Abstract
Protein quality control is an essential process for cellular survival. When protein damage occurs, a series of coordinated response mechanisms repair or degrade damaged proteins to avoid the accumulation of toxic protein aggregates and restore proteostasis. However, the amount of misfolded proteins increases during aging overwhelming the mechanisms responsible for protein quality control, thus leading to the development of several age-dependent neurodegenerative disorders. Interestingly, targeted expression of proteins causative of these diseases in flies reproduces the pathological behaviors seen in humans. This remarkable conservation provides a valuable experimental tool to elucidate the complex mechanisms associated with the maintenance of proteostasis. In this chapter, we summarize how Drosophila has contributed to understand the roles of the heat shock response, the unfolded protein response, autophagy and the ubiquitin proteasome system in brain aging and neurodegeneration associated with protein-misfolding disorders. In addition, we describe fundamental contributions of the fly system to the design of new therapeutic strategies for these devastating disorders.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- Protein quality control
- Proteostasis
- Misfolded proteins
- Neurodegenerative disorders
- Heat shock response
- Autophagy
1 Introduction
Protein quality control or proteostasis refers to the cellular processes involved in the biogenesis, folding, maturation, distribution, and degradation of proteins in different cellular compartments. These activities are critical for proper cell function because they maintain active proteins and organelles, and also prevent the accumulation of toxic unfolded/misfolded proteins . When protein damage occurs, a series of coordinated response mechanisms repair or degrade damaged proteins to avoid the accumulation of toxic protein aggregates that can, ultimately, lead to cell death. Protein toxicity can be the result of different alterations in protein homeostasis associated with aberrant folding and aggregation, leading to the so-called proteinopathies or protein misfolding disorders. The most common causes of these disorders include missense mutations, abnormal post-translational modifications, protein overexpression, and exogenous stressors, including temperature and chemicals among others. Most of the cellular machinery implicated in protein quality control is essential for cell survival , highlighting the importance of protein homeostasis. This is particularly true for neurons , because of their weak capacity to regenerate and their long life span. It is, thus, obvious that maintaining protein homeostasis is critical for healthy brain aging and longevity.
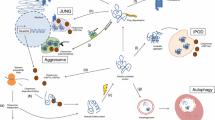
A complex network of conserved cellular processes controls the quality of the proteome and restores proteostasis following the aberrant accumulation of misfolded proteins . This protective network can be grouped into mechanisms that detect and respond to protein unfolding/misfolding, which include the heat shock response (HSR) and the unfolded protein response (UPR), and mechanisms responsible for abnormal protein degradation, which comprises autophagy and the ubiquitin proteasome system (UPS). All four processes are highly integrated to prevent the cellular toxicity associated with the accumulation of protein aggregates (Fig. 8.1).
Impact of aging and disease in cellular proteostasis . Under physiologic conditions, the heat shock response (HSR), the unfolded protein response (UPR), and two catalytic pathways (autophagy and UPS) cooperate to prevent the aggregation and toxicity of misfolded proteins . Heat shock proteins (HSPs) act as the first line of defense by binding misfolded substrates, promoting their refolding, and directing their degradation through chaperone-mediated autophagy (CMA). In the ER, accumulation of misfolded proteins activates the three UPR pathways that alter gene expression to restore proteostasis, including the up-regulation of ER associated degradation (ERAD). When misfolded proteins escape HSPs, they accumulate and aggregate in the cytosol, where become substrates for degradation by autophagy and the UPS. This complex, coordinated system maintains cellular proteostasis for decades and is critical for the function of post-mitotic neurons . However, protein misfolding disorders and aging place the proteostasis machinery under stress by overproducing misfolded proteins and reducing the cellular response to insults. Misfolded proteins saturate the response systems and prevent the regular turnover of old proteins and organelles. Aging is known to weaken gene expression of all stress-response pathways, thus dampening the ability to protect the cell. The combination of both stressors may be at the heart of multiple late-onset neurodegenerative disorders
A particular group of human disorders has played a critical role in the characterization of protein quality control pathways in neuronal survival, the so-called protein-misfolding diseases or proteinopathies. A large number of human diseases are associated with protein misfolding, which can affect specific organs (brain, muscle, eye, skin) or have systemic effects (Valastyan and Lindquist 2014). Currently, more than 30 different human diseases are linked to aberrant protein accumulation, including the highly prevalent Alzheimer’s disease (AD), Parkinson’s disease (PD), and amyotrophic lateral sclerosis (ALS), which have a mostly sporadic etiology. A large number of rare genetic conditions also have protein aggregation as the main trigger, including Huntington’s disease (HD) and several spinocerebellar ataxias. These disorders have distinct clinical presentations, affect different parts of the brain, and are caused by the misfolding and aggregation of unrelated proteins, which can be either wild type or mutant. A common thread to these diverse disorders, though, is that they all have late onset presentation. This link is revealing because in the genetic diseases the triggering protein is expressed throughout the lifetime, but several decades go by without clinical signs. This observation suggests that the cellular machinery that controls protein homeostasis prevents protein aggregation and neuronal cell death for several decades. Then, in middle to old age, the same machinery seems to be overwhelmed by the accumulation of misfolded proteins and an age-related decrease in the expression of key factors of the quality control pathways. Overall, the tight connection of aging with protein-misfolding diseases enables the study of the cellular machinery that keeps protein quality in check during normal brain physiology and aging. In this chapter we summarize the proteostasis mechanisms, and how Drosophila has contributed to understand their roles in brain aging and neurodegeneration associated with protein-misfolding disorders.
Several model systems have been employed to investigate the pathways and protein interactions involved in proteostasis, including in vitro and cell culture paradigms. The small fruit fly Drosophila melanogaster has proven a useful tool for the study of multiple human disorders, protein misfolding diseases and aging. Around 75 % of human genes associated with human diseases have a homologous in Drosophila. Moreover, the low cost and easy handling make this small fly an excellent model organism. Another advantage of Drosophila is the large number of genetic tools available for gene manipulation (loss-of-function and gain-of-function alleles), tissue specific gene expression with the UAS/GAL4 system, temporal control of gene expression with Gal80TS, and the recent introduction of markers of neuronal architecture and function (del Valle Rodriguez et al. 2011).
Drosophila has also contributed significantly to understand human protein misfolding diseases. A large number of these diseases have been modeled in flies because the proteins causative of these diseases in human exhibit the same aberrant behavior in flies and, thus, misfold, aggregate, and induce toxicity in the Drosophila brain and eyes (Rincon-Limas et al. 2012). Interestingly, Drosophila models have played a critical role in unraveling the cellular mechanisms underlying disease pathogenesis. Indeed, unbiased genetic screens have revealed the involvement of unsuspected genes and pathways. Moreover, candidate approaches allowed the efficient testing of hypotheses in vivo, confirming the role of suspected pathways, including chaperones and autophagy . Finally, these models have also been used to examine the effectiveness of pharmacological modulators of suspected pathways, providing in vivo information that can shorten the path to the translation of effective therapies in humans.
2 Molecular Chaperones: The Cell First Line of Defense
Molecular chaperones are ubiquitous and highly conserved proteins with a main role in protein homeostasis . They recognize and bind unfolded and misfolded proteins to prevent aggregation and promote refolding. Chaperones were first identified through their transcriptional response to heat stress in Drosophila, but they can respond to several stresses such as cold, ischemia, hypoxia , oxidative stress , proteotoxic stress, chemicals, and heavy metals. Heat shock proteins (HSPs) are the major group of molecular chaperones inside the cell, and include several families with distinct localization in organelles and cellular compartments, and different functions (Kim et al. 2013).
