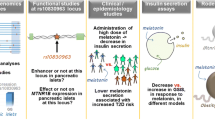
Abstract
The discovery that variants in the melatonin receptor 2 (MTNR1B) gene were associated with glucose levels, insulin secretion, and risk for type 2 diabetes (T2D) in genome-wide association studies (GWAS) reinforced the previously suggested link between glucose homeostasis and circadian rhythmicity. Diurnal secretion of melatonin has reported to be altered in people with diabetes and rodent models of T2D. The proposed underlying mechanisms by which altered melatonin signaling could predispose to progression to T2D and gestational diabetes mellitus (GDM) involve altered expression of MTNR1B in pancreatic beta cells, leading to impaired insulin secretion, consequent increased fasting glucose concentrations, and eventually overt T2D. Thus blocking the inhibition of insulin secretion may have potential clinical implications, and these effects could be more pronounced in individuals carrying risk genotypes. Finally, given that melatonin could emerge as an attractive treatment for a variety of conditions including pregnancies associated with GDM, preeclampsia, and intrauterine growth retardation, pharmacogenetic studies are warranted to determine treatment response and side effects according to genotype.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- Insulin Secretion
- GDMgestational Diabetes Mellitus
- Impaired Fasting Glucose
- Pancreatic Beta Cell
- Fast Plasma Glucose Level
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
1 MTNR1B as a Primary Genome-Wide Association Signal for Fasting Plasma Glucose Levels
Type 2 diabetes (T2D) is a complex genetic metabolic disorder which has developed into a major health problem responsible for early morbidities (including severe micro- and/or macro-vascular complications) and mortality, with a burden increasing globally, particularly in developing countries (World Health Organization 2013). T2D results from progressive dysfunction of insulin secretion from pancreatic beta cells on the background of resistance to insulin action (American Diabetes Association 2014a). Since 2007, exponential progress has been made in identifying genetic determinants of T2D through the use of cutting-edge DNA microarrays allowing for large-scale genome-wide association studies (GWAS) and their meta-analyses. After analyzing T2D as a binary disorder in case–control GWAS, a logical next step was to investigate new variants contributing to the variation of metabolic quantitative traits linked to the pathophysiological processes leading to T2D, as this may provide new insights into the etiological mechanisms of this highly complex disorder. Among such quantitative traits, fasting plasma glucose (FPG) levels were of major interest as hyperglycemia in the fasting state remains one of the criteria used by the American Diabetes Association (ADA) to define T2D (American Diabetes Association 2014b). Furthermore, elevated FPG levels within the range specified by the ADA was known to be an independent risk factor for T2D (Tirosh et al. 2005) and for cardiovascular and all-cause mortality (Barr et al. 2007). Of note, approximately one-third of the variation of FPG was shown to be genetic (Watanabe et al. 1999; Pilia et al. 2006).
In 2008, via GWAS, two independent groups identified a new genome-wide association signal with FPG levels closed to G6PC2 and ABCB11 genes, in nondiabetic European participants (Bouatia-Naji et al. 2008; Chen et al. 2008). Of note, the single nucleotide polymorphism (SNP; rs560887) in G6PC2 which significantly contributed to variation of FPG levels was not associated with T2D risk (Bouatia-Naji et al. 2008) (see Chap. 17).
One year later, using larger sample sizes in GWAS meta-analyses which markedly increased the statistical power, a new locus associated with FPG levels was identified in MTNR1B (encoding melatonin receptor 2 [MT2]) in nondiabetic European individuals (Bouatia-Naji et al. 2009; Prokopenko et al. 2009). The association signal was subsequently refined by genotyping eight SNPs in strong linkage disequilibrium (r 2 > 0.70) within the association block (Sparsø et al. 2009). This study showed that rs10830963, located in the middle of the unique MTNR1B intron, carried most of the effect on FPG variation (Sparsø et al. 2009). Of note, rs10830963 was the strongest SNP associated with FPG levels in the first and most recent publications by the Meta-Analyses of Glucose and Insulin-related traits Consortium (MAGIC) in 36,610 and 133,010 nondiabetic individuals of European descent, respectively (Fig. 19.1) (Prokopenko et al. 2009; Scott et al. 2012). The MTNR1B locus was shown to be robustly associated with FPG levels in nondiabetic individuals from other various ethnicities including African-Americans, Koreans, Japanese, Sri Lankans, Han Chinese, and Indian Asians (Rönn et al. 2009; Chambers et al. 2009; Liu et al. 2010; Takeuchi et al. 2010; Kan et al. 2010; Ramos et al. 2011; Kim et al. 2011; Rasmussen-Torvik et al. 2012), suggesting a putatively causal role of the MTNR1B gene in the association with FPG levels. Furthermore, studies in adolescents and children suggested that variants in MTNR1B most likely influence FPG levels early from childhood onward (Barker et al. 2011).
Regional plots showing the association between SNPs and FPG levels at the MTNR1B locus in the lastly published MAGIC study performed in 133,010 nondiabetic individuals of European descent (Scott et al. 2012)
2 Effect of MTNR1B Genetic Variants on T2D Risk
In addition to the variation of FPG levels, the MTNR1B locus was found to be significantly associated with T2D risk (Bouatia-Naji et al. 2009; Lyssenko et al. 2009; Prokopenko et al. 2009), demonstrating that studies of continuous glycemic phenotypes in nondiabetic individuals can successfully complement the genetic analyses of T2D as a dichotomous trait. Dupuis et al. demonstrated a genome-wide significant association between MTNR1B SNP rs10830963 and T2D risk (Dupuis et al. 2010), which was confirmed by the Diabetes Genetics Replication And Meta-analysis (DIAGRAM) consortium (Voight et al. 2010). The association between MTNR1B locus and T2D risk was also reported in individuals from various ethnicities including Han Chinese, Japanese, and Indian Asians (Rönn et al. 2009; Chambers et al. 2009; Kan et al. 2010; Ling et al. 2011; Ohshige et al. 2011; Zhao et al. 2014), even though evidence of heterogeneity due to ethnicity was found in two meta-analyses: the association seemed to be stronger in Europeans, compared with East or South Asians (Xia et al. 2012; Wang et al. 2013).
The causal link between the MTNR1B locus and T2D risk has been demonstrated by a large-scale exon resequencing study of MTNR1B in combination with systematic functional investigations of each identified non-synonymous variant (Bonnefond et al. 2012). The sequencing of MTNR1B coding exons in 7632 Europeans, including 2186 individuals with T2D, identified 40 non-synonymous variants of which 36 variants were very rare (with a minor allele frequency below 0.1 %) and significantly associated with T2D, while the four frequent or rare non-synonymous variants (with a minor allele frequency above 1 %) did not contribute to T2D risk (Bonnefond et al. 2012). After functional investigations (including proper surface expression study, melatonin binding study, and ERK or Gi pathway study of each mutant), Bonnefond et al. demonstrated that 13 partial- or total-loss-of-function very rare variants in combination yield more than fivefold increased risk for T2D, while the 23 neutral very rare variants did not exhibit any effect on T2D (Bonnefond et al. 2012). This study established a firm functional link between the MTNR1B gene and T2D risk, highlighting that the discovery of functional coding mutations in a previously associated locus can help establish a specific gene as the molecular cause of the association signal.
