Abstract
To classify each stage for a progressing disease such as Alzheimer’s disease is a key issue for the disease prevention and treatment. In this study, we derived structural brain networks from diffusion-weighted MRI using whole-brain tractography since there is growing interest in relating connectivity measures to clinical, cognitive, and genetic data. Relatively little work has used machine learning to make inferences about variations in brain networks in the progression of the Alzheimer’s disease. Here we developed a framework to utilize generalized low rank approximations of matrices (GLRAM) and modified linear discrimination analysis for unsupervised feature learning and classification of connectivity matrices. We apply the methods to brain networks derived from DWI scans of 41 people with Alzheimer’s disease, 73 people with EMCI, 38 people with LMCI, 47 elderly healthy controls and 221 young healthy controls. Our results show that this new framework can significantly improve classification accuracy when combining multiple datasets; this suggests the value of using data beyond the classification task at hand to model variations in brain connectivity.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
- Mild Cognitive Impairment
- Linear Discriminant Analysis
- Brain Network
- Health Control
- Structural Brain Network
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
1 Introduction
Alzheimer’s disease is by far the leading form of dementia. There is no cure for the disease, which worsens as it progresses, and eventually leads to death. According to the studies of Alzheimer’s Disease Neuroimaging Initiative (ADNI) and other large-scale multicenter studies, this disease has been described into four stages: health control (HC); early mild cognitive impairment (EMCI), late mild cognitive impairment (LMCI) and Alzheimer’s disease (AD) [1–3]. HC means there is no sign/clue that subject have any cognition impairment, while EMCI and LMCI are the middle stages in time for disease detection. AD is the last stage when there is clearly clue that disease has been onset. Defining at-risk stages of this disease is crucial for predementia detection, which in turn is the requirement for future predementia treatment. In literature, the Alzheimer’s disease multiple stages’ classification is mainly based on subjective questionnaire [1, 4]. Here we adopted machine learning method to explore multiple stages’ automatic classification using diffusion-weighted MRI (DW-MRI).
DW-MRI is a non-invasive brain imaging technique, sensitive to aspects of the brain’s white matter microstructure that are not typically detectable with standard anatomical MRI [5]. With DWI, anisotropic water diffusion can be tracked along the direction of axons using tractography methods. When tractography is applied to the entire brain, one can reconstruct major fiber bundles and describe connectivity patterns in the brain’s anatomical network [6]. Brain networks and topological measures derived from them have been shown to be highly associated with aspects of brain function and clinical measures of disease burden [7]. Some studies have begun to apply machine learning techniques to identify network features that differentiate people with various neurological and psychiatric disorders from matched HC [8]. However, most studies focus only on identifying abnormal connectivity patterns in a single disease, compared to controls, and not intermediate stages of the disease, using only using one dataset to do so. While this may improve our understanding of the outcome of the disease, when applying the same analysis to a new disease or a new dataset, the model must be re-trained and re-evaluated. Often, disease effects (or effects of other predictors on brain networks) are subtle and may not be detected in one dataset alone, or may show conflicting results across datasets. In this light, consortia such as Enhancing Neuro Imaging Genetics through Meta-Analysis (Enigma) have been formed to jointly analyze over 20,000 brain scans from patients and controls scanned at over 100 sites worldwide to meta-analyze effects on the brain [9]. This allows researchers to compare effect sizes obtained with different imaging protocols and scanners, but also across different diseases. The notion of who qualifies as a healthy control may also depend on the dataset and may not represent the healthy population at large. If multiple datasets are used to model normal variation, then arguably diagnostic classification may be improved without retraining new models for every disease and every new dataset.