HSPs contain a substrate-binding domain (SBD) that recognizes short hydrophobic stretches on unfolded/misfolded client proteins and this interaction is the first key step to prevent their aggregation. In fact, in the absence of chaperones, proteins with exposed hydrophobic stretches self-associate creating aggregates that can be unstructured or highly ordered into amyloid fibers (Douglas and Cyr 2009). Some HSPs contain ATPase domains and cooperate with ATP-independent co-chaperones to efficiently bind and refold substrates. HSPs are classified based on their molecular weight, ranging from small HSPs with less than 30 kDa to large proteins over 100 kDa. Each family comprises several members with different tissue and subcellular distribution (Voisine et al. 2010).
2.1 The Hsp70/Hsp40 System
The Hsp70 family is among the most conserved proteins in the animal world. Their main role is to promote the correct protein folding of both nascent and mature proteins by recognizing exposed hydrophobic residues on unfolded or misfolded substrates, and prevent their aggregation. Structurally, Hsp70 chaperones have two major functional domains: an N-terminal nucleotide binding domain (NBD) that binds ATP and hydrolyzes it to ADP and a C-terminal SBD that interacts with exposed linear hydrophobic segments. In its ATP-bound state, Hsp70 binds substrates with low affinity. ATP hydrolysis to ADP forces a conformational change in the SBD that increases the affinity for the substrate. Then, interaction with a nucleotide exchange factor restores ATP binding of Hsp70, which forces the release of the client protein, completing the Hsp70 cycle. Although Hsp70 exerts a potent refolding activity in vitro, the Hsp40 family regulates Hsp70 cycling in vivo. Hsp40s are ATP-independent chaperones characterized by a J-domain that interacts with the NBD of Hsp70 and stimulates ATP hydrolysis, thus strengthening the interaction with the client protein . In vitro studies suggest that Hsp40 first binds client proteins and presents them to the SBD of Hsp70 while stimulating ATP hydrolysis in the NDB domain, therefore enhancing Hsp70 refolding efficiency (Young 2010).
The characterization of several brain disorders linked to protein misfolding and deposition suggested that HSPs should be involved in the pathogenesis and could also play a role in therapy. Initially, a study in cell culture showed that overexpressed Hsp70 co-localized with mutant Ataxin1 (Atx1-82Q) in nuclear inclusions, which pointed to a stress response against pathogenic protein aggregation. To determine whether increased Hsp70 activity could harbor a protective role against protein aggregates, Warrick and cols. overexpressed HSPA1L, a human inducible Hsp70, in Drosophila showing aberrant eye morphology linked to mutant Ataxin3 (Atx3tr-78Q). Co-expression of HSPA1L completely suppressed the aberrant phenotype caused by Atx3tr-78Q, indicating the critical role of Hsp70 in the toxicity of this aggregate-prone polyglutamine allele (Warrick et al. 1999). Moreover, co-expression of Atx3tr-78Q and a constitutive Drosophila Hsp70 (Heat shock cognate 4, Hsc4) carrying the K71S substitution that inactivates the ATPase activity enhanced neurotoxicity , suggesting that the endogenous activity of Hsp70 mitigates the toxicity of Atx3tr-78Q. A prediction from these results was that Hsp70 overexpression would exhibit the same protective activity against other polyglutamine expansions, since the same mutation was responsible for the aberrant protein folding in a different protein. Spinal-bulbar muscular atrophy (SBMA) is linked to a polyglutamine expansion in the androgen receptor (AR) and expression of the mutant allele in flies induces neurotoxicity. As expected, co-expression of HSPA1L rescued the toxicity of AR-108Q, confirming the beneficial effect of Hsp70 against related misfolded substrates (Chan et al. 2000).
The next question in the field was whether Hsp70 could exert the same neuroprotective activity against other toxic amyloids unrelated to the polyglutamine expansions. The same lab addressed this question in a Drosophila model expressing α-synuclein, the most abundant protein in Lewy bodies, typically found aggregating in PD patients. HSPA1L not only co-localized with α-synuclein aggregates but also increased the survival rate of dopaminergic cells (Auluck et al. 2002). In contrast, co-expression of α-synuclein and dominant-negative Hsc4 resulted in increased dopaminergic cell death without changing the number of Lewy body-like aggregates (Auluck et al. 2002). Additionally, the prion protein (PrP), a membrane-anchored glycoprotein widely expressed in the brain, leads to aggressive vacuolar degeneration when misfolds. We asked whether Hsp70 could exert a protective activity against PrP misfolding in vivo either by indirectly maintaining intracellular homeostasis or directly by exiting the cell and interacting with PrP. To our surprise, we found that Hsp70 overexpression protected against PrP toxicity, reduced PrP levels, and inhibited PrP misfolding (Fernandez-Funez et al. 2009). Since we found that Hsp70 co-immunoprecipitated with PrP, Hsp70 seems to mediate its protective effects by binding directly to PrP at the membrane. This ability of Hsp70 and other chaperones to exit the cell has been described previously, but is associated with stressful conditions (Pittet et al. 2002; Fleshner and Johnson 2005). Thus, HSPA1L is beneficial against a wide variety of misfolded proteins that accumulate in the nucleus, cytosol, and the extracellular space.
With Hsp70 working as a potent suppressor of toxicity, the potential protective activity of other chaperones emerged over the next few years. Two independent screens for modifiers of Atx1-82Q and 65Q-only toxicity in flies identified the protective activity of dDnaJ-1/Hsp40 overexpression (Fernandez-Funez et al. 2000; Kazemi-Esfarjani and Benzer 2000). Interestingly, Hsp40 overexpression altered the distribution of nuclear inclusions of Atx1-82Q, which appeared to coalesce into a single compact aggregate per nucleus, suggesting a link between aggregate distribution and toxicity (Fernandez-Funez et al. 2000). Further prove of the protective activity of Hsp70 came from overexpression of the Drosophila orthologue of human Hdj-1/Hsp40 in fly models of polyglutamine toxicity. Flies overexpressing dHdj1 strongly suppressed Atx3tr-78Q toxicity, but only partially suppressed the toxicity of mutant Huntingtin (Htt-120Q). On the other hand, elimination of the J-domain or mutation of the SBD enhanced the toxicity of Atx3tr-78Q, suggesting that mutant dHdj1 behaves as a dominant-negative by blocking the protective activity of endogenous factors, i.e., Hsp70. However, expression of a second orthologue, dHdj2, partially rescued Atx3tr-78Q and did not rescue Htt-120Q, indicating that Hsp40s exhibit substrate selectivity. Moreover, a coordinated overexpression of both, dHdj1 and Hsp70 showed an even stronger protection (Chan et al. 2000; Bonini 2002). This different function supported the theory that chaperones from the same family can recognize different substrates and, in consequence, exert different functions (Bonini 2002). Interestingly, these studies failed to observe changes in nuclear inclusions, which suggested a decrease in protein neurotoxicity without reductions on protein aggregation (Chan et al. 2000).
Subsequent studies demonstrated that, besides Hsp70 , other chaperones cooperate with Hsp40 to mitigate protein toxicity. Kuo and collaborators described a chaperone capable of enhancing the protective effect of DNAJ-1, Hsp70bc, an orthologue of human Hsp110. Despite the structural and functional conservation with Hsp70, Hsp70bc did not rescue polyQ toxicity on its own. However, Hsp70bc enhanced the protective activity of Hsp40, whereas inactivation of its ATPase domain led to a complete loss of such protective function (Kuo et al. 2013), demonstrating the cooperative activity of HSPs.