3 Effect of MTNR1B Variants on Early Phase Insulin Secretion
The mechanisms by which variation at MTNR1B raises FPG and T2D risk were explored by the Diabetes Genetics Initiative GWAS for insulin secretion (Lyssenko et al. 2009). Lyssenko et al. demonstrated that the glucose-raising allele of the MTNR1B rs10830963 was associated with impaired early insulin release to both oral (insulinogenic and disposition index) and intravenous (first phase insulin response) glucose loads (Lyssenko et al. 2009). In addition, the risk allele carriers showed deterioration in insulin secretion over time as compared to non-risk allele carriers (Lyssenko et al. 2009). The same study showed that in the risk allele carriers, elevated FPG levels and reduced phase insulin response to glucose were translated into 11 % increased risk for future T2D in two large prospective studies of more than 18,000 individuals, of whom 2201 developed T2D during a mean follow-up period of 23.5 years (Lyssenko et al. 2009). Very recently, a largest to date GWAS meta-analyses for dynamic measurements of insulin secretion during an oral glucose tolerance test (OGTT) in more than 10,000 nondiabetic individuals confirmed MTNR1B rs10830963 as the strongest signal for first phase insulin secretion (Prokopenko et al. 2014). Importantly, in the Diabetes Prevention Program, the association of MTNR1B rs10830963 with impaired early insulin release persisted at 1 year despite adjustment for the baseline trait, suggesting a progressive deterioration of the effect at this locus (Florez et al. 2012).
Notably, Lyssenko et al. demonstrated that MTNR1B mRNA was expressed in human pancreatic islets and more specifically that nondiabetic individuals carrying the risk allele and patients with T2D display increased expression of the receptor in pancreatic islets (Lyssenko et al. 2009). These observations were further confirmed by a large gene-expression analysis of human pancreatic islets (Taneera et al. 2012). Exogenously administered melatonin has been shown to inhibit insulin secretion in rodents (Bailey et al. 1974). In line with these observations, Lyssenko et al. showed that melatonin inhibited insulin release in response to glucose in INS-1 rat beta cells (Lyssenko et al. 2009).
Of note, MTNR1B rs10830963 significantly increased the risk of isolated impaired fasting glucose but not the risk of isolated impaired glucose tolerance (Sparsø et al. 2009), and the same SNP was shown to impact the rate of progression from normal fasting glucose to impaired fasting glucose, but not the rate of progression from impaired fasting glucose to T2D (Walford et al. 2012). In the GLACIER study which is a population-based prospective cohort study from northern Sweden, MTNR1B rs10830963 was reported to increase risk of developing impaired fasting glucose during 10-year follow-up but to be protective of worsening glucose tolerance (Renström et al. 2011), which confirmed a strong effect of MTNR1B locus on impaired fasting glucose, independently of impaired glucose tolerance measured 2 h after a meal.
In addition to its effects on insulin secretion and isolated impaired fasting glucose, variants in the MTNR1B gene were also reported to be associated with hepatic insulin resistance (Sparsø et al. 2009; Vangipurapu et al. 2011). Notably, the risk carriers of MTNR1B were a clear outlier when known hyperbolic relationship between insulin secretion and the degree of insulin sensitivity was plotted, showing a strong insulin-resistant phenotype for the given impairment in insulin secretion (Jonsson et al. 2013). An association with insulin sensitivity could involve effects of MTNR1B on energy expenditure. In this vein, it has been recently demonstrated that MTNR1B variant could modify effects of dietary fat intake on changes in energy expenditure during a 2-year period (Mirzaei et al. 2014). Additionally, this could involve insular-incretin axis as MTNR1B variants were shown to be associated with incretin-stimulated insulin secretion (Simonis-Bik et al. 2010).
4 Effect of MTNR1B Variants on the Risk for Gestational Diabetes Mellitus
In addition to the risk of T2D, there is compelling robust evidence for an association of the MTNR1B locus with gestational diabetes mellitus (GDM ) in several ethnic populations (Kwak et al. 2012; Vlassi et al. 2012; Huopio et al. 2013). A GWAS in a Korean population consisting of 468 women with GDM and 1242 nondiabetic women reported a variant near MTNR1B, SNP rs10830962, to be associated with GDM at a genome-wide significance level (Kwak et al. 2012). In a smaller study from Greece, SNP rs10830963 also conferred association with GDM (Vlassi et al. 2012), while in one study of Chinese pregnant women, several variants in the MTNR1B gene were associated with elevated glucose concentrations (Liao et al. 2012). An independent Chinese study demonstrated reduced beta-cell function as measured with HOMA-B index (Wang et al. 2011). Recently, a large Finnish study showed that the risk genotypes of the MTNR1B rs10830963 were associated with GDM risk, increased fasting plasma glucose, and reduced insulin secretion (Huopio et al. 2013). Notably, melatonin crosses the human placenta easily and rapidly (Okatani et al. 1998) and has been suggested to play an important role during pregnancy through its antioxidant properties but also as a regulator of normal growth and development of fetal organs (Reiter et al. 2014). The underlying mechanisms were attributed to the melatonin effect on the epigenetic modifications of genes implicated in placental development, fetal growth, and intrauterine programming (Korkmaz et al. 2012). However, there have been no any studies thus far on whether variants in the melatonin receptor(s) could contribute to these effects.
5 Melatonin Secretion and Physiological Functions
Melatonin (N-acetyl-5-methoxytryptamine), also known as the hormone of darkness, is an indoleamine synthesized from the amino acid tryptophan via serotonin. Tryptophan is converted by 5-tryptophan hydroxylase to 5-hydroxytryptophan, which then in the pineal gland is converted through hydroxylation and decarboxylation into serotonin. Serotonin is subsequently acetylated by arylalkylamine-N-acetyltransferase (AANAT, also called serotonin N-acetyltransferase), the rate-limiting step in melatonin biosynthesis, and, finally, through a methylation reaction converted into melatonin by acetylserotonin O-methyltransferase (ASMT) (Axelrod and Weissbach 1960). Melatonin is a key mediator of the entrainment of biological rhythms in the body, a “zeitgeber” (German: “time giver”), in which biological rhythmic secretion was initially linked to regulation of seasonal reproduction in photoperiodic species (Hoffman and Reiter 1965).
