–422. doi:10.3390/genes3030409
Piskurek O, Okada N (2007) Poxviruses as possible vectors for horizontal transfer of retroposons from reptiles to mammals. Proc Natl Acad Sci USA 104(29):12046–12051. doi:10.1073/pnas.0700531104
Roy AM, Carroll ML, Nguyen SV, Salem AH, Oldridge M, Wilkie AO, Batzer MA, Deininger PL (2000) Potential gene conversion and source genes for recently integrated alu elements. Genome Res 10(10):1485–1495
Ruiz-González I, Xu J, Wang X, Burghardt RC, Dunlap K, Bazer FW (2014) Exosomes, endogenous retroviruses and toll-like receptors: pregnancy recognition in ewes. Reproduction
Ruiz-González I, Minten M, Wang X, Dunlap K, Bazer FW (2015) Involvement of TLR7 and TLR8 in conceptus development and establishment of pregnancy in Sheep. Reproduction
Shimotohno K, Temin HM (1981) Evolution of retroviruses from cellular movable genetic elements. Cold Spring Harb Symp Quant Biol 45(Pt 2):719–730
Skog J, Würdinger T, van Rijn S, Meijer DH, Gainche L, Sena-Esteves M, Curry WT Jr, Carter BS, Krichevsky AM, Breakefield XO (2008) Glioblastoma microvesicles transport rna and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol 10(12):1470–1476. doi:10.1038/ncb1800
Sormacheva I, Smyshlyaev G, Mayorov V, Blinov A, Novikov A, Novikova O (2012) Vertical evolution and horizontal transfer of cr1 non-ltr retrotransposons and tc1/mariner dna transposons in lepidoptera species. Mol Biol Evol 29(12):3685–3702. doi:10.1093/molbev/mss181
Speek M (2001) Antisense promoter of human l1 retrotransposon drives transcription of adjacent cellular genes. Mol Cell Biol 21(6):1973–1985. doi:10.1128/MCB.21.6.1973-1985.2001
Spencer TE, Bazer FW (1995) Temporal and spatial alterations in uterine estrogen receptor and progesterone receptor gene expression during the estrous cycle and early pregnancy in the ewe. Biol Reprod 53(6):1527–1543
Stankiewicz P, Lupski JR (2002) Genome architecture, rearrangements and genomic disorders. Trends Genet 18(2):74–82
Startek M, Szafranski P, Gambin T, Campbell IM, Hixson P, Shaw CA, Stankiewicz P, Gambin A (2015) Genome-wide analyses of line-line-mediated nonallelic homologous recombination. Nucleic Acids Res. doi:10.1093/nar/gku1394
Swergold GD (1990) Identification, characterization, and cell specificity of a human line-1 promoter. Mol Cell Biol 10(12):6718–6729
Tanaka T, Hosokawa M, Vagin VV, Reuter M, Hayashi E, Mochizuki AL, Kitamura K, Yamanaka H, Kondoh G, Okawa K, Kuramochi-Miyagawa S, Nakano T, Sachidanandam R, Hannon GJ, Pillai RS, Nakatsuji N, Chuma S (2011) Tudor domain containing 7 (tdrd7) is essential for dynamic ribonucleoprotein (rnp) remodeling of chromatoid bodies during spermatogenesis. Proc Natl Acad Sci USA 108(26):10579–10584. doi:10.1073/pnas.1015447108
Tarlinton RE, Meers J, Young PR (2006) Retroviral invasion of the koala genome. Nature 442(7098):79–81. doi:10.1038/nature04841
Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO (2007) Exosome-mediated transfer of mrnas and micrornas is a novel mechanism of genetic exchange between cells. Nat Cell Biol 9(6):654–659. doi:10.1038/ncb1596
Villarroya-Beltri C, Gutiérrez-Vázquez C, Sánchez-Cabo F, Pérez-Hernández D, Vázquez J, Martin-Cofreces N, Martinez-Herrera DJ, Pascual-Montano A, Mittelbrunn M, Sánchez-Madrid F (2013) Sumoylated hnrnpa2b1 controls the sorting of mirnas into exosomes through binding to specific motifs. Nat Commun 4:2980. doi:10.1038/ncomms3980
Vlachogiannis G, Niederhuth CE, Tuna S, Stathopoulou A, Viiri K, de Rooij DG, Jenner RG, Schmitz RJ, Ooi SKT (2015) The dnmt3 l add domain controls cytosine methylation establishment during spermatogenesis. Cell Rep. doi:10.1016/j.celrep.2015.01.021
Walsh AM, Kortschak RD, Gardner MG, Bertozzi T, Adelson DL (2013) Widespread horizontal transfer of retrotransposons. Proc Natl Acad Sci USA 110:1012
Watanabe T, Totoki Y, Toyoda A, Kaneda M, Kuramochi-Miyagawa S, Obata Y, Chiba H, Kohara Y, Kono T, Nakano T, Surani MA, Sakaki Y, Sasaki H (2008) Endogenous sirnas from naturally formed dsrnas regulate transcripts in mouse oocytes. Nature 453(7194):539–543. doi:10.1038/nature06908
Watanabe T, Cheng EC, Zhong M, Lin H (2014) Retrotransposons and pseudogenes regulate mrnas and lncrnas via the pirna pathway in the germline. Genome Res. doi:10.1101/gr.180802.114
Webster KE, O’Bryan MK, Fletcher S, Crewther PE, Aapola U, Craig J, Harrison DK, Aung H, Phutikanit N, Lyle R, Meachem SJ, Antonarakis SE, de Kretser DM, Hedger MP, Peterson P, Carroll BJ, Scott HS (2005) Meiotic and epigenetic defects in dnmt3l-knockout mouse spermatogenesis. Proc Natl Acad Sci USA 102(11):4068–4073. doi:10.1073/pnas.0500702102
Yamamoto Y, Watanabe T, Hoki Y, Shirane K, Li Y, Ichiiyanagi K, Kuramochi-Miyagawa S, Toyoda A, Fujiyama A, Oginuma M, Suzuki H, Sado T, Nakano T, Sasaki H (2013) Targeted gene silencing in mouse germ cells by insertion of a homologous dna into a pirna generating locus. Genome Res 23(2):292–299. doi:10.1101/gr.137224.112
Yuan A, Farber EL, Rapoport AL, Tejada D, Deniskin R, Akhmedov NB, Farber DB (2009) Transfer of micrornas by embryonic stem cell microvesicles. PLoS ONE 4(3):e4722. doi:10.1371/journal.pone.0004722
Zhou Y, Mishra B (2005) Quantifying the mechanisms for segmental duplications in mammalian genomes by statistical analysis and modeling. Proc Natl Acad Sci USA 102(11):4051–4056. doi:10.1073/pnas.0407957102
Acknowledgments
The authors wish to thank R. Daniel Kortschak and Joy M. Raison for helpful discussions and advice.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2015 Springer International Publishing Switzerland
About this chapter
Cite this chapter
Adelson, D.L., Buckley, R.M., Ivancevic, A.M., Qu, Z., Zeng, L. (2015). Retrotransposons: Genomic and Trans-Genomic Agents of Change. In: Pontarotti, P. (eds) Evolutionary Biology: Biodiversification from Genotype to Phenotype. Springer, Cham. https://doi.org/10.1007/978-3-319-19932-0_4
Download citation
DOI: https://doi.org/10.1007/978-3-319-19932-0_4
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-19931-3
Online ISBN: 978-3-319-19932-0
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)