Another interesting member of the Hsp40 family of chaperones are the cysteine-string proteins (CSPs). CSPs contain the characteristic N-terminal J domain that recognize and bind to Hsc70/Hsp70 proteins promoting their ATPase activity (Miller et al. 2003; Zinsmaier 2010). Under physiological conditions, CSPs participate on the maintenance of synaptic function and structure by regulating the Soluble NSF Attachment Protein Receptor (SNARE) complex assembly and modulating presynaptic Ca2+ channels activity (Zinsmaier 2010). In addition, CSP chaperone activity could impede neurodegeneration by preventing toxic accumulation of misfolded synaptic proteins. However, when abnormal protein aggregation occurs in the neurons , sequestration of CSPs by the toxic aggregates compromise CSP function, reducing synapsis and triggering neurodegeneration (Miller et al. 2003; Fernandez-Chacon et al. 2004; Zinsmaier 2010). Recently, Wang et al. described that overexpression of Hip, a co-chaperone that enhanced and stabilized binding of Hsp70 to its substrates, promotes client protein’s ubiquitination and poly Q-Androgen receptor clearance. In this regard, YM-1, a synthetic co-chaperone similar to Hip, led to similar results when supplied to a Drosophila model of SBMA. Thus, allosteric activators of Hsp70 can be used as new therapeutic approaches against the protein aggregation that occurs during age-depending neurodegenerative disorders (Wang et al. 2013).
2.2 Hsp90
Hsp90 is the most conserved chaperone involved in the folding, stability and maturation of the structural integrity of proteins. Hsp90 contains an N-terminal ATPase domain, a SBD in the middle, and a C-terminal domain for dimerization (Pearl and Prodromou 2006). Hsp90 assists in protein folding and stabilization in coordination with Hsp70 and Hsp40, but it seems to have a higher affinity for aberrant proteins (Waza et al. 2006; Luo et al. 2008; Koren et al. 2009). Hsp90 plays a key regulatory role in the heat shock response due to its binding to Heat shock factor 1 (HSF1), a transcription factor that regulates the expression of Hsp70 and other HSPs. Hsp70 and Hsp40 also bind HSF1, but do not inhibit its transcriptional activity. Under stress, Hsp90 releases HSF1, which forms trimers, relocates to the nucleus, and induces the expression of HSPs (Morimoto 2008; Luo 2010, 2013). Hsp90 inhibitors have been developed to reverse the permissive role of Hsp90 in tumorigenesis. Several geldanamycin derivatives show lower toxicity and improved activity, including 17-allylamino-17-demethoxygeldanamycin (17-AAG) and 17-(dimethylaminoethylamino)-17-demethoxygeldanamycin (17-DMAG). However, these Hsp90 inhibitors still have limitations for clinical use due to their toxicity.
Quite surprisingly, the potential benefit of Hsp90 refolding activity against amyloids has not been tested in animal models, so far. In fact, Hsp90 has been targeted for inhibition as a strategy to induce the protective activity of HSF1 and its downstream targets, including Hsp70 . Administration of Hsp90 inhibitors in the media activates HSF-1 in Drosophila, which leads to suppression of Atx3tr-78Q, α-synuclein, and Htt-120Q (Auluck et al. 2002; Fujikake et al. 2008). This recovery correlates with an increase in Hsp70 and Hsp40 transcription, demonstrating for the first time the use of chaperones as therapeutic targets in protein-misfolding diseases. We had shown before the beneficial effect of Hsp70 overexpression against PrP neurotoxicity and wanted to find out whether the Hsp90 inhibitors also showed this protective activity. We found that the Hsp90 inhibitors geldanamycin and 17-DMAG had no effect on the accumulation of PrP. This was somehow expected because the protective effect of Hsp70 on PrP is weak due to their presence in different compartments. We argued that boosting Hsp70 induction with a second compound would enhance the activity of 17-DMAG and result in direct effects on PrP accumulation. We found that the glucocorticoid dexamethasone acted as an Hsp70 activator by increasing the transcriptional activity of HSF1 by stabilizing its binding to the transcriptional machinery. As predicted, the combination of 17-DMAG and dexamethasone significantly increased the levels of inducible Hsp70 and decreased PrP steady-state levels (Zhang et al. 2014). This treatment reduced final levels of pathogenic PrP, and improved locomotor activity without the potential deleterious effects observed when adding either drug in high doses.
2.3 Small Heat Shock Proteins (SHSPs)
Small HSPs (sHSPs) comprise all chaperones under 30 kDa with an 80 amino acid α-crystallin domain responsible for intra- and inter-molecular interactions. The sHSPs family includes 10 members in mammals (referred to as HSPB1 to 10) and four in Drosophila (Hsp22, Hsp23, Hsp26, and Hsp27) that exert different functions, although most inhibit protein aggregation and increase the clearance of abnormally folded proteins.
HSPBs dimerize through their α-crystallin domains and form oligomers that recognize and inhibit the aggregation of unfolded proteins (Carra et al. 2008, 2009). Although all HSPBs share structural similarities, they exert different activities in vivo. For instance, human HspB7 and HspB8 exhibited the strongest protective function against Atx3tr-78Q toxicity in flies. Cell culture assays showed that this protective activity was due to a high anti-aggregation function that does not require the refolding activity. Interestingly, the anti-aggregation activity associated with HSPB7 and HSPB8 is exerted through different mechanisms and is substrate-dependent (Vos et al. 2010). In contrast, HSPB1, HSPB4, and HSPB5 display milder protective effects against Atx3tr-78Q with little anti-aggregation activity, but a strong refolding effect. Regarding Drosophila sHsps, Hsp27 is the only one that improves paraquat-induced movement disorder and mild toxicity caused by a short polyQ tract (47 residues); however, it does not alleviate the severe toxicity caused by a long polyQ repeat (121 residues) (Liao et al. 2008). These data support the hypothesis that HSPBs have different functions and substrate selectivity to maintain protein homeostasis (Vos et al. 2010).
2.4 Heterologous Hsp104
The strong protective effect of classic HSPs against misfolded protein has sparked a renewed interest in new chaperones with therapeutic applications. Hsp104 is a yeast chaperone that has recently gain attention because of its ability to efficiently disaggregate amyloids, although it shows poor ability to prevent aggregation of unfolded substrates. Hsp104 forms a hexameric complex with two AAA+-ATPase sites per monomer (NBD1 and 2) and a large central cavity for the interaction with aggregated substrates.
The interest on Hsp104 relies on its unusual ability to disaggregate large protein aggregates, which has been tested in different animal models including Drosophila and mice . Recently, Hsp104 demonstrated different activities against closely related substrates: Hsp104 suppressed the toxicity of Atx3tr-78Q without affecting its aggregation, but enhanced the toxicity of full length Atx3-78Q (Cushman-Nick et al. 2014). These opposite effects were explained by the interaction of Hsp104 with different domains that modulate Atx3 conformation. Another interesting finding in this study is that Hsp104, contrary to Hsp70, reduces Atx3tr-78Q aggregation after it has begun. By using the conditional Gene Switch system in Drosophila (Roman et al. 2001), the authors induced Hsp104 or Hsp70 at day 7 in flies that expressed Atx3tr-78Q constitutively in the eye. While Hsp70 had no effect, Hsp104 mitigated the degeneration associated with pre-existing Atx3tr-78Q aggregates (Cushman-Nick et al. 2014). These promising results suggest that this heterologous HSP could have therapeutic applications against proteinopathies after the onset of pathogenic protein-induced degeneration.
2.5 NMNAT
NAD synthase nicotinamide mononucleotide adenylyltransferase (NMNAT) is a protein that acts both as NAD synthase and chaperone in Drosophila (Zhai et al. 2006). NMNAT delays axonal degeneration and protects against protein toxicity in Drosophila models of spinocerebellar ataxia type 1 (SCA1), suggesting that NMNAT is required for neuronal maintenance and neuroprotection . Presumably, NMNAT reduces aggregation and promotes protein degradation through a proteasome-mediated pathway (Zhai et al. 2008). In addition, NMNAT promotes ubiquitination and clearance of toxic tau species in Drosophila, and when overexpressed suppresses tau-related phenotypes (Ali et al. 2012). Moreover, in fly brains overexpressing poly-glutamine expanded proteins, NMNAT appears up-regulated and co-localizes with Hsp70 in protein aggregates; however, both proteins act independently and in an additive manner. These data suggest that NMNAT acts as a stress-response chaperone to maintain and protect neuronal cells (Zhai et al. 2008; Jaiswal et al. 2012).