The hormone was traditionally thought to emanate from rhythmicity pinealocytes located in the pineal gland. This endocrine organ is located in the midline of the brain, just above the posterior commissure at the dorsal edge of the third ventricle, and receives a rich supply of adrenergic innervation from the superior cervical ganglion. However, melatonin remains detectable after pinealectomy in some species (Axelrod and Weissbach 1960), leading to the realization that the hormone is produced in neuroendocrine cells of both central (pineal gland, retina, Harderian glands) and peripheral origins, comprising enterochromaffin cells dispersed in a number of organs, e.g., the gastrointestinal tract, pancreas, and many more (Kvetnoy et al. 1997). Melatonin is also produced by numerous non-endocrine cells, e.g., immune cells. Thus, while substantial local biosynthesis also occurs in retina and in some other organs, e.g., the gastrointestinal tract, the diurnal rhythm of the circulating melatonin in blood exclusively accounts for its secretion from the pineal gland.
Light has a dual effect on the production of melatonin. First, it entrains the circadian clock , making melatonin production occur during the night. The duration of melatonin production varies with time over the year, because the onset and offset of melatonin secretion are controlled by the clock and can move closer (summer) or apart (winter) (Illnerová and Sumová 1997). In fact, in the absence of light, and in blind people, the melatonin rhythm persists following a circadian rhythm and cycle length governed by the suprachiasmatic nucleus (SCN) (Klerman et al. 2002). Light at night can also have an acute suppressive effect on melatonin production. In rodents, light at night decreases melatonin within minutes, while RNA levels remain high for hours. Circulating levels of melatonin peak at 80–100 pg/ml in the middle of the night and drop to 10–20 pg/ml during day time; the half-life of the hormone in the circulation is less than 20 min.
The hormone exerts its effects both through activation of its receptors (melatonin receptor 1 [MT1], MT2, and the orphan GPR50 receptor that can regulate the function of MT1 and MT2 through receptor heterodimerization) (Boutin et al. 2005; Jockers et al. 2008) but also via receptor-independent mechanisms such as its capacity to act as an antioxidant (Hardeland 2005). These effects can either be through the circulating levels of the hormone or in a more autocrine/paracrine fashion near target tissues (Kvetnoy et al. 1997; Peschke 2008).
Melatonin is widely known to affect the CNS , where it alters hormone release and phase shifts neuronal firing both in the 24 h rhythm and seasonal changes (Dubocovich and Markowska 2005). The phase-advancing effects of melatonin are taken advantage of treating insomnia or limiting jet lag when traveling across time zones (Arendt et al. 1997; Zhdanova and Wurtman 1997; Arendt 2006). Recently, the effects of melatonin in synergizing maternal and fetal circadian rhythms during pregnancy, the development of normal placentation, and its potential beneficial properties in the treatment of compromised pregnancies associated with GDM, preeclampsia, and intrauterine growth retardation have been emphasized (Reiter et al. 2014).
In the periphery, melatonin promotes vasoconstriction through MT1 and vasodilation through MT2 (Masana et al. 2002). In the adrenal cortex, it lowers cortisol secretion, an action shared with insulin (Weitzman et al. 1971; Peschke 2008). Interestingly, human adipocytes, a major target tissue for insulin, express MT2 and have been shown to reduce the expression of the insulin-dependent glucose transporter GLUT4 after melatonin stimulation (Brydon et al. 2001). In muscle cells, melatonin stimulates glucose uptake by phosphorylation of the insulin receptor substrate-1 (IRS-1) through suggested MT2 signaling (Ha et al. 2006). MT2 is also expressed in hepatocytes, and melatonin injections elevated glucose release from the liver in mice (Poon et al. 2001). Melatonin receptors are also widely expressed in the gut and could thus have an effect on incretin hormones like glucose insulinotropic polypeptide (GIP) and glucagon-like peptide 1 (GLP-1) (Chen et al. 2011).
6 Melatonin Receptors: A Putative Role in Pancreatic Islets?
The genetic studies described above have strongly suggested a putatively direct role of MTNR1B variants in the dysfunction of insulin secretion from pancreatic beta cells, leading to T2D. The discovery that genetic variants in the MTNR1B gene were associated with FPG levels, insulin secretion, and risk for T2D reinforced the previously suggested link between glucose homeostasis and circadian rhythmicity. It has been previously observed that plasma insulin levels exhibit a circadian rhythm and that disturbance in the oscillation affects plasma glucose and hormone levels (Scheer et al. 2009). Diurnal secretion of melatonin has reported to be altered in patients and rodent models of T2D and that expression of the melatonin receptor is increased (Peschke et al. 2006b). Importantly, it has recently been demonstrated that lower nocturnal melatonin secretion was independently associated with a higher risk of developing T2D in nondiabetic women (McMullan et al. 2013). However, the specific role of the melatonin receptors in these observations remains an open question. How did cell studies and animal models indicate a putative role of melatonin receptors in pancreatic islets and T2D?
6.1 Melatonin Receptors Are G-Protein-Coupled Receptors (GPCRs)
Melatonin receptors belong to the GPCR superfamily which represents the largest family of cell membrane receptors in humans with approximately 800 GPCRs (>1 % of the total protein-coding human genome) (Jassal et al. 2010). GPCRs are characterized by seven alpha-helical transmembrane domains which are connected by intra- and extracellular loops of various lengths. They sense extracellular signals (including photons, metabolites, amino acids, lipids, odor molecules, hormones, neurotransmitters, peptides) and activate various intracellular signaling pathways, through the interaction with G proteins (including Gs proteins which activate the adenylyl cyclase pathway, Gi/o proteins which inhibit this pathway, Gq/11 proteins which activate the phospholipase C pathway, and G12/13 proteins which are involved in the activation of small G proteins and cytoskeleton rearrangements) and/or beta-arrestin (Kristiansen 2004; Venkatakrishnan et al. 2013).
Melatonin receptors constitute a subfamily of rhodopsin-like (type A or 1) GPCRs which include three members: MT1, MT2, and the orphan GPR50 receptor (Jockers et al. 2008; Jassal et al. 2010). They share specific short amino acid sequences and approximately 55 % overall sequence homology (Reppert et al. 1994, 1995). Melatonin receptors have been neglected for a long time by academic research due to the lack of specific pharmacological tools, apparently mild phenotypes of knockout mice, and poorly defined functions. Notably, rodent animal models are limited for the study of the link between melatonin and metabolism, as rodents are nocturnal animals (therefore the circadian rhythms of food intake and metabolism are shifted by 12 h with respect to humans, despite the nocturnal secretion of melatonin by humans and rodents). Furthermore, some mouse strains used in laboratory are known to exhibit very low or even undetectable circulating melatonin levels due to impaired melatonin production within the pineal gland (Kennaway et al. 2002). Therefore, in the studies of melatonin and its receptors, the extrapolation of results obtained from rodents to humans must be done cautiously (Karamitri et al. 2013).
