Abstract
Graphs are becoming increasingly larger, with datasets having millions of vertices and billions (or even trillions) of edges. As a result, the ability to fit the entire graph into the main memory of a single machine faces challenges in common hardware, even more so in edge/IoT-like devices (i.e., more energy efficient but also more resource constrained). Reading the graph from secondary storage may pose in itself significant overhead, negatively impacting query performance and storage requirements. It thus becomes relevant to explore techniques to optimize the storage of graphs, specially in memory, in a way that circumvents space limitations, while avoiding compromising the performance of processing.
We observe that current graph storage systems manage the graph representation by storing graphs in an uncompressed format, either: i) in a shared architecture which leads to a higher space overhead and the inability to represent the graph entirely in main memory, or ii) in a distributed architecture, where the graph dataset is partitioned over a cluster of machines with each one storing in main memory only a fragment (shard) of the (uncompressed) graph. We present PK-Graph, our proposal which extends a distributed graph processing system, highly used in academia and industry (Spark GraphX), in order to deploy the use of a compressed graph representation, with added support for dynamic updatable graphs (not currently supported in GraphX). Our experimental results show that PK-Graph can achieve up to 50% lower graph memory usage, while maintaining competitive performance in executing typical graph operations used in common applications.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Graphs are now more relevant than ever and their importance will continue to expand [38], as well continuing to grow in size, having millions of vertices and billions (or even trillions) of edges in some cases [9, 15], stimulating the need for novel and more efficient storage and representation solutions due to increasing space requirements. Fitting an entire graph in the main memory of a single multiprocessor machine becomes challenging if the graph is very large. This may lead to a significant overhead by having to read the graph from secondary storage. Thus, it is relevant to try to minimize the storage requirements of the graph, for efficiency and viability, without degrading access time and ideally even improving it. Current solutions store graphs in uncompressed format [16, 18, 19, 22, 24, 39, 42, 44]. By using a lossless graph compression technique, it is possible to store the graph in a compressed format that can reside in the main memory of a single resource-rich machine [6, 28, 43], achieving equal or better performance when accessing the graph, thus motivating the employment of compression techniques in graph storage.
A relevant use-case when working with graph-based data is the ability to modify it as a dynamic graph, where it is possible to add or remove vertices and edges. For this use-case, using basic compression techniques would require converting the graph to an uncompressed format before modifying it, which would imply limitations in required storage and obtained speed. Popular graph algorithms, such as PageRank [31], mutate attributes stored in the vertices and edges of graphs as part of their logic. As a consequence, the ability to use compressed graph representations which support graph-changing operations without having to decompress becomes very important.
Existing solutions focus on partitioning graphs based on their edges to achieve better work distribution among computing nodes [24, 44]. This leads to edges being assigned to unique partitions and vertices, while being replicated throughout various partitions. In a worst-case scenario, a vertex needs to be replicated throughout all partitions. This approach is used because the number of edges is typically much higher than the number of vertices, leading to smaller storage requirements when replicating vertices. We focus on addressing several shortcomings that current solutions present, such as: i) not being able to store large graphs completely in main memory, requiring access to secondary storage which is much slower; ii) storing graphs in an uncompressed format, potentially leading to higher resource consumption and comparatively worse processing performance than compressed representations; iii) immutable graphs that do not support removing or adding vertices/edges, requiring the entire graph to be re-constructed when adding new elements.
Herein we present our design, implementation and evaluation of PK-Graph, an extension to the storage component of the Spark GraphX distributed graph processing system, incorporating the \(k^2\)-tree lossless compressed graph representation to improve space-efficiency. Our solution was designed with the goal of achieving performance within the same order of magnitude of the uncompressed version of the system and with the goal of supporting dynamic graphs, with mutation of attributes and addition/removal of graph elements. This paper is structured as follows. Section 2 addresses relevant state-of-the-art in graph processing systems, graph databases, and optimized graph representations. In Sect. 3 we present the architecture of PK-Graph. Section 4 describes the evaluation methodology and the results obtained for our implementation. Section 5 concludes by summarizing our findings and mentioning future vectors of research.
2 Related Work
Distributed Graph Processing Systems. They focus on scalable iteration of potentially large input graphs in order to execute algorithms over them. Their approach consists in partitioning the graph throughout a cluster of processors, where each processor stores only a fraction of the total graph in main memory. They maintain serialized graph formats in secondary storage, at penalty, and only if the graph is too large. These systems do not typically require fine-grained access to the vertices and/or edges of the graph and instead iterate all elements of the graph or a subset of them.