2.6 Engineered Chaperones: Secreted Hsp70
One of the limitations of HSPs is that small amounts leak outside the cell under stressful conditions. However, several proteinopathies are characterized by protein aggregates in the extracellular space, where limited chaperone activity exists. We devised a new approach to increase the chaperone activity in the extracellular space by designing a secreted form of Hsp70 (secHsp70). This new chaperone consists in fusing the signal peptide of the human Immunoglobulin heavy chain V–III to HSPA1L. SecHsp70 can be detected in cell media and in the lumen of the eye imaginal disc , confirming the functionality of the signal peptide. Moreover, the activity of Hsp70 outside the cell neutralizes the toxicity of Aβ42 in a Drosophila model of AD, including life span extension. We confirmed that secHsp70 binds Aβ42, but this interaction has no effect on Aβ42 steady-state levels or aggregation. We propose that the protective activity of secHsp70 is mediated by masking key neurotoxic Aβ42 epitopes, thus preventing the interaction of Aβ42 with cellular substrates that mediate pathogenesis, including receptors and channels. This ability of secHsp70 to bind misfolded extracellular substrates may also prove useful to prevent the prion-like spread of different oligomeric assemblies proposed to mediate pathogenesis, including synucleinopathies, tauopathies, and TDP-43 proteinopathies (Fernandez-Funez et al. under review).
2.7 Chaperone Dynamics During Aging
Aggregation of damaged or unfolded proteins is associated with an impairment in the mechanisms involved in protein quality control associated with aging, and is a risk factor for the development of several age-dependent diseases. Chaperone proteins are the first cell defense to assure correct protein folding and ameliorate protein aggregation. However, during aging, protein aggregation is enhanced, which can be partly associated with a decrease in the expression and function of HSPs, or defects in capacity of HSPs (Soti and Csermely 2003). In Drosophila, expression of hsp22 appears up-regulated during aging, most likely as a response to stressors only present later in life; and, at normal temperatures, transient heat induced overexpression of Hsp70 increases life span (Tatar et al. 1997; King and Tower 1999). Furthermore, overexpression of Drosophila Hsp22, Hsp23 and Hsp26 resulted in a mean increase of life span in flies exposed to mild heat stress and paraquat-induced oxidative stress (Morrow and Tanguay 2003; Liao et al. 2008). However, as stated above, only HSP27 has the ability to ameliorate paraquat-induced movement disorder (Liao et al. 2008).
Interestingly, although expression of HSPs is positively-correlated with life span under mild stressors , under constant stress HSP expression can be an indicative of susceptibility and failure in homeostasis (Yang and Tower 2009).
3 ER Stress and Unfolded Protein Response
The endoplasmic reticulum (ER) is an essential organelle responsible for the translocation and folding of ER-resident proteins, membrane proteins, and secreted proteins. A complex protein network in the ER lumen regulates post-translational modifications to ensure correct protein function (Hetz et al. 2011). Alterations in ER homeostasis due to protein accumulation in the lumen results in a condition termed ER stress, which results in the activation of the unfolded protein response (UPR). Under normal circumstances, UPR controls abnormal protein aggregation by attenuating protein synthesis, promoting protein refolding, and enhancing abnormal protein degradation. However, chronic disruption in ER proteostasis can trigger cell death leading to neurodegeneration (Matus et al. 2011). Recent evidence indicates that chronic ER stress is involved in diverse diseases, including neurodegenerative conditions, cancer , and diabetes (Koumenis 2006; Lipson et al. 2006; Hetz and Glimcher 2009).
Three key ER stress sensors regulate the UPR: protein kinase RNA-like ER kinase (PERK), activating transcription factor 6 (ATF6), and inositol-requiring enzyme 1 (IRE1) (Hetz and Mollereau 2014). PERK is a type I ER transmembrane protein with a cytosolic kinase domain. When misfolded proteins accumulate in the ER, the cytosolic domain of PERK dimerizes and autophosphorylates, which leads to eIF2a phosphorylation and inactivation (Harding et al. 1999). EIF2a inactivation causes a decrease in the general protein synthesis rate and increases the translation of the transcription factor ATF4, which regulates expression of UPR genes involved in amino acid metabolism , autophagy , and resistance to oxidative stress (Hetz et al. 2011). The PERK signaling cascade is reversible once ER homeostasis is reestablished, recovering the normal translation rate.
ATF6 is a type II ER transmembrane protein that comprises a bZIP transcriptional motif in the cytosolic domain. Under normal conditions, the ATF6 is retained at the ER membrane. Under ER stress conditions, ATF6 migrates to the Golgi apparatus, where two protease cleavages release the cytosolic fragment. Cleaved ATF6, then, enters the nucleus to activate the transcription of several ER-chaperones and endoplasmic reticulum-associated protein degradation (ERAD)-associated genes among others. ATF6 response initiates early during the UPR and activates expression of unspliced X-box binding protein 1 (XBP1), which accumulates in the ER becoming available for the later IRE1 response.
IRE1 is the most conserved branch of the UPR, with homologues in plants, C. elegans, Drosophila, and mammals . IRE1 is a type I transmembrane protein with a luminal domain responsible for detecting ER stress and a cytosolic domain with protein kinase and endonuclease activities (Rasheva and Domingos 2009). Upon ER stress, IRE1 oligomerizes and autophosphorylates, activating an alternative splicing in XBP1 mRNA . Spliced XBP1 (XBP1s) is a potent transcription factor that regulates the expression of genes associated with protein folding , ERAD, protein translocation into the ER, and lipid synthesis among other processes. During the recovery phase of ER stress, the unspliced XBP1 acts as a negative regulator of XBP1s, which inhibits transcription of target genes (Hetz et al. 2011).
Under physiological circumstances, the UPR is activated as a protective mechanism against ER stress caused by abnormal protein aggregation associated with cellular differentiation and exposure to stressors . ER stress markers also appear up-regulated in several models of neurodegeneration as well as in postmortem brains of several neurodegenerative diseases suggesting a pathogenic role of UPR. While a transitory response can be beneficial for cell survival, a chronic UPR can lead to apoptotic mechanisms promoting neurodegeneration (Halliday and Mallucci 2014).
In recent years, Drosophila has contributed to elucidate the role of UPR against protein toxicity in vivo. Retinitis pigmentosa (RP) is a degenerative eye condition caused by an accumulation of misfolded mutant Rhodopsin 1 (Rh1*) in the ER. In flies, accumulation of misfolded Rh1* in the ER elevates the expression of XBP1s. Accordingly, reduction of XBP1 increases the retinal degeneration induced by Rh1*, which indicates that XBP1 and its target genes are protective (Ryoo and Steller 2007). Moreover, the authors describe that disruption of the ERAD pathway leads to an increase in Rh1 protein levels in Drosophila. Consistently, the ER stress caused by Rh1 is reduced when some subunits of the ERAD machinery are overexpressed (Kang and Ryoo 2009). Interestingly, UPR is also implicated in neurodegenerative diseases. In a fly model of tauopathy, UPR is increased due to phosphorylation of tau, and when levels of XBP1 are reduced, the ER stress response decreases promoting tau toxicity and neurodegeneration (Loewen and Feany 2010). In addition, we recently described that a fly model expressing Aβ42 displays ER stress-mediated neurodegeneration and that overexpression of XBP1s suppresses this phenotype (Casas-Tinto et al. 2013). Accordingly, XBP1 loss-of-function exacerbates Aβ42-induced toxicity. This protective effect of XBP1s was mediated by the down-regulation of ryanodine calcium channels in the ER, which prevents the release of pro-apoptotic levels of calcium in the cytoplasm (Casas-Tinto et al. 2013). Another study with Drosophila identified mild ER stress (“preconditioning”) as a neuroprotective mechanism against human α-synuclein toxicity (Fouillet et al. 2012). These authors demonstrated that α-synuclein-expressing flies treated with a mild dose of tunicamycin, a chemical inducer of UPR, display activation of the IRE1-XBP1 pathway in the brain, improvement of locomotor function and protection of dopaminergic neurons . In addition, the authors found that this “preconditioned” mild ER stress mediates neuronal survival by blocking apoptosis in vivo. Interestingly, this ER-mediated protection was lost upon autophagy impairment suggesting that autophagic clearance contributes to the ER-mediated protection (Fouillet et al. 2012). These data agree with the hypothesis that UPR functions as neuroprotective mechanism when ER stress is not prolonged in time.