6.2 Distribution of MT1 and MT2 in Humans
Melatonin receptors are widely expressed throughout the human body. Notably, both MTNR1A and MTNR1B are expressed in the brain (in particular the SCN which is the master circadian pacemaker, the hippocampus, and the thalamus) and in the retina (Reppert et al. 1995; Jockers et al. 2008). Interestingly, insulin-target tissues express MTNR1B: adipocytes, muscle cells, hepatocytes, and the gut. Furthermore, both MTNR1A mRNA and MTNR1B mRNA have been detected in human islets (Ramracheya et al. 2008). The same study showed that MTNR1A mRNA was primarily detected in pancreatic alpha cells and the level of MTNR1B mRNA was globally much lower than the level of MTNR1A mRNA in human islets (Ramracheya et al. 2008). This last result was not confirmed by Lyssenko et al., who did not detect any differences in the levels of MT1 and MT2 in human islets (Lyssenko et al. 2009). MT2 was predominantly observed in human pancreatic beta cells, while MT1 was mostly detected in a population of peripherally located beta cells (Lyssenko et al. 2009).
6.3 Melatonin Receptor Signaling in Pancreatic Beta Cells
Most of the existing literature (based on rodent models) has shown that melatonin inhibits insulin secretion from pancreatic beta cells (Peschke 2008). However, in human pancreatic islets, a study demonstrated that melatonin stimulates insulin secretion, without affecting cAMP levels (Ramracheya et al. 2008).
Melatonin receptors can actually modulate insulin secretion from pancreatic beta cells via different signaling pathways which can lead to opposite effects (Fig. 19.2). First, activated melatonin receptors coupled with Gαi proteins inhibit cAMP production (via adenylyl cyclase [AC]) and activation of cAMP-dependent protein kinase A (PKA) and therefore decrease insulin granules exocytosis (Fig. 19.2) (Kemp et al. 2002; Peschke et al. 2002, 2006a). Furthermore, activated MT2 receptor coupled with Gαi proteins can decrease cGMP levels (via guanylate cyclase), leading to reduced insulin secretion (Fig. 19.2) (Stumpf et al. 2008, 2009). In addition, activated melatonin receptors coupled with Gαq/11 enhance phospholipase C (PLC), increasing the release of Ca2+ from intracellular stores (endoplasmic reticulum) by the stimulation of inositol 1,4,5-triphosphate (IP3) receptor, which induces insulin granules exocytosis (Fig. 19.2) (Bach et al. 2005). All these results were obtained from rodent models which can have some limitations. More investigations based on human pancreatic islets would be of major interest for the field. Of note, melatonin receptors (in particular MT1) may also play a role in pancreatic alpha cells, which could regulate glucose homeostasis via the glucagon. However, the mechanisms are not clear as there are some discrepancies between studies, and investigations based on human islets are also lacking (Karamitri et al. 2013).
Signal transduction of melatonin in pancreatic beta cells. AC, adenylyl cyclase; ATP, adenosine triphosphate; cAMP, cyclic adenosine monophosphate; cGMP, cyclic guanosine monophosphate; DAG, diacylglycerol; IP3, inositol 1,4,5-triphosphate; PIP2, phosphatidylinositol 4,5-bisphosphate; PKA, protein kinase A; PLC, phospholipase C
7 Dysfunction of Circadian Rhythm and Metabolic Disorders
Organisms, ranging from bacteria to mammals, possess accurate interna l biological rhythms that time various daily events from photosynthesis in plants to sleep/wakefulness in humans (Takahashi et al. 2008). In mammals, many physiological processes are actually regulated by these inherent clocks, including glucose homeostasis, body temperature, feeding behavior, and hormone release (Takahashi et al. 2008). The circadian system (defined as its approximately 24 h cycles in the absence of environmental clues) represents the hierarchy of these multiple biological clocks, which is controlled by the SCN known as the “master clock” or “master circadian pacemaker,” located in the anterior hypothalamus (Mohawk et al. 2012). The SCN is synchronized by the daily light–dark cycle in mammals, and in turn, it regulates output pathways controlling various biological functions and overt rhythms (e.g., hormone release, feeding, body temperature) which can synchronize peripheral biological clocks including circadian rhythms in pancreatic islets (Takahashi et al. 2008). Of note, these peripheral clocks can feed back and interfere with the master clock SCN (Mohawk et al. 2012).
At the molecular level, circadian clocks are mostly controlled by an autoregulatory transcriptional feedback loop (the “core circadian clock”) involving the activator transcription factors CLOCK and BMAL1 and their target genes period (PER1, PER2, PER3) and cryptochrome (CRY1, CRY2) whose gene products form proteins that complex, translocate into the nucleus, and repress CLOCK and BMAL1-mediated transcription until the degradation of the Per/Cry repressor complex (Takahashi et al. 2008; Mohawk et al. 2012). This loop takes roughly 24 h and operates in most cells of the body to control either ubiquitous or tissue-specific physiological functions. Of note, there are additional (less essential) loops involving other clock genes or proteins (e.g., Rev-erbα, Rorα) which influence the core circadian clock. All these feedback loops interact with electrical and metabolic oscillations which modulate physiological function (Evans and Davidson 2013).
Compelling evidence has linked a disturbed circadian rhythm to metabolic syndrome, including T2D and obesity (Bass and Takahashi 2010; Shi et al. 2013). First, in the early 2000s, an outstanding study showed that a significant part of the transcriptome follows circadian rhythms in the mouse and that most of the signaling pathways regulated by these circadian clocks are involved in fundamental metabolism (Panda et al. 2002). Furthermore, several mutant animal models for clock genes (Clock, Bmal1, Rev-erbα, Rev-erbβ) have been shown to present with metabolic disorders including obesity, diabetic phenotypes (insulin resistance, hyperglycemia, impaired pancreatic function), hypertension, hyperlipidemia, and/or hepatic steatosis (Turek et al. 2005; Takahashi et al. 2008; Lamia and Evans 2010; Marcheva et al. 2010; Cho et al. 2012). Importantly, two studies demonstrated that pancreatic islets (probably pancreatic beta cells) possess self-sustained circadian oscillations (Marcheva et al. 2010; Sadacca et al. 2011). This intrinsic pancreatic circadian clock is required for normal insulin secretion and glucose homeostasis (Sadacca et al. 2011) and, when disrupted, leads to hypoinsulinemia and diabetes (Marcheva et al. 2010). In humans, a considerable number of epidemiological studies have reported that shift work is associated with increased risk for metabolic syndrome (including T2D and obesity) and cardiovascular disease, relative to day work (Kivimäki et al. 2011; Pan et al. 2011; Vyas et al. 2012; Buxton et al. 2012; Monk and Buysse 2013). Three main factors were known to explain these negative effects: circadian misalignment, sleep deprivation, and exposure to light at night (Evans and Davidson 2013). However, lastly, Eve Van Cauter’s group has reported that circadian misalignment may increase risk of T2D and obesity-related phenotypes, independently of sleep deprivation (Leproult et al. 2014). Finally, some genetic studies reported nominal or genome-wide significant associations between common SNPs near clock genes (CRY2, PER3, ARNTL [BMAL1], CLOCK, NR1D1 [REV-ERBΑ]) and risk of T2D/obesity or variation of related metabolic traits (including FPG, lipid levels) (Woon et al. 2007; Dupuis et al. 2010; Garaulet et al. 2010, 2013; Below et al. 2011; Kelly et al. 2012; Goumidi et al. 2013). Interestingly, a functional study showed that expression of PER2, PER3, and CRY2 genes was significantly decreased in pancreatic islets of patients with T2D, and this low expression was positively correlated with increased levels of HbA1c and decreased insulin secretion (Stamenkovic et al. 2012).