When pooling scans from patients with a variety of diseases, or at different stages of disease progression, machine learning techniques can classify the data into diagnostic groups. This may involve feature extraction, dimension reduction, model training and testing. For example, principal component analysis (PCA) uses an orthogonal linear transformation to convert observations of potentially correlated variables into a new set of linearly uncorrelated principal components (PC). New datasets can then be classified into groups based on PC-projected features. Linear discriminant analysis (LDA) can also be used for dimensionality reduction and classification. It finds a linear combination of features that optimally separates two or more classes. LDA and PCA both use linear combinations of variables to model the data. LDA models the differences between classes within the data, but PCA seeks components that have the highest variance possible under the constraint that they are orthogonal to (i.e., uncorrelated with) the preceding components [10]. These dimensionality reduction methods assume that the data form a vector space. Here, each subject’s data is modeled as a vector and the collection of subjects is modeled as a single data matrix. Each column of the data matrix corresponds to one subject and each row corresponds to a feature. There are disadvantages of this vector model, as it overlooks spatial relations within the data. To overcome this, generalized low rank approximations of matrices (GLRAM) has been proposed to use a lower dimension 2D matrix to obtain more compact representations of original data with limited loss of information [11].
In this study, we combined two different datasets collected with both standard T1-weighted MRI and DW-MRI and created connectivity networks for all study participants. Both datasets had scanned healthy controls; one had also scanned patients with Alzheimer’s disease and patients with early and advanced signs of mild cognitive impairment (early MCI and late MCI respectively). We merged this data hypothesizing that we could automatically classify the scans into four groups (HC, EMCI, LMCI, and AD) using brain networks as the raw features. We used GLRAM to first reduce the dimensionality, and then applied LDA in the PCA subspace to classify the data. Classification of data from multiple sites and scanners will help us to understand differences in disease progression, ideally unconfounded by scanner differences.
2 Subjects and Methods
2.1 Data Description
Table 1 summarizes the two datasets used in this study. For all datasets, participants were scanned with both DW-MRI and standard T1-weighted structural MRI.
The first dataset included 221 healthy young adults. Images were acquired with a 4T Bruker Medspec MRI scanner, using single-shot echo planar imaging with the following parameters: TR/TE = 6,090/91.7 ms, 23 cm FOV, and a 128×128 acquisition matrix. Each 3D volume consisted of 55 2-mm axial slices, with no gap, and 1.79×1.79 mm2 in-plane resolution. Hundred and five image volumes were acquired per subject: 11 with T2-weighted b0 volumes and 94 diffusion-weighted volumes (b = 1,159 s/mm2).
The second dataset was from ADNI2, the second stage of the Alzheimer’s disease neuroimaging initiative (ADNI), publically available online (http://adni.loni.usc.edu). This dataset has 199 subjects, which includes 47 healthy elderly controls, 111 with mild cognitive impairment (MCI) and 41 with Alzheimer’s disease (AD). Images were acquired with 3T GE Medical Systems scanners at 14 sites across North America. Each 3D volume consisted of 2.7 mm isotropic voxels with a 128×128 acquisition matrix. Forty six image volumes were acquired per subject: 5 T2-weighted b0 images and 41 diffusion-weighted volumes (b = 1,000 s/mm2).
2.2 Proposed Framework
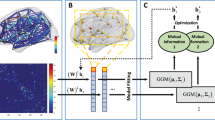
First, we first used GLRAM to create dimensionality-reduced matrices for each subject. These new matrices were used as input to LDA on PCA for model training. Adaptive 1-nearest neighbor classification (A-1NNC) was used to label the test cases. The framework’s flowchart is shown in Fig. 1.
2.2.1 Brain Network Computation
In this study, we used the subjects’ structural networks as features for classification. To compute the brain networks, FreeSurfer (http://freesurfer.net/) was run on the T1-weighted images to automatically segment the cortex into 68 unique regions (34 per hemisphere). This segmentation was dilated with an isotropic box kernel of 5 mm to ensure cortical labels would intersect with the white matter tissue in areas of reliable tractography for the connectivity analysis. We registered the T1-weighted intensity image to the fractional anisotropy (FA) image from the DWI data. The resultant transformations were used to transform the dilated cortical segmentations into the DWI space.