3.1 UPR Impairment During Aging
It is clear that a better resistance to stress leads to an increase in life span, which correlates with a better prevention against different harmful insults (Salminen and Kaarniranta 2010). However, the amount of misfolded proteins increases in the ER lumen during aging suggesting an involvement of ER stress in the aging process (Salminen and Kaarniranta 2010). Indeed, while the UPR is strong in young animals, it is compromised during aging and the expression of several ER proteins is reduced (Naidoo 2009). It is not surprising then, that some neurodegenerative disorders that involved accumulation of misfolded proteins present impaired UPR (Brown and Naidoo 2012). As it occurs with other protein quality control systems, duration and strength of the stressor have different consequences in the cell and life span of the organisms. Mild doses of a stressor can lead to a better stress tolerance, whereas a chronic stress exposure leads to cellular degeneration. This is especially true in the ER, where abnormal protein accumulation lead to a sustained UPR that triggers cell apoptosis.
4 Autophagy
Autophagy is a catabolic process for degrading and recycling organelles and misfolded proteins inside double membrane vesicles called autophagosomes. The fusion of the autophagosome with lysosomes allows the degradation of the contents of the vesicle, producing fresh building materials or energy. Autophagy occurs at basal levels in healthy cells but can be up-regulated by starvation , cellular stress, and other stimuli. This basic cellular process is critical for cell survival and renewal, but it is particularly important for quality control in neurons due to the limited regenerating capacity of the nervous system.
4.1 Macroautophagy and Protein Misfolding Diseases
In the last decade, several studies have shown presence of autophagy vesicles in neurodegenerative disorders such as HD or PD, pointing to a protective role of this mechanism against neurodegeneration . Drosophila has been a useful tool to study this process. Ravikumar and colleges showed that TOR (target of rapamycin, a key regulator of autophagy) is sequestered in polyQ aggregates and promotes autophagy (Ravikumar et al. 2004). These authors proposed that rapamycin, a negative regulator of TOR, would increase autophagy and protect against the toxicity of Htt-120Q. Flies expressing Htt-120Q in the eye fed with rapamycin experienced an increase in the number of photoreceptors (Ravikumar et al. 2004). In fact, rapamycin also reduces toxicity caused by wild type and mutant tau in Drosophila presumably because of autophagy degradation of insoluble tau (Ravikumar and Rubinsztein 2006). Moreover, overexpression of TSC1 and TSC2, which are negative regulators of TOR, inhibit neuronal degeneration caused by Htt-120Q (Wang et al. 2009a). Overexpression of the autophagy gene 1 (Atg1) also suppresses photoreceptor cell death in the Htt-120Q flies due to an induction of autophagy (Wang et al. 2009a).
In another study, inhibition of the small GTPase Rab5, which regulates endosome trafficking, enhances the toxicity of Htt-120Q, whereas overexpression enhances autophagosome synthesis, suppression of aggregation, and suppression of Htt-120Q toxicity (Ravikumar et al. 2008). These data suggest that besides the role of Rab5 in endocytosis, it also functions in the early stages of autophagosome formation (Ravikumar and Rubinsztein 2006; Ravikumar et al. 2008). On the other hand, a screen for modifiers of Atx3tr-78Q neurodegeneration identified genes that affect protein accumulation through autophagy confirming the protective effect of this process over protein deposition (Bilen and Bonini 2007).
However, not all aggregate-prone proteins are degraded by autophagy. P. Salvaterra’s group found that expression of Aβ1-42 induces the formation of autophagic vesicles that compromise cell viability and led to neurological deficits, whereas Aβ1-40 did not show changes in autophagy (Ling et al. 2009). This suggests that, depending of the substrate, autophagy can begin as a protective mechanism but due to a decrease in its degradation function induces abnormal autophagy vesicles leading to neurodegeneration (Ling et al. 2009). In a posterior study, Ling and Salvaterra observed that this deterioration in the autophagy system shifts normal brain aging into pathological aging. The authors used temporal-dependent paradigms in flies and found an early protective effect of autophagy in life that becomes progressively deleterious during normal brain aging (Ling and Salvaterra 2010). Under these paradigms, expression of Aβ1-42 exacerbates the dysfunction of the autophagy system increasing the ratio of neurodegeneration (Ling and Salvaterra 2010). This observation agrees with the fact that age is a risk factor in the onset of AD.
4.2 Interaction Between Autophagy and Chaperones
Chaperone-mediated autophagy (CMA) is a process that selectively removes damaged or unfolded cytosolic proteins in order to maintain proteostasis (Cuervo and Wong 2014). Although CMA occurs under normal conditions, its activity is increased under stress conditions. In CMA, misfolded proteins are targeted on the cytosol by chaperones, which recognize a specific amino acid sequence, and, then, transported to the lysosomal membrane. Once there, the protein enters the lysosome by crossing the membrane through a translocation complex. CMA selective degradation increases over time in the presence of a stressor as a protective mechanism. In this regard, its specific substrate selection avoids the elimination of proteins or organelles that are essential for the cell survival , and preferentially targets non-essential proteins. Interestingly, CMA is impaired in several human diseases including neurodegenerative disorders as PD, AD or PolyQ diseases. Aberrant proteins such as alpha-synuclein or leucine-rich repeat kinase 2 (LRRK2) can interact with lysosome-associated membrane protein 2A (LAMP-2A), the receptor for chaperone-mediated autophagy , with high affinity but are unable to translocate to the lysosome lumen. This interaction not only fails to eliminate these aberrant or damaged proteins, but also affects the degradation of other CMA substrates by saturating the system (Cuervo et al. 2004; Orenstein et al. 2013). In the case of tauopathies, it seems that tau binds successfully LAMP-2A but undergoes a partial translocation to the lysosome lumen releasing a toxic tau fragment to the cytosol, therefore, increasing toxicity and cell death (Wang et al. 2009b; Cuervo and Wong 2014).
It is clear that chaperones play an important role against protein misfolding (Kim et al. 2013). However, some proteins cannot be refolded and need to be directed to degradation by an independent system. Some chaperones lack refolding activity and, instead, act as part of the autophagy system to target unfolded proteins to degradation by the lysosome. For instance, two members of HSPB family, HspB7 and HspB8, protect against polyQ toxicity in an autophagy-mediated manner (Vos et al. 2010). These findings suggest that autophagy acts downstream of the sHSP response. In support of this idea, overexpression of autophagy related 7 gene (Atg7) is sufficient to revert Hsp27 knockdown-mediated shortened life span in Drosophila. Conversely, knockdown of Atg7 blocks Hsp27-mediated long life span (Chen et al. 2012).