Therefore, it is highly probable that dysfunction of central and/or peripheral circadian clocks leads to T2D and other cardiometabolic studies. However, the specific involvement of the melatonin and its receptors into this dysfunction remains an open question.
8 Clinical and Pharmacological Implications
Taken together, as most observations available support an inhibitory effect of melatonin on insulin secretion, selective blocking of the melatonin ligand-receptor system in islets would be an attractive potential pharmacological target for the treatment of T2D. An individual carrying a risk variant may thus be more sensitive to the inhibitory melatonin effect than an individual without the risk allele, with a normal level of MTNR1B expression in islets. Such a restraining effect of melatonin fits with the impairment of early phase insulin secretion that was observed in risk carriers (Lyssenko et al. 2009). Thus, assessment of plasma levels of melatonin during different stages of glucose intolerance (NGT, IFG, and T2D) but also during pregnancy in GDM women in risk and non-risk genotype carriers of the MTNR1B gene deserves future studies to support or reject this notion. Furthermore, it remains to be elucidated whether ascribed protective properties of melatonin during pregnancy could be altered in the individuals carrying the risk allele in the MTNR1B gene. Finally, whether the ascribed effects of melatonin on epigenetic intrauterine programming contribute to the early defects in abnormal glucose metabolism and risk for T2D later in life warrants further investigation.
9 Conclusions
The proposed mechanisms by which altering melatonin signaling could predispose to progression to T2D and putatively GDM involve altering expression of MTNR1B in pancreatic beta cells leading to impaired insulin secretion. As a result, this leads to increased fasting glucose concentrations and eventually overt T2D. Thus, it would be of great clinical value to evaluate the combined risk for T2D but also for GDM of a model comprising MTNR1B common and rare functional variants in the gene, together with melatonin metabolites reflective of the activity of key enzymes in the melatonin pathway: in an intriguing finding that supports this notion, Illig et al. have shown that the tryptophan to phenylalanine ratio is affected by genetic variants in the MTNR1B gene (Illig et al. 2010). Whether this observation is reproducible and clinically translatable awaits the integration of independent genomic, metabolomic, and prospective data sets.
References
American Diabetes Association (2014a) Standards of medical care in diabetes--2014. Diabetes Care 37(Suppl 1):S14–S80. doi:10.2337/dc14-S014
American Diabetes Association (2014b) Diagnosis and classification of diabetes mellitus. Diabetes Care 37(Suppl 1):S81–S90. doi:10.2337/dc14-S081
Arendt J (2006) Does melatonin improve sleep? Efficacy of melatonin. BMJ 332:550. doi:10.1136/bmj.332.7540.550
Arendt J, Skene DJ, Middleton B et al (1997) Efficacy of melatonin treatment in jet lag, shift work, and blindness. J Biol Rhythms 12:604–617
Axelrod J, Weissbach H (1960) Enzymatic O-methylation of N-acetylserotonin to melatonin. Science 131:1312
Bach AG, Wolgast S, Mühlbauer E, Peschke E (2005) Melatonin stimulates inositol-1,4,5-trisphosphate and Ca2+ release from INS1 insulinoma cells. J Pineal Res 39:316–323. doi:10.1111/j.1600-079X.2005.00253.x
Bailey CJ, Atkins TW, Matty AJ (1974) Melatonin inhibition of insulin secretion in the rat and mouse. Horm Res 5:21–28
Barker A, Sharp SJ, Timpson NJ et al (2011) Association of genetic Loci with glucose levels in childhood and adolescence: a meta-analysis of over 6,000 children. Diabetes 60:1805–1812. doi:10.2337/db10-1575
Barr ELM, Zimmet PZ, Welborn TA et al (2007) Risk of cardiovascular and all-cause mortality in individuals with diabetes mellitus, impaired fasting glucose, and impaired glucose tolerance: the Australian Diabetes, Obesity, and Lifestyle Study (AusDiab). Circulation 116:151–157. doi:10.1161/CIRCULATIONAHA.106.685628
Bass J, Takahashi JS (2010) Circadian integration of metabolism and energetics. Science 330:1349–1354. doi:10.1126/science.1195027
Below JE, Gamazon ER, Morrison JV et al (2011) Genome-wide association and meta-analysis in populations from Starr County, Texas, and Mexico City identify type 2 diabetes susceptibility loci and enrichment for expression quantitative trait loci in top signals. Diabetologia 54:2047–2055. doi:10.1007/s00125-011-2188-3
Bonnefond A, Clément N, Fawcett K et al (2012) Rare MTNR1B variants impairing melatonin receptor 1B function contribute to type 2 diabetes. Nat Genet 44:297–301. doi:10.1038/ng.1053
Bouatia-Naji N, Rocheleau G, Van Lommel L et al (2008) A polymorphism within the G6PC2 gene is associated with fasting plasma glucose levels. Science 320:1085–1088. doi:10.1126/science.1156849
Bouatia-Naji N, Bonnefond A, Cavalcanti-Proença C et al (2009) A variant near MTNR1B is associated with increased fasting plasma glucose levels and type 2 diabetes risk. Nat Genet 41:89–94. doi:10.1038/ng.277
Boutin JA, Audinot V, Ferry G, Delagrange P (2005) Molecular tools to study melatonin pathways and actions. Trends Pharmacol Sci 26:412–419. doi:10.1016/j.tips.2005.06.006
Brydon L, Petit L, Delagrange P et al (2001) Functional expression of MT2 (Mel1b) melatonin receptors in human PAZ6 adipocytes. Endocrinology 142:4264–4271. doi:10.1210/endo.142.10.8423
Buxton OM, Cain SW, O’Connor SP et al (2012) Adverse metabolic consequences in humans of prolonged sleep restriction combined with circadian disruption. Sci Transl Med 4:129ra43. doi:10.1126/scitranslmed.3003200
Chambers JC, Zhang W, Zabaneh D et al (2009) Common genetic variation near melatonin receptor MTNR1B contributes to raised plasma glucose and increased risk of type 2 diabetes among Indian Asians and European Caucasians. Diabetes 58:2703–2708. doi:10.2337/db08-1805