DWI images were corrected for eddy current distortions using FSL [12]. Then we used an optimized global probabilistic tractography method [13] to generate whole brain tractography for each subject. We combined the cortical segmentation and tractography to compute a connectivity matrix for each subject. The matrices were 68×68 in dimension, corresponding to the 68 segmented cortical regions. Each cell value of the matrix represented the number of fibers that intersected pairs of cortical regions. We normalized the matrix by the total number of fibers per subject. This symmetric 68×68 matrix served as the input for our classification.
2.2.2 Data Normalization
Some form for data normalization is critical especially when working with data from different cohorts or projects, covering a wide age range. So directly pooling two datasets may introduce bias, if the proportion of controls depends on the scanner used or scanning site. To account for these confounds, we used generalized linear regression to adjust each value in the brain connectivity matrix for age, sex and scanning site. Then we further normalized the residual after regression to yield centered, scaled data, which served as the input for next step. This normalization used a Z-transformation based on the standardized statistic Z = (X-mean(X))/std(X), where X is one feature vector within each dataset. For our connectivity matrix, X is element (i,j) for all subjects in each dataset.
2.2.3 GLRAM
The purpose of GLRAM, proposed in [11], is similar to singular value decomposition (SVD) but has lower computational cost; it finds a lower rank 2D matrix Di to approximate the original 2D matrix Ai, realizing the following function:
Here, Ai is each subject’s raw brain network, N is the total number of subjects, Di is the reduced representation of Ai; and L and R are transformation matrices on the left and right side, respectively. F is the Frobenius norm. Details of how to solve this cost function optimization problem are in [11].
2.2.4 LDA on the PCA Subspace
PCA finds linear projections that maximize the scatter of all projected samples. Mathematically, given a set of N subjects \(X = \left \{x_{1},x_{2},\ldots x_{N}\right \}\), where each subject belongs to one of C classes X1, X2, …XC, we plan to map xi to yi where y i ∈ R m and m¡n. To do this, we define a linear transformation W to satisfy yi = WTxi (i = 1,2,…N). In PCA, the optimal projection Wopt−pca is defined as:
Here μ ∈ R n is the mean value of all samples. And \(W_{\mathit{opt-pca}} = \left \{w_{i} \in R^{n}\left \vert i = 1..m\right.\right \}\) is the set of eigenvectors of ST corresponding to the m largest eigenvalues. Once eigenvectors are determined, all data can be projected into this eigenspace for classification. However, PCA is not optimal for classification as the dimensions that model the greatest amount of variance in the data are not typically the ones that best differentiate groups. In other words, the discriminant dimensions could be thrown out or intermixed during PCA.
LDA seeks a projection to maximize the ratio of the determinant of the between-class scatter matrix (SB) of the projected data to the determinant of the within-class scatter matrix (SW) of the projected data. However, the within-class scatter matrix SW in LDA is typically singular. This is because the number of subjects is often much smaller than the number of variables in the data. To overcome the complication of a singular SW, we adopted the solution in [14]. In short, C is the number of classes, so we first adopted PCA to reduce the dimension of the feature space to N-C, and then we applied the standard LDA to reduce the dimension to C-1, so the transformation Wopt is given by:
Where \(\upmu\) i is the mean vector of class Xi, and Ni is the number of samples in class Xi. Also, Wopt−pca can be computed using Eq. 2.
2.2.5 Adaptive 1-NNC
We classified the subject’s class membership based on the Euclidean distance using 1-nearest neighbor classification (1-NNC). 1-NNC is designed to assign an object to the same class as its single nearest neighbor. Adaptive 1-NNC (A-1NNC) is a variation of 1-NNC. The test objects’ class membership is still decided based on the class membership of the single nearest-neighbor used for training, but once a new test object’s class membership has been determined, it is grouped into training group to enhance the membership class affinity.
2.3 Experimental Procedure
The detailed procedure is described as follows:
-
1.