Apache Spark [20] and Apache Flink [45] are known examples of generic distributed processing systems, based on dataflow programming. Although they are generic, graph-specific libraries have been built over them, such as GraphX [44] on Spark and Gelly on Flink. There are also systems designed with an ab initio architectural focus on graph processing such as Apache Giraph [42], implementing a vertex-centric approach known as think-like-a-vertex (TLAV), where a user-defined function is applied in the context of each vertex. This model first debuted in Google Pregel [26]. Other approaches exist regarding the unit of computation when expressing graph-processing logic. The computational unit may also be the edge, in which case the system is said to be edge-centric, known as think-like-an-edge (TLEV). This approach was popularized with the X-Stream [37] and Chaos [36] graph processing systems (they are no longer maintained or developed). Other approaches exist, such as defining the unit of computation as a part of the graph, but they are outside this scope.
In terms of dynamism, systems such as Spark and Flink typically only allow for applying changes to the graph (updating attributes or adding/removing vertices/edges), by transforming an existing graph into a new one [3]. This a functional programming aspect of the dataflow-based computation of these systems, and even if the systems provide primitives to reuse or cache data between dataflow jobs to keep changing and using a graph, that does not necessarily lead to an improvement in these sequences of graph changes [10]. In the literature there are other efficient graph processing systems such as GraphBolt [27], PowerLyra [8] and GraphTau [17], among others. While presenting innovative distributed graph processing techniques, as far as we know they typically do not have an active development community or were tailor-made for specific experiments.
Graph Databases. Graph database systems are akin to typical relational databases, but have specialized formats to efficiently store graphs. These systems also focus on fine-grained access to the vertices and edges of a graph, allowing for complex queries to be made while not necessarily needing to traverse the entire graph for each query. As such, the storage of the graph is made to be very space efficient but also to allow for very low latency when performing queries. Throughout these databases we find graph storage location approaches such as: storing in the file system, potentially a distributed one like HDFS [41] or S3 [32]; in key-value stores, where the vertices and edges are stored by mapping their identifier to their attributes, or in NoSQL databases adapted to store graph data.
The database can also be distributed, with the graph stored across multiple machines, or centralized, with the entire graph stored in a single machine. In some cases, where specialized hardware is available, centralized systems may have similar or even better performance than distributed ones (such as Ringo [33] and Mosaic [25]). An example of a relevant graph database is Neo4j [16], a native graph database platform used to store, query, analyze and manage highly connected data expressed in the property graph model. It provides its own query language Cypher [14], with data stored on disk as linked lists of fixed-size records. Properties are stored as a linked list of property records, each holding a key and value and pointing to the next property.
Compact Graph Representations. Compressed graph representations are employed to reduce the computational space complexity of graphs, lowering their storage requirements and enabling their processing with hardware that is less powerful. In the context of this work, we focus on computational representations that are directly compatible with or enabling components of the property graph model [2], which allows for attributes to be held in the elements of the graph. Depending on the relationship between the representation design and its implementation, it is possible to store element attributes in a compressed form together with the rest of the graph structure. There are factors that influence the design of a representation. Whether the graph is directed or undirected has an influence on the representation. If the graph is directed, then twice the number of edges (of an equivalent undirected graph) would be necessary, as each undirected edge may be represented with two edges with opposing directions (between the same two vertices). For example, another factor influencing the representation is tied to the potential need of representing more than one edge between the same two vertices (multi-graph) or not (simple graph). In the context of dynamism, if compressed graph representations allow mutating graphs, they are also known as compact representations.
Some of the most well-known compressed representations are the WebGraph [5] framework and the \(k^2\)-tree [6]). The WebGraph [5] framework uses mathematical analysis and information theory [30] to represent the graph (in a lossless way) with lower complexity (using traits such as vertex ordering [4]). WebGraph enabled the exploration of many graph datasets, enabling researchers to analyze them and obtain statistics using files with smaller sizes. It is implemented in Java and does not support mutating the graph, which limits the scope of its applicability.
Some more recent work in compressed graph representations includes g-Sum [34], a graph summarization approach for large social networks that minimizes the Reconstruction Error (RE) of the representation, allowing for a more accurate summarization and improving its usefulness. Another recent work [21] presents MoSSo, an algorithm for incremental lossless graph summarization. This work provides a novel approach in the efficient and lossless summarization of fully dynamic graphs. However, this representation is not suitable for distributed processing systems like Spark GraphX since the graph would need to be partitioned throughout various executors. Furthermore, the summarization is not intended to allow for the iteration of all edges/vertices of the graph, instead it focuses on specifically handling the processing of individual changes to the underlying graph.
Hornet [7] is a data structure for efficient computation of dynamic sparse graphs and matrices using GPUs. It is platform-independent and implements its own memory allocation operation instead of standard function calls. The implementation uses an internal data manager which makes use of block arrays to store adjacency lists, a bit tree for finding and reclaiming empty memory blocks and \(B^{+}\) trees to manage them. It was evaluated using an NVIDIA Tesla GPU and experiments targeted the update rates it supports, algorithms such as breadth-first search (BFS) and sparse matrix-vector multiplication. While a relevant mark in the literature, it is GPU-focused.