Chen W-M, Erdos MR, Jackson AU et al (2008) Variations in the G6PC2/ABCB11 genomic region are associated with fasting glucose levels. J Clin Invest 118:2620–2628. doi:10.1172/JCI34566
Chen C-Q, Fichna J, Bashashati M et al (2011) Distribution, function and physiological role of melatonin in the lower gut. World J Gastroenterol 17:3888–3898. doi:10.3748/wjg.v17.i34.3888
Cho H, Zhao X, Hatori M et al (2012) Regulation of circadian behaviour and metabolism by REV-ERB-α and REV-ERB-β. Nature 485:123–127. doi:10.1038/nature11048
Dubocovich ML, Markowska M (2005) Functional MT1 and MT2 melatonin receptors in mammals. Endocrine 27:101–110. doi:10.1385/ENDO:27:2:101
Dupuis J, Langenberg C, Prokopenko I et al (2010) New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet 42:105–116. doi:10.1038/ng.520
Evans JA, Davidson AJ (2013) Health consequences of circadian disruption in humans and animal models. Prog Mol Biol Transl Sci 119:283–323. doi:10.1016/B978-0-12-396971-2.00010-5
Florez JC, Jablonski KA, McAteer JB et al (2012) Effects of genetic variants previously associated with fasting glucose and insulin in the Diabetes Prevention Program. PLoS ONE 7, e44424. doi:10.1371/journal.pone.0044424
Garaulet M, Corbalán MD, Madrid JA et al (2010) CLOCK gene is implicated in weight reduction in obese patients participating in a dietary programme based on the Mediterranean diet. Int J Obes 34:516–523. doi:10.1038/ijo.2009.255
Garaulet M, Smith CE, Gomez-Abellán P et al (2013) REV-ERB-ALPHA circadian gene variant associates with obesity in two independent populations: Mediterranean and North American. Mol Nutr Food Res. doi:10.1002/mnfr.201300361
Goumidi L, Grechez A, Dumont J et al (2013) Impact of REV-ERB alpha gene polymorphisms on obesity phenotypes in adult and adolescent samples. Int J Obes 37:666–672. doi:10.1038/ijo.2012.117
Ha E, Yim S-V, Chung J-H et al (2006) Melatonin stimulates glucose transport via insulin receptor substrate-1/phosphatidylinositol 3-kinase pathway in C2C12 murine skeletal muscle cells. J Pineal Res 41:67–72. doi:10.1111/j.1600-079X.2006.00334.x
Hardeland R (2005) Antioxidative protection by melatonin: multiplicity of mechanisms from radical detoxification to radical avoidance. Endocrine 27:119–130
Hoffman RA, Reiter RJ (1965) Pineal gland: influence on gonads of male hamsters. Science 148:1609–1611
Huopio H, Cederberg H, Vangipurapu J et al (2013) Association of risk variants for type 2 diabetes and hyperglycemia with gestational diabetes. Eur J Endocrinol 169:291–297. doi:10.1530/EJE-13-0286
Illig T, Gieger C, Zhai G et al (2010) A genome-wide perspective of genetic variation in human metabolism. Nat Genet 42:137–141. doi:10.1038/ng.507
Illnerová H, Sumová A (1997) Photic entrainment of the mammalian rhythm in melatonin production. J Biol Rhythms 12:547–555
Jassal B, Jupe S, Caudy M et al (2010) The systematic annotation of the three main GPCR families in Reactome. Database (Oxford) 2010:baq018. doi:10.1093/database/baq018
Jockers R, Maurice P, Boutin JA, Delagrange P (2008) Melatonin receptors, heterodimerization, signal transduction and binding sites: what’s new? Br J Pharmacol 154:1182–1195. doi:10.1038/bjp.2008.184
Jonsson A, Ladenvall C, Ahluwalia TS et al (2013) Effects of common genetic variants associated with type 2 diabetes and glycemic traits on α- and β-cell function and insulin action in humans. Diabetes 62:2978–2983. doi:10.2337/db12-1627
Kan MY, Zhou DZ, Zhang D et al (2010) Two susceptible diabetogenic variants near/in MTNR1B are associated with fasting plasma glucose in a Han Chinese cohort. Diabet Med 27:598–602. doi:10.1111/j.1464-5491.2010.02975.x
Karamitri A, Renault N, Clement N et al (2013) Minireview: toward the establishment of a link between melatonin and glucose homeostasis: association of melatonin MT2 receptor variants with type 2 diabetes. Mol Endocrinol 27:1217–1233. doi:10.1210/me.2013-1101
Kelly MA, Rees SD, Hydrie MZI et al (2012) Circadian gene variants and susceptibility to type 2 diabetes: a pilot study. PLoS ONE 7, e32670. doi:10.1371/journal.pone.0032670
Kemp DM, Ubeda M, Habener JF (2002) Identification and functional characterization of melatonin Mel 1a receptors in pancreatic beta cells: potential role in incretin-mediated cell function by sensitization of cAMP signaling. Mol Cell Endocrinol 191:157–166
Kennaway DJ, Voultsios A, Varcoe TJ, Moyer RW (2002) Melatonin in mice: rhythms, response to light, adrenergic stimulation, and metabolism. Am J Physiol Regul Integr Comp Physiol 282:R358–R365. doi:10.1152/ajpregu.00360.2001
Kim YJ, Go MJ, Hu C et al (2011) Large-scale genome-wide association studies in East Asians identify new genetic loci influencing metabolic traits. Nat Genet 43:990–995. doi:10.1038/ng.939
Kivimäki M, Batty GD, Hublin C (2011) Shift work as a risk factor for future type 2 diabetes: evidence, mechanisms, implications, and future research directions. PLoS Med 8, e1001138. doi:10.1371/journal.pmed.1001138
Klerman EB, Shanahan TL, Brotman DJ et al (2002) Photic resetting of the human circadian pacemaker in the absence of conscious vision. J Biol Rhythms 17:548–555
Korkmaz A, Rosales-Corral S, Reiter RJ (2012) Gene regulation by melatonin linked to epigenetic phenomena. Gene 503:1–11. doi:10.1016/j.gene.2012.04.040
Kristiansen K (2004) Molecular mechanisms of ligand binding, signaling, and regulation within the superfamily of G-protein-coupled receptors: molecular modeling and mutagenesis approaches to receptor structure and function. Pharmacol Ther 103:21–80. doi:10.1016/j.pharmthera.2004.05.002
Kvetnoy I, Sandvik AK, Waldum HL (1997) The diffuse neuroendocrine system and extrapineal melatonin. J Mol Endocrinol 18:1–3
Kwak SH, Kim S-H, Cho YM et al (2012) A genome-wide association study of gestational diabetes mellitus in Korean women. Diabetes 61:531–541. doi:10.2337/db11-1034
Lamia KA, Evans RM (2010) Metabolism: tick, tock, a beta-cell clock. Nature 466:571–572. doi:10.1038/466571a
Leproult R, Holmbäck U, Van Cauter E (2014) Circadian misalignment augments markers of insulin resistance and inflammation, independently of sleep loss. Diabetes. doi:10.2337/db13-1546