Altogether, these data suggests that the heat shock protein response functions upstream of autophagy and that the association between these pathways can be due to CMA or chaperone-assisted selective autophagy (CASA).
4.3 Autophagy and Aging
During normal aging, the expression of several autophagy genes is reduced in Drosophila. A decrease in autophagy is associated with an increase of neuronal damage and a reduced life span, however maintaining the expression of autophagy-related genes promotes longevity and prevents the age-dependent damage. For instance, when the Atg8a gene (autophagy-related 8a) is mutated in flies, life span is shorter and sensitivity to oxidative stress is higher (Simonsen et al. 2008). Interestingly, a raise in Atg8a expression in old fly brains results in an increase over 50 % in life span and promotes resistance to oxidative stress (Simonsen et al. 2008). Similarly, expression of Atg7, an essential factor for longevity of flies, has been consistently associaed with the regulation of aging (Hara et al. 2006; Chen et al. 2012). Furthermore, mutations in Atg5 and Atg7 genes, which are required for autophagosome formation, lead to increased neurodegeneration (Hara et al. 2006). These data support the hypothesis that autophagy plays an important role in regulation of aging and that its activation can be neuroprotective.
5 UPS and Interaction Between Protein Degradation Pathways
Protein quality control is a complex mechanism that needs a tight collaboration between several processes to avoid misfolded protein aggregation and cell death. When chaperones fail to unfold and refold abnormal proteins, degradation pathways are activated to prevent protein toxicity and ultimately cell death. The UPS is one of the pathways involved in the aberrant protein degradation (Hershko and Ciechanover 1998). In fact, a wide range of mutations in UPS-associated genes are associated with accumulation of proteasome substrates and, therefore, with neurodegeneration (Ciechanover and Brundin 2003). Interestingly, compromising the proteasome pathway in a Drosophila model of SBMA enhances degeneration and decreases poly(Q) protein solubility (Chan et al. 2002). Moreover, the protective activity of SCA3 observed against polyQ neurotoxicity in vivo requires the ubiquitin-associated activities of the protein and is dependent upon proteasome function (Warrick et al. 2005). In addition, reduction of the 26S proteasome activity in flies is associated with age-related accumulation of proteins and the duration of life span (Tonoki et al. 2009).
Recent evidences suggest a compensatory regulation between autophagy, UPR and UPS. For instance, in a fly model of SBMA, expression of histone deacetylase 6 (HDAC6), a microtubule-associated deacetylase that interacts with polyubiquitinated proteins, rescues degeneration associated with UPS impairment through autophagy (Pandey et al. 2007). Similarly, in a fly model for Gaucher’s disease, activation of UPR leads to accumulation of parkin substrates triggering cell death most likely through an attenuation of normal autophagy (Maor et al. 2013). In addition, Drosophila proteasome mutants show increased autophagy, whereas autophagy mutants display increased ubiquinated protein agreggation (Chang and Neufeld 2010; Jaiswal et al. 2012). Moreover, flies treated with Bortezomib—a selective proteasome inhibitor—exhibit UPR induction that leads to autophagy activation to counterbalance UPS impairment (Velentzas et al. 2013). All these data illustrate the complexity of the regulatory mechanisms that coexist in the cell to maintain proteostasis during aging in an attempt to prevent protein aggregation and cell death.
6 Concluding Remarks
The functional similarities between Drosophila and mammalian proteomes make it possible to model human protein misfolding disorders with results that are relevant to human physiology. In fact, Drosophila has been behind many of the fundamental advances that have occurred in this field during the last 15 years. While fruit flies cannot replace the need for studying mammalian models, the studies outlined here illustrate their potential to untangle the molecular associations between protein aggregation, neurodegeneration and aging. In addition, these studies illustrate how Drosophila has contributed to understand a number of complex processes utilized by the cell to maintain protein homeostasis in the brain. It is clear now that promoting a correct refolding of misfolded proteins by overexpressing chaperones, enhancing the activity of the UPR, or promoting protein clearance with autophagy or the ubiquitin proteasome system reduces protein toxicity and improves life span. However, it is important to keep a balance among the pathways involved in proteostasis, otherwise a sustained stimulation can overwhelm the systems and contribute to potentiate cell death. Altogether, these important discoveries demonstrate the relevance of Drosophila as a model to study protein quality control in late-onset neurodegenerative diseases and its potential to design new therapeutic strategies for these devastating disorders. We anticipate, therefore, that the power of Drosophila genetics will further extend our understanding of neuronal proteostasis in new and unexpected directions in the years to come.
References
Ali YO, Ruan K, Zhai RG (2012) NMNAT suppresses tau-induced neurodegeneration by promoting clearance of hyperphosphorylated tau oligomers in a Drosophila model of tauopathy. Hum Mol Genet 21(2):237–250
Auluck PK, Chan HY, Trojanowski JQ, Lee VM, Bonini NM (2002) Chaperone suppression of alpha-synuclein toxicity in a Drosophila model for Parkinson’s disease. Science 295(5556):865–868
Bilen J, Bonini NM (2007) Genome-wide screen for modifiers of ataxin-3 neurodegeneration in Drosophila. PLoS Genet 3(10):1950–1964
Bonini NM (2002) Chaperoning brain degeneration. Proc Natl Acad Sci USA 99(Suppl 4):16407–16411
Brown MK, Naidoo N (2012) The endoplasmic reticulum stress response in aging and age-related diseases. Front Physiol 3:263
Carra S, Seguin SJ, Landry J (2008) HspB8 and Bag3: a new chaperone complex targeting misfolded proteins to macroautophagy. Autophagy 4(2):237–239
Carra S, Brunsting JF, Lambert H, Landry J, Kampinga HH (2009) HspB8 participates in protein quality control by a non-chaperone-like mechanism that requires eIF2{alpha} phosphorylation. J Biol Chem 284(9):5523–5532
Casas-Tinto S, Zhang Y, Sanchez-Garcia J, Gomez-Velazquez M, Rincon-Limas DE, Fernandez-Funez P (2013) The ER stress factor XBP1s prevents amyloid-beta neurotoxicity. Hum Mol Genet 20(11):2144–2160
Chan HY, Warrick JM, Gray-Board GL, Paulson HL, Bonini NM (2000) Mechanisms of chaperone suppression of polyglutamine disease: selectivity, synergy and modulation of protein solubility in Drosophila. Hum Mol Genet 9(19):2811–2820