Another recent example is the \(k^2\)-tree [6], an optimized compressed graph representation that takes advantage of sparse adjacency matrices by recursively decomposing them. Figure 1 shows one such tree. The tree represents the structure of the graph’s adjacency matrix, where each node in the tree is represented by a single bit: 1 for internal nodes and 0 for leaf nodes, except in the last level where all nodes are leaves and represent the bit values in the adjacency matrix. Different implementations (C/C++) of the \(k^2\)-tree exist, and although the original one did not support graph mutability, more recent implementations allow the graph to be mutated, either by directly using dynamic bit vectors (which suffers a performance bottleneck on compressed dynamic indexing [29, 30]), or more recently, by using techniques to provide dynamic behavior on underlying static collections [11], achieving competitive performance compared to other implementations [12].
3 PK-Graph: Architecture
While many graph processing systems are available, many were released solely to assess and validate specific scientific ideas. From our analysis of graph processing systems, we find value in attributes such as the pace of development of the systems as well as active communities with which it is possible to engage to discuss ideas or troubleshoot development challenges that are found. While Flink and Spark are prime candidates with these attributes, Spark was chosen to implement our contribution, as its design implementation already has some concern for some form of data reuse (such as its cache() operator).
In Spark, data storage is handled by its Resilient Distributed Dataset (RDD) construct. It represents an immutable collection of elements which may undergo transformations (e.g. map(), filter()) defined in the functional programming paradigm, and they can be processed in distributed fashion by splitting elements into various partitions and having different machines in the cluster process different partitions.
Our solution extends Spark’s GraphX graph library to make use of a recent dynamic, compact and competitive \(k^2\)-tree implementation [11, 12], allowing for a compressed representation of property graphs in main memory. PK-Graph is built into a JAR file which must be coupled with GraphX’s own JAR in order to use it. GraphX provides an abstraction over graphs, containing views of: a) vertices; b) edges; c) edge triplets which correspond to the union of an edge with its corresponding source and destination vertices. All views are partitioned according to user criteria (with default strategies also offered). GraphX implements this abstraction by replicating the vertices in the edge partitions, thus efficiently performing a join between an edge and its corresponding vertices. This abstraction is static and does not allow the addition/removal of vertices/edges. It is possible to update the attributes of either vertices or edges, but because Spark’s RDD is immutable, updating the graph becomes a challenge (within the same dataflow job). Our solution provides the same three views while maintaining a compressed and fully dynamic representation of the graph, capable of adding new edges or vertices as well as updating their attributes.
3.1 Overall Architecture
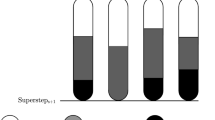
Figure 2 shows a diagram of the architecture overview of our system and how it integrates with the GraphX platform. The main classes of the GraphX implementation are shown in blue and the main classes of our system are in green. The Graph class provides an interface for all basic graph operations, primitives used to implement graph algorithms and access to the underlying vertex and edge RDDs. All graph operations are executed in a lazy and distributed fashion, by propagating them throughout a cluster of computing nodes and aggregating the result in the driver program. Figure 3 shows an example of how a graph operation can be distributed throughout a cluster.
Vertices: Representation. The VertexRDD class provides an interface for vertex-specific RDDs, containing operations to iterate and transform the underlying vertices of the graph. The VertexRDDImpl contains the default GraphX implementation of the VertexRDD class. Our solution incorporates the \(k^2\)-tree data structure to optimize operation on edges, with the vertex functionality of PK-Graph remaining unchanged from what GraphX provides.
The vertex partitions (where the actual vertices are stored) are implemented by the ShippableVertexPartition that keeps them in a format ready to be shipped to their corresponding edge partitions. Each vertex partition keeps track of the routing information for each of its vertices, later to be used to determine to which edge partition the vertices are shipped. The mask bitset keeps track of all active vertices in the partition. The vertex operations of a partition are only applied to the active vertices. To access the vertices of a partition, all set bits in the mask are iterated, retrieving the corresponding vertex identifier and attribute (see Algorithm 3.1).

Edges: Representation. The EdgeRDD class provides an interface for edge-specific RDDs and contains operations to iterate and transform the underlying edges of the graph. Our solution extends this abstraction with the PKEdgeRDD class, providing a specific implementation of the edge partitions (PKEdgePartition) using the compressed \(k^2\)-tree data structure to store the edges of the graph (K2Tree). The edge partitions are stored in the PKEdgePartition class, which provides operations to iterate and transform the underlying edges. The actual edges are stored in the K2Tree class, which implements the \(k^2\)-tree compressed data structure. In our modified edge partitioning, every operation in the edge partition creates a new instance with copies of the previous data and any modifications applied. This is done in order to offer the same expected semantics of GraphX when changing the elements of an RDD.