Liao S, Liu Y, Tan Y et al (2012) Association of genetic variants of melatonin receptor 1B with gestational plasma glucose level and risk of glucose intolerance in pregnant Chinese women. PLoS ONE 7, e40113. doi:10.1371/journal.pone.0040113
Ling Y, Li X, Gu Q et al (2011) A common polymorphism rs3781637 in MTNR1B is associated with type 2 diabetes and lipids levels in Han Chinese individuals. Cardiovasc Diabetol 10:27. doi:10.1186/1475-2840-10-27
Liu C, Wu Y, Li H et al (2010) MTNR1B rs10830963 is associated with fasting plasma glucose, HbA1C and impaired beta-cell function in Chinese Hans from Shanghai. BMC Med Genet 11:59. doi:10.1186/1471-2350-11-59
Lyssenko V, Nagorny CLF, Erdos MR et al (2009) Common variant in MTNR1B associated with increased risk of type 2 diabetes and impaired early insulin secretion. Nat Genet 41:82–88. doi:10.1038/ng.288
Marcheva B, Ramsey KM, Buhr ED et al (2010) Disruption of the clock components CLOCK and BMAL1 leads to hypoinsulinaemia and diabetes. Nature 466:627–631. doi:10.1038/nature09253
Masana MI, Doolen S, Ersahin C et al (2002) MT(2) melatonin receptors are present and functional in rat caudal artery. J Pharmacol Exp Ther 302:1295–1302
McMullan CJ, Schernhammer ES, Rimm EB et al (2013) Melatonin secretion and the incidence of type 2 diabetes. JAMA 309:1388–1396. doi:10.1001/jama.2013.2710
Mirzaei K, Xu M, Qi Q et al (2014) Variants in glucose- and circadian rhythm-related genes affect the response of energy expenditure to weight-loss diets: the POUNDS LOST Trial. Am J Clin Nutr 99:392–399. doi:10.3945/ajcn.113.072066
Mohawk JA, Green CB, Takahashi JS (2012) Central and peripheral circadian clocks in mammals. Annu Rev Neurosci 35:445–462. doi:10.1146/annurev-neuro-060909-153128
Monk TH, Buysse DJ (2013) Exposure to shift work as a risk factor for diabetes. J Biol Rhythms 28:356–359. doi:10.1177/0748730413506557
Ohshige T, Iwata M, Omori S et al (2011) Association of new loci identified in European genome-wide association studies with susceptibility to type 2 diabetes in the Japanese. PLoS ONE 6, e26911. doi:10.1371/journal.pone.0026911
Okatani Y, Okamoto K, Hayashi K et al (1998) Maternal-fetal transfer of melatonin in pregnant women near term. J Pineal Res 25:129–134
Pan A, Schernhammer ES, Sun Q, Hu FB (2011) Rotating night shift work and risk of type 2 diabetes: two prospective cohort studies in women. PLoS Med 8, e1001141. doi:10.1371/journal.pmed.1001141
Panda S, Antoch MP, Miller BH et al (2002) Coordinated transcription of key pathways in the mouse by the circadian clock. Cell 109:307–320
Peschke E (2008) Melatonin, endocrine pancreas and diabetes. J Pineal Res 44:26–40. doi:10.1111/j.1600-079X.2007.00519.x
Peschke E, Mühlbauer E, Musshoff U et al (2002) Receptor (MT(1)) mediated influence of melatonin on cAMP concentration and insulin secretion of rat insulinoma cells INS-1. J Pineal Res 33:63–71
Peschke E, Bach AG, Mühlbauer E (2006a) Parallel signaling pathways of melatonin in the pancreatic beta-cell. J Pineal Res 40:184–191. doi:10.1111/j.1600-079X.2005.00297.x
Peschke E, Frese T, Chankiewitz E et al (2006b) Diabetic Goto Kakizaki rats as well as type 2 diabetic patients show a decreased diurnal serum melatonin level and an increased pancreatic melatonin-receptor status. J Pineal Res 40:135–143. doi:10.1111/j.1600-079X.2005.00287.x
Pilia G, Chen W-M, Scuteri A et al (2006) Heritability of cardiovascular and personality traits in 6,148 Sardinians. PLoS Genet 2, e132. doi:10.1371/journal.pgen.0020132
Poon AM, Choy EH, Pang SF (2001) Modulation of blood glucose by melatonin: a direct action on melatonin receptors in mouse hepatocytes. Biol Signals Recept 10:367–379
Prokopenko I, Langenberg C, Florez JC et al (2009) Variants in MTNR1B influence fasting glucose levels. Nat Genet 41:77–81. doi:10.1038/ng.290
Prokopenko I, Poon W, Mägi R et al (2014) A central role for GRB10 in regulation of islet function in man. PLoS Genet 10, e1004235. doi:10.1371/journal.pgen.1004235
Ramos E, Chen G, Shriner D et al (2011) Replication of genome-wide association studies (GWAS) loci for fasting plasma glucose in African-Americans. Diabetologia 54:783–788. doi:10.1007/s00125-010-2002-7
Ramracheya RD, Muller DS, Squires PE et al (2008) Function and expression of melatonin receptors on human pancreatic islets. J Pineal Res 44:273–279. doi:10.1111/j.1600-079X.2007.00523.x
Rasmussen-Torvik LJ, Guo X, Bowden DW et al (2012) Fasting glucose GWAS candidate region analysis across ethnic groups in the Multiethnic Study of Atherosclerosis (MESA). Genet Epidemiol 36:384–391. doi:10.1002/gepi.21632
Reiter RJ, Tan DX, Korkmaz A, Rosales-Corral SA (2014) Melatonin and stable circadian rhythms optimize maternal, placental and fetal physiology. Hum Reprod Update 20:293–307. doi:10.1093/humupd/dmt054
Renström F, Shungin D, Johansson I et al (2011) Genetic predisposition to long-term nondiabetic deteriorations in glucose homeostasis: ten-year follow-up of the GLACIER study. Diabetes 60:345–354. doi:10.2337/db10-0933
Reppert SM, Weaver DR, Ebisawa T (1994) Cloning and characterization of a mammalian melatonin receptor that mediates reproductive and circadian responses. Neuron 13:1177–1185
Reppert SM, Godson C, Mahle CD et al (1995) Molecular characterization of a second melatonin receptor expressed in human retina and brain: the Mel1b melatonin receptor. Proc Natl Acad Sci USA 92:8734–8738
Rönn T, Wen J, Yang Z et al (2009) A common variant in MTNR1B, encoding melatonin receptor 1B, is associated with type 2 diabetes and fasting plasma glucose in Han Chinese individuals. Diabetologia 52:830–833. doi:10.1007/s00125-009-1297-8
Sadacca LA, Lamia KA, deLemos AS et al (2011) An intrinsic circadian clock of the pancreas is required for normal insulin release and glucose homeostasis in mice. Diabetologia 54:120–124. doi:10.1007/s00125-010-1920-8
Scheer FAJL, Hilton MF, Mantzoros CS, Shea SA (2009) Adverse metabolic and cardiovascular consequences of circadian misalignment. Proc Natl Acad Sci USA 106:4453–4458. doi:10.1073/pnas.0808180106