Chan HY, Warrick JM, Andriola I, Merry D, Bonini NM (2002) Genetic modulation of polyglutamine toxicity by protein conjugation pathways in Drosophila. Hum Mol Genet 11(23):2895–2904
Chang YY, Neufeld TP (2010) Autophagy takes flight in Drosophila. FEBS Lett 584(7):1342–1349
Chen SF, Kang ML, Chen YC, Tang HW, Huang CW, Li WH, Lin CP, Wang CY, Wang PY, Chen GC, Wang HD (2012) Autophagy-related gene 7 is downstream of heat shock protein 27 in the regulation of eye morphology, polyglutamine toxicity, and lifespan in Drosophila. J Biomed Sci 19:52
Ciechanover A, Brundin P (2003) The ubiquitin proteasome system in neurodegenerative diseases: sometimes the chicken, sometimes the egg. Neuron 40(2):427–446
Cuervo AM, Wong E (2014) Chaperone-mediated autophagy: roles in disease and aging. Cell Res 24(1):92–104
Cuervo AM, Stefanis L, Fredenburg R, Lansbury PT, Sulzer D (2004) Impaired degradation of mutant alpha-synuclein by chaperone-mediated autophagy. Science 305(5688):1292–1295
Cushman-Nick M, Bonini NM, Shorter J (2014) Hsp104 suppresses polyglutamine-induced degeneration post onset in a Drosophila MJD/SCA3 model. PLoS Genet 9(9):e1003781
del Valle Rodriguez A, Didiano D, Desplan C (2011) Power tools for gene expression and clonal analysis in Drosophila. Nat Methods 9(1):47–55
Douglas PM, Cyr DM (2009) Interplay between protein homeostasis networks in protein aggregation and proteotoxicity. Biopolymers 93(3): 229–236
Fernandez-Chacon R, Wolfel M, Nishimune H, Tabares L, Schmitz F, Castellano-Munoz M, Rosenmund C, Montesinos ML, Sanes JR, Schneggenburger R, Sudhof TC (2004) The synaptic vesicle protein CSP alpha prevents presynaptic degeneration. Neuron 42(2):237–251
Fernandez-Funez P, Nino-Rosales ML, de Gouyon B, She WC, Luchak JM, Martinez P, Turiegano E, Benito J, Capovilla M, Skinner PJ, McCall A, Canal I, Orr HT, Zoghbi HY, Botas J (2000) Identification of genes that modify ataxin-1-induced neurodegeneration. Nature 408(6808):101–106
Fernandez-Funez P, Casas-Tinto S, Zhang Y, Gomez-Velazquez M, Morales-Garza MA, Cepeda-Nieto AC, Castilla J, Soto C, Rincon-Limas DE (2009) In vivo generation of neurotoxic prion protein: role for hsp70 in accumulation of misfolded isoforms. PLoS Genet 5(6):e1000507
Fleshner M, Johnson JD (2005) Endogenous extra-cellular heat shock protein 72: releasing signal(s) and function. Int J Hyperth 21(5):457–471
Fouillet A, Levet C, Virgone A, Robin M, Dourlen P, Rieusset J, Belaidi E, Ovize M, Touret M, Nataf S, Mollereau B (2012) ER stress inhibits neuronal death by promoting autophagy. Autophagy 8(6):915–926
Fujikake N, Nagai Y, Popiel HA, Okamoto Y, Yamaguchi M, Toda T (2008) Heat shock transcription factor 1-activating compounds suppress polyglutamine-induced neurodegeneration through induction of multiple molecular chaperones. J Biol Chem 283(38):26188–26197
Halliday M, Mallucci GR (2014) Targeting the unfolded protein response in neurodegeneration: a new approach to therapy. Neuropharmacology 76(Pt A):169–174
Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, Yokoyama M, Mishima K, Saito I, Okano H, Mizushima N (2006) Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature 441(7095):885–889
Harding HP, Zhang Y, Ron D (1999) Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature 397(6716):271–274
Hershko A, Ciechanover A (1998) The ubiquitin system. Annu Rev Biochem 67:425–479
Hetz C, Glimcher LH (2009) Fine-tuning of the unfolded protein response: assembling the IRE1alpha interactome. Mol Cell 35(5):551–561
Hetz C, Mollereau B (2014) Disturbance of endoplasmic reticulum proteostasis in neurodegenerative diseases. Nat Rev Neurosci 15(4):233–249
Hetz C, Martinon F, Rodriguez D, Glimcher LH (2011) The unfolded protein response: integrating stress signals through the stress sensor IRE1alpha. Physiol Rev 91(4):1219–1243
Jaiswal M, Sandoval H, Zhang K, Bayat V, Bellen HJ (2012) Probing mechanisms that underlie human neurodegenerative diseases in Drosophila. Annu Rev Genet 46:371–396
Kang MJ, Ryoo HD (2009) Suppression of retinal degeneration in Drosophila by stimulation of ER-associated degradation. Proc Natl Acad Sci USA 106(40):17043–17048
Kazemi-Esfarjani P, Benzer S (2000) Genetic suppression of polyglutamine toxicity in Drosophila. Science 287(5459):1837–1840
Kim YE, Hipp MS, Bracher A, Hayer-Hartl M, Hartl FU (2013) Molecular chaperone functions in protein folding and proteostasis. Annu Rev Biochem 82:323–355
King V, Tower J (1999) Aging-specific expression of Drosophila hsp22. Dev Biol 207(1):107–118
Koren J 3rd, Jinwal UK, Lee DC, Jones JR, Shults CL, Johnson AG, Anderson LJ, Dickey CA (2009) Chaperone signalling complexes in Alzheimer’s disease. J Cell Mol Med 13(4):619–630
Koumenis C (2006) ER stress, hypoxia tolerance and tumor progression. Curr Mol Med 6(1):55–69
Kuo Y, Ren S, Lao U, Edgar BA, Wang T (2013) Suppression of polyglutamine protein toxicity by co-expression of a heat-shock protein 40 and a heat-shock protein 110. Cell Death Dis 4:e833
Liao PC, Lin HY, Yuh CH, Yu LK, Wang HD (2008) The effect of neuronal expression of heat shock proteins 26 and 27 on lifespan, neurodegeneration, and apoptosis in Drosophila. Biochem Biophys Res Commun 376(4):637–641
Ling D, Salvaterra PM (2010) Brain aging and Abeta(1)(-)(4)(2) neurotoxicity converge via deterioration in autophagy-lysosomal system: a conditional Drosophila model linking Alzheimer’s neurodegeneration with aging. Acta Neuropathol 121(2):183–191
Ling D, Song HJ, Garza D, Neufeld TP, Salvaterra PM (2009) Abeta42-induced neurodegeneration via an age-dependent autophagic-lysosomal injury in Drosophila. PLoS ONE 4(1):e4201
Lipson KL, Fonseca SG, Urano F (2006) Endoplasmic reticulum stress-induced apoptosis and auto-immunity in diabetes. Curr Mol Med 6(1):71–77
Loewen CA, Feany MB (2010) The unfolded protein response protects from tau neurotoxicity in vivo. PLoS ONE 5(9):e13084
Luo W, Rodina A, Chiosis G (2008) Heat shock protein 90: translation from cancer to Alzheimer’s disease treatment? BMC Neurosci 9(Suppl 2):S7
Luo W, Sun W, Taldone T, Rodina A, Chiosis G (2010) Heat shock protein 90 in neurodegenerative diseases. Mol Neurodegener 5:24
Luo D, Bu Y, Ma J, Rajput S, He Y, Cai G, Liao DF, Cao D (2013) Heat shock protein 90-alpha mediates aldo-keto reductase 1B10 (AKR1B10) protein secretion through secretory lysosomes. J Biol Chem 288(51):36733–36740
Maor G, Rencus-Lazar S, Filocamo M, Steller H, Segal D, Horowitz M (2013) Unfolded protein response in Gaucher disease: from human to Drosophila. Orphanet J Rare Dis 8:140
Matus S, Glimcher LH, Hetz C (2011) Protein folding stress in neurodegenerative diseases: a glimpse into the ER. Curr Opin Cell Biol 23(2):239–252
Miller LC, Swayne LA, Chen L, Feng ZP, Wacker JL, Muchowski PJ, Zamponi GW, Braun JE (2003) Cysteine string protein (CSP) inhibition of N-type calcium channels is blocked by mutant huntingtin. J Biol Chem 278(52):53072–53081
Morimoto RI (2008) Proteotoxic stress and inducible chaperone networks in neurodegenerative disease and aging. Gene Dev 22(11):1427–1438