Construct the brain network for each subject in both datasets.
-
2.
Data Normalization to get input matrix A.
-
3.
Group subjects into four classes: HC, EMCI, LMCI and AD.
-
4.
Divide each class into three parts by randomization: training (80 %), optimizing (10 %) and testing (10 %).
-
5.
Pick up training dataset Atrain
-
6.
Set the initial dimension size to run GLRAM on Atrain to get Ltrain, Rtrain and Dtrain = D1, D2, …DN for each class (using Eq. 1)
-
7.
Transfer Dtrain into vector xi and form matrix Xtrain = { x1, x2, …xN }
-
8.
Run LDA in PCA subspace to get Wopt (using Eq. 3) and get the projected data Ytrain = y1, y2, …yN = WoptX
-
9.
Then the projection of the optimizing dataset Aoptimizing can be generated using Eq. 4.
-
10.
Use A-1NNC classification to assign Y\(_{\mathit{optimizing}}^{}\) s class based on Ytrain and compute the accuracy by comparing the assigned membership to ground truth
-
11.
Then adjust the parameter in Step 6, re-run steps 6–10 to find the optimal parameter for the dimension of L and R in Eq. 1 that achieves best accuracy
-
12.
Use this optimal parameter achieved in Step 11, and use the test dataset to test our framework and get final grade
-
13.
Repeat steps 4–12 (100 times) and compute the area under the curve (AUC) for overall classification accuracy, as well as for the accuracy of each class. The higher the AUC, the better the model performance.
3 Results and Discussion
Before we ran classification experiments, we first studied the effects of pooling datasets 1 and 2 together. A testable null hypothesis is that the feature set in the dataset 1 and dataset 2 are independent random samples drawn from Normal distributions with equal means and equal but unknown variances. As each subject’s brain network is symmetric and has dimension 68×68, we have 68×67/2 = 2,278 features per subject. Thus we adopted false discovery rate (FDR) to account for the multiple comparisons (FDR q = 0.05). Figure 2 shows the FDR-corrected P map from a Student’s t-test between dataset 1 (all HC) and HC from dataset 2. Our results showed that by using our proposed normalization methods, there are no detectable differences between the HCs in dataset 1 and dataset 2. Given this information, we pooled data bettering an effort to boost statistical power.
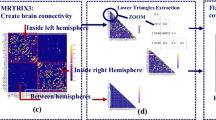
An FDR-corrected P map (on a log scale) for the null hypothesis asserting that features in datasets 1 and 2 are independent random samples drawn from Normal distributions with equal means and equal but unknown variances. All P values larger than the critical FDR threshold have been set to zero. The top left map is for the raw brain networks (generated in Sect. 2.2.1). The top right map is for residual brain networks after the effects of age and sex are removed (regression on age, sex and data label, described in Sect. 2.2.2). The bottom map is for normalized brain networks generated as in Sect. 2.2.2
Then we compared our proposed method with the other three methods including: direct PCA, LDA in the PCA subspace, and GLRAM only. Table 2 shows the AUC comparison for the 4-class (HC, EMCI, LMCI and AD) classification results using dataset 2 only and then also using both datasets for defining the PCs. The results indicated that our proposed framework performed better than other methods. As shown in Table 2, PCA showed the poorest performance, which is reasonable as PCA emphasizes the data variance, which is not necessarily useful for classification. Also, GLRAM performed better than LDA. The possible explanation could be that our features were the full brain networks, which emphasized the connections between the nodes. So there may be some 2D spatial information in the features that are ignored in the vector space model (LDA). Moreover, HC classification accuracy improved when adding dataset 1, suggesting the advantage of pooling data, so long as appropriate normalization is applied.
4 Conclusion
Here we presented a novel framework using GLRAM and modified LDA to reduce the dimension of a 68×68 element structural brain connectivity network. We then used Adaptive-1NNC to classify patients with different stages of Alzheimer’s disease versus healthy controls. Our proposed method outperformed classical classification methods, but incorporating healthy controls from additional datasets also improved classification.