Scott RA, Lagou V, Welch RP et al (2012) Large-scale association analyses identify new loci influencing glycemic traits and provide insight into the underlying biological pathways. Nat Genet 44:991–1005. doi:10.1038/ng.2385
Shi S, Ansari TS, McGuinness OP et al (2013) Circadian disruption leads to insulin resistance and obesity. Curr Biol 23:372–381. doi:10.1016/j.cub.2013.01.048
Simonis-Bik AM, Nijpels G, van Haeften TW et al (2010) Gene variants in the novel type 2 diabetes loci CDC123/CAMK1D, THADA, ADAMTS9, BCL11A, and MTNR1B affect different aspects of pancreatic beta-cell function. Diabetes 59:293–301. doi:10.2337/db09-1048
Sparsø T, Bonnefond A, Andersson E et al (2009) G-allele of intronic rs10830963 in MTNR1B confers increased risk of impaired fasting glycemia and type 2 diabetes through an impaired glucose-stimulated insulin release: studies involving 19,605 Europeans. Diabetes 58:1450–1456. doi:10.2337/db08-1660
Stamenkovic JA, Olsson AH, Nagorny CL et al (2012) Regulation of core clock genes in human islets. Metabolism 61:978–985. doi:10.1016/j.metabol.2011.11.013
Stumpf I, Mühlbauer E, Peschke E (2008) Involvement of the cGMP pathway in mediating the insulin-inhibitory effect of melatonin in pancreatic beta-cells. J Pineal Res 45:318–327. doi:10.1111/j.1600-079X.2008.00593.x
Stumpf I, Bazwinsky I, Peschke E (2009) Modulation of the cGMP signaling pathway by melatonin in pancreatic beta-cells. J Pineal Res 46:140–147. doi:10.1111/j.1600-079X.2008.00638.x
Takahashi JS, Hong H-K, Ko CH, McDearmon EL (2008) The genetics of mammalian circadian order and disorder: implications for physiology and disease. Nat Rev Genet 9:764–775. doi:10.1038/nrg2430
Takeuchi F, Katsuya T, Chakrewarthy S et al (2010) Common variants at the GCK, GCKR, G6PC2-ABCB11 and MTNR1B loci are associated with fasting glucose in two Asian populations. Diabetologia 53:299–308. doi:10.1007/s00125-009-1595-1
Taneera J, Lang S, Sharma A et al (2012) A systems genetics approach identifies genes and pathways for type 2 diabetes in human islets. Cell Metab 16:122–134. doi:10.1016/j.cmet.2012.06.006
Tirosh A, Shai I, Tekes-Manova D et al (2005) Normal fasting plasma glucose levels and type 2 diabetes in young men. N Engl J Med 353:1454–1462. doi:10.1056/NEJMoa050080
Turek FW, Joshu C, Kohsaka A et al (2005) Obesity and metabolic syndrome in circadian Clock mutant mice. Science 308:1043–1045. doi:10.1126/science.1108750
Vangipurapu J, Stančáková A, Pihlajamäki J et al (2011) Association of indices of liver and adipocyte insulin resistance with 19 confirmed susceptibility loci for type 2 diabetes in 6,733 non-diabetic Finnish men. Diabetologia 54:563–571. doi:10.1007/s00125-010-1977-4
Venkatakrishnan AJ, Deupi X, Lebon G et al (2013) Molecular signatures of G-protein-coupled receptors. Nature 494:185–194. doi:10.1038/nature11896
Vlassi M, Gazouli M, Paltoglou G et al (2012) The rs10830963 variant of melatonin receptor MTNR1B is associated with increased risk for gestational diabetes mellitus in a Greek population. Hormones (Athens) 11:70–76
Voight BF, Scott LJ, Steinthorsdottir V et al (2010) Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat Genet 42:579–589. doi:10.1038/ng.609
Vyas MV, Garg AX, Iansavichus AV et al (2012) Shift work and vascular events: systematic review and meta-analysis. BMJ 345, e4800
Walford GA, Green T, Neale B et al (2012) Common genetic variants differentially influence the transition from clinically defined states of fasting glucose metabolism. Diabetologia 55:331–339. doi:10.1007/s00125-011-2353-8
Wang Y, Nie M, Li W et al (2011) Association of six single nucleotide polymorphisms with gestational diabetes mellitus in a Chinese population. PLoS ONE 6, e26953. doi:10.1371/journal.pone.0026953
Wang H, Liu L, Zhao J et al (2013) Large scale meta-analyses of fasting plasma glucose raising variants in GCK, GCKR, MTNR1B and G6PC2 and their impacts on type 2 diabetes mellitus risk. PLoS ONE 8, e67665. doi:10.1371/journal.pone.0067665
Watanabe RM, Valle T, Hauser ER et al (1999) Familiality of quantitative metabolic traits in Finnish families with non-insulin-dependent diabetes mellitus. Finland-United States Investigation of NIDDM Genetics (FUSION) Study investigators. Hum Hered 49:159–168, doi: 22865
Weitzman ED, Fukushima D, Nogeire C et al (1971) Twenty-four hour pattern of the episodic secretion of cortisol in normal subjects. J Clin Endocrinol Metab 33:14–22. doi:10.1210/jcem-33-1-14
Woon PY, Kaisaki PJ, Bragança J et al (2007) Aryl hydrocarbon receptor nuclear translocator-like (BMAL1) is associated with susceptibility to hypertension and type 2 diabetes. Proc Natl Acad Sci USA 104:14412–14417. doi:10.1073/pnas.0703247104
World Health Organization (2013) http://www.who.int/mediacentre/factsheets/fs312/. Fact sheet N°312
Xia Q, Chen Z-X, Wang Y-C et al (2012) Association between the melatonin receptor 1B gene polymorphism on the risk of type 2 diabetes, impaired glucose regulation: a meta-analysis. PLoS ONE 7, e50107. doi:10.1371/journal.pone.0050107
Zhao Q, Xiao J, He J et al (2014) Cross-sectional and longitudinal replication analyses of genome-wide association Loci of type 2 diabetes in han chinese. PLoS ONE 9, e91790. doi:10.1371/journal.pone.0091790
Zhdanova IV, Wurtman RJ (1997) Efficacy of melatonin as a sleep-promoting agent. J Biol Rhythms 12:644–650
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2016 Springer International Publishing Switzerland
About this chapter
Cite this chapter
Bonnefond, A., Lyssenko, V. (2016). From Association to Function: MTNR1B . In: Florez, J. (eds) The Genetics of Type 2 Diabetes and Related Traits. Springer, Cham. https://doi.org/10.1007/978-3-319-01574-3_19
Download citation
DOI: https://doi.org/10.1007/978-3-319-01574-3_19
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-01573-6
Online ISBN: 978-3-319-01574-3
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)