Morrow G, Tanguay RM (2003) Heat shock proteins and aging in Drosophila melanogaster. Semin Cell Dev Biol 14(5):291–299
Naidoo N (2009) The endoplasmic reticulum stress response and aging. Rev Neurosci 20(1):23–37
Orenstein SJ, Kuo SH, Tasset I, Arias E, Koga H, Fernandez-Carasa I, Cortes E, Honig LS, Dauer W, Consiglio A, Raya A, Sulzer D, Cuervo AM (2013) Interplay of LRRK2 with chaperone-mediated autophagy. Nat Neurosci 16(4):394–406
Pandey UB, Nie Z, Batlevi Y, McCray BA, Ritson GP, Nedelsky NB, Schwartz SL, DiProspero NA, Knight MA, Schuldiner O, Padmanabhan R, Hild M, Berry DL, Garza D, Hubbert CC, Yao TP, Baehrecke EH, Taylor JP (2007) HDAC6 rescues neurodegeneration and provides an essential link between autophagy and the UPS. Nature 447(7146):859–863
Pearl LH, Prodromou C (2006) Structure and mechanism of the Hsp90 molecular chaperone machinery. Annu Rev Biochem 75:271–294
Pittet JF, Lee H, Morabito D, Howard MB, Welch WJ, Mackersie RC (2002) Serum levels of Hsp 72 measured early after trauma correlate with survival. J Trauma 52(4):611–617
Rasheva VI, Domingos PM (2009) Cellular responses to endoplasmic reticulum stress and apoptosis. Apoptosis 14(8):996–1007
Ravikumar B, Rubinsztein DC (2006) Role of autophagy in the clearance of mutant huntingtin: a step towards therapy? Mol Aspects Med 27(5–6):520–527
Ravikumar B, Vacher C, Berger Z, Davies JE, Luo S, Oroz LG, Scaravilli F, Easton DF, Duden R, O’Kane CJ, Rubinsztein DC (2004) Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat Genet 36(6):585–595
Ravikumar B, Imarisio S, Sarkar S, O’Kane CJ, Rubinsztein DC (2008) Rab5 modulates aggregation and toxicity of mutant huntingtin through macroautophagy in cell and fly models of Huntington disease. J Cell Sci 121(Pt 10):1649–1660
Rincon-Limas DE, Jensen K, Fernandez-Funez P (2012) Drosophila models of proteinopathies: the little fly that could. Curr Pharm Des 18(8):1108–1122
Roman G, Endo K, Zong L, Davis RL (2001) P[Switch], a system for spatial and temporal control of gene expression in Drosophila melanogaster. Proc Natl Acad Sci USA 98(22):12602–12607
Ryoo HD, Steller H (2007) Unfolded protein response in Drosophila: why another model can make it fly. Cell Cycle 6(7):830–835
Salminen A, Kaarniranta K (2010) ER stress and hormetic regulation of the aging process. Ageing Res Rev 9(3):211–217
Simonsen A, Cumming RC, Brech A, Isakson P, Schubert DR, Finley KD (2008) Promoting basal levels of autophagy in the nervous system enhances longevity and oxidant resistance in adult Drosophila. Autophagy 4(2):176–184
Soti C, Csermely P (2003) Aging and molecular chaperones. Exp Gerontol 38(10):1037–1040
Tatar M, Khazaeli AA, Curtsinger JW (1997) Chaperoning extended life. Nature 390(6655):30
Tonoki A, Kuranaga E, Tomioka T, Hamazaki J, Murata S, Tanaka K, Miura M (2009) Genetic evidence linking age-dependent attenuation of the 26S proteasome with the aging process. Mol Cell Biol 29(4):1095–1106
Valastyan JS, Lindquist S (2014) Mechanisms of protein-folding diseases at a glance. Dis Model Mech 7(1):9–14
Velentzas PD, Velentzas AD, Mpakou VE, Antonelou MH, Margaritis LH, Papassideri IS, Stravopodis DJ (2013) Detrimental effects of proteasome inhibition activity in Drosophila melanogaster: implication of ER stress, autophagy, and apoptosis. Cell Biol Toxicol 29(1):13–37
Voisine C, Pedersen JS, Morimoto RI (2010) Chaperone networks: tipping the balance in protein folding diseases. Neurobiol Dis 40(1):12–20
Vos MJ, Zijlstra MP, Kanon B, van Waarde-Verhagen MA, Brunt ER, Oosterveld-Hut HM, Carra S, Sibon OC, Kampinga HH (2010) HSPB7 is the most potent polyQ aggregation suppressor within the HSPB family of molecular chaperones. Hum Mol Genet 19(23):4677–4693
Wang T, Lao U, Edgar BA (2009a) TOR-mediated autophagy regulates cell death in Drosophila neurodegenerative disease. J Cell Biol 186(5):703–711
Wang Y, Martinez-Vicente M, Kruger U, Kaushik S, Wong E, Mandelkow EM, Cuervo AM, Mandelkow E (2009b) Tau fragmentation, aggregation and clearance: the dual role of lysosomal processing. Hum Mol Genet 18(21):4153–4170
Wang AM, Miyata Y, Klinedinst S, Peng HM, Chua JP, Komiyama T, Li X, Morishima Y, Merry DE, Pratt WB, Osawa Y, Collins CA, Gestwicki JE, Lieberman AP (2013) Activation of Hsp70 reduces neurotoxicity by promoting polyglutamine protein degradation. Nat Chem Biol 9(2):112–118
Warrick JM, Chan HY, Gray-Board GL, Chai Y, Paulson HL, Bonini NM (1999) Suppression of polyglutamine-mediated neurodegeneration in Drosophila by the molecular chaperone HSP70. Nat Genet 23(4):425–428
Warrick JM, Morabito LM, Bilen J, Gordesky-Gold B, Faust LZ, Paulson HL, Bonini NM (2005) Ataxin-3 suppresses polyglutamine neurodegeneration in Drosophila by a ubiquitin-associated mechanism. Mol Cell 18(1):37–48
Waza M, Adachi H, Katsuno M, Minamiyama M, Tanaka F, Sobue G (2006) Alleviating neurodegeneration by an anticancer agent: an Hsp90 inhibitor (17-AAG). Ann N Y Acad Sci 1086:21–34
Yang J, Tower J (2009) Expression of hsp22 and hsp70 transgenes is partially predictive of Drosophila survival under normal and stress conditions. J Gerontol A Biol Sci Med Sci 64(8):828–838
Young JC (2010) Mechanisms of the Hsp70 chaperone system. Biochem Cell Biol 88(2):291–300
Zhai RG, Cao Y, Hiesinger PR, Zhou Y, Mehta SQ, Schulze KL, Verstreken P, Bellen HJ (2006) Drosophila NMNAT maintains neural integrity independent of its NAD synthesis activity. PLoS Biol 4(12):e416
Zhai RG, Zhang F, Hiesinger PR, Cao Y, Haueter CM, Bellen HJ (2008) NAD synthase NMNAT acts as a chaperone to protect against neurodegeneration. Nature 452(7189):887–891
Zhang Y, Casas-Tinto S, Rincon-Limas DE, Fernandez-Funez P (2014) Combined pharmacological induction of Hsp70 suppresses prion protein neurotoxicity in Drosophila. PLoS ONE 9(2):e88522
Zinsmaier KE (2010) Cysteine-string protein’s neuroprotective role. J Neurogenet 24(3):120–132
Acknowledgments
We apologize to all authors whose work could not be cited due to space constraints. This work was supported in part by NIH grant R21NS081356 to DER-L, and by a McKnight Brain Institute Research Development Award and start-up funds from the UF Department of Neurology to PF-F and DER-L.
Author information
Authors and Affiliations
Corresponding authors
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2015 Springer International Publishing Switzerland
About this chapter
Cite this chapter
de Mena, L., Fernandez-Funez, P., Rincon-Limas, D.E. (2015). Protein Quality Control in Brain Aging: Lessons from Protein Misfolding Disorders in Drosophila . In: Vaiserman, A., Moskalev, A., Pasyukova, E. (eds) Life Extension. Healthy Ageing and Longevity, vol 3. Springer, Cham. https://doi.org/10.1007/978-3-319-18326-8_8
Download citation
DOI: https://doi.org/10.1007/978-3-319-18326-8_8
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-18325-1
Online ISBN: 978-3-319-18326-8
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)