As our proposed framework is based on some elementary approaches (such as PCA and LDA), we compared these methods to ours, instead of other more complex approaches. In future work, we will try more sophisticated approaches. As an innovation, most current studies focus on one type disease vs. HC, while our target is for a more complicated (realistic) situation and we know there are ways to improve the proposed framework. Our current results indicate that our approach is promising.
References
Jessen, F., Wolfsgruber, S., Wiese, B., et al.: AD dementia risk in late MCI, in early MCI, and in subjective memory impairment. Alzheimers Dement. 10, 76–83 (2014)
Mitchell, A.J.: CSF phosphorylated tau in the diagnosis and prognosis of mild cognitive impairment and Alzheimer’s disease: a meta-analysis of 51 studies. J Neurol. Neurosurg. Psychiatry 80, 966–975 (2009)
Aisen, P.S., Petersen, R.C., Donohue, M.C., et al.: Clinical core of the Alzheimer’s disease neuroimaging initiative: progress and plans. Alzheimers Dement. 6, 239–246 (2010)
Winblad, B., Palmer, K., Kivipelto, M., et al.: Mild cognitive impairment–beyond controversies, towards a consensus: report of the international working group on mild cognitive impairment. J. Intern. Med. 256, 240–246 (2004)
LeBihan, D.: IVIM method measures diffusion and perfusion. Diagn. Imaging (San Franc) 12, 133–136 (1990)
Sporns, O.: The human connectome: a complex network. Ann. N.Y. Acad. Sci. 1224, 109–125 (2011)
Ajilore, O., Lamar, M., Leow, A., et al.: Graph theory analysis of cortical-subcortical networks in late-life depression. Am. J. Geriatr. Psychiatry 22, 195–206 (2014)
Zhang, J., Cheng, W., Wang, Z., et al.: Pattern classification of large-scale functional brain networks: identification of informative neuroimaging markers for epilepsy. PLoS One 7, e36733 (2012)
Thompson, P.M., Stein, J.L., Medland, S.E., et al.: The ENIGMA consortium: large-scale collaborative analyses of neuroimaging and genetic data. Brain Imaging Behav. 8, 153–162 (2014)
Wang, J., Zhou, L.:Research on magnetoencephalography-brain computer interface based on the PCA and LDA data reduction. Sheng Wu Yi Xue Gong Cheng Xue Za Zhi 28, 1069–1074 (2011)
Ye, J.: Generalized low rank approximations of matrices. Mach. Learn. 61, 167–191 (2005)
Jenkinson, M., Beckmann, C.F., Behrens, T.E., et al.: FSL. Neuroimage 62, 782–790 (2012)
Aganj, I., Lenglet, C., Jahanshad, N., et al.: A Hough transform global probabilistic approach to multiple-subject diffusion MRI tractography. Med. Image Anal. 15, 414–425 (2011)
Belhumeur, P.N., Hespanha, J.P., Kriegman, D.J.: Eigenfaces vs. fisherfaces: recognition using class specific linear projection. IEEE Trans. Pattern Anal. Mach. Intell. 19, 711–720 (1997)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2014 Springer International Publishing Switzerland
About this paper
Cite this paper
Zhan, L. et al. (2014). Multiple Stages Classification of Alzheimer’s Disease Based on Structural Brain Networks Using Generalized Low Rank Approximations (GLRAM). In: O'Donnell, L., Nedjati-Gilani, G., Rathi, Y., Reisert, M., Schneider, T. (eds) Computational Diffusion MRI. Mathematics and Visualization. Springer, Cham. https://doi.org/10.1007/978-3-319-11182-7_4
Download citation
DOI: https://doi.org/10.1007/978-3-319-11182-7_4
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-11181-0
Online ISBN: 978-3-319-11182-7
eBook Packages: Mathematics and StatisticsMathematics and Statistics (R0)