The dynamic operations (addEdges and removeEdges) can add or remove edges from the partition. Although they are dynamic operations, the edge partition does not need to be mutable, since a new instance of the PKEdgePartition class is returned as a result of these operations.
As stated previously, the edge partition uses a \(k^2\)-tree compressed data structure to store the edges of the graph. This data structure is capable of representing the edges of a graph in a very space-efficient format. Our architecture only requires that the implementation of this structure provides a method to access and iterate its edges. This will require iterating the \(k^2\)-tree in a depth-first fashion and calculating the line and column in the adjacency matrix of each edge. Each line and column will correspond to local vertex identifiers, which then will need to be efficiently mapped to global identifiers, as well as determining for each edge its corresponding attribute. Algorithm 3.2 shows an example in pseudo-code of a possible implementation to access the edges of an edge partition by iterating its corresponding \(k^2\)-tree.
In a similar fashion to the GraphX system, our solution also uses a simple wrapper over an edge RDD, provided by the PKReplicatedVertexView to handle the shipping of vertices to the underlying edge partitions. This class stores the underlying PKEdgeRDD instance and keeps track of whether the view includes the attributes of both the source and destination vertices or if these are only partially shipped, since in some cases these may be unnecessary.
Dataflow Operations. The GraphX API offers dataflow operators to manipulate the graph. We list the most relevant ones here.
The updateVertices operation receives an iterator referencing cached vertices in the partition that should be updated with new attributes. The reverse operation reverses all edges in the partition by switching the source vertices with the destination vertices. This operation is directly used by the graph abstraction to perform its own reverse operation. The map operation applies a user function to all edges stored in the partition. The filter operation filters both the vertices of an edge and the actual edge according to the user defined predicates. The innerJoin operation performs an inner join between two edge partitions. The aggregateMessages operation is the primitive used to implement all popular graph algorithms. It implements a Pregel-like messaging system to exchange messages between the vertices of a graph. Each vertex is capable of sending a message through an edge to another vertex. These messages are then aggregated and merged at each vertex and collected after all messages have been sent.
The GraphX computing model also has the ability to maintain only some vertices in an active state, with only the active vertices able to receive messages. Active vertex information is stored in each edge partition and the non-active vertices are skipped when aggregating messages. The activeness requirements can then be specified as a parameter of the aggregateMessages function.
3.2 Dynamism
The DynamicGraph interface exposes various functions to both add and remove vertices and edges from a graph. However, since the underlying Spark RDDs are immutable, some partitions of the graph will need to be rebuilt, or at the very least a new copy of them will need to be made. This does not necessarily mean that the entire graph will need to be rebuilt, only the partitions which we are transforming. Thus, adding or removing both vertices and edges will require determining the partitions affected, and only transforming these.
The addVertices and addEdges functions add new vertices and edges, respectively, to the graph, returning a new graph instance in the process. The removeVertices and removeEdges functions remove the given vertices and edges from the graph, also returning a new graph instance in the process. Both of these functions work very similarly to applying a filter over the graph, with the slight optimization that only either the vertices or the edges of a graph are affected, instead of always having to filter both. All dynamic functions receive RDD instances as parameters to allow for these operations to be distributed throughout a computing cluster. We note that the impact of PK-Graph and the \(k^2\)-tree data structure is focused only on the addEdges and removeEdges functions.
3.3 Partitioning
Because GraphX processes the graph data in a distributed fashion, our solution will also need to address the problem of how to partition the graph to allow for spatial and computational efficiency. The input graph is represented by two RDDs provided by the user, one representing the vertices and another representing the edges (similar to the GraphX implementation). For the case of edges, our solution will interpret them as an edge adjacency matrix that will be partitioned using a 2D partitioning scheme [1] that splits the adjacency matrix into several sub-matrices of equal size, each assigned to a unique partition.
In case the number of partitions is not a perfect square, the last column will have a different number of rows than the others. One problem with this distribution is that it leads to poor work balance since, given a sparse adjacency matrix, some partitions will have many more edges than others. To overcome this, we shuffle the vertex locations in order to evenly distribute them through all partitions.
Like GraphX’s implementation, our solution will also replicate the vertices in the edge partitions to provide an efficient way to join the edges with their respective vertices. Using this distribution we guarantee that any vertex is replicated at most \(2 \times \sqrt{|P|}\) times, where |P| is the number of partitions of the adjacency matrix, since any vertex is represented by a line and a corresponding column in the matrix, and every line and column intersect at most \(\sqrt{|P|}\) partitions.
The described partitioning scheme is applied by default, with no configuration required for the edges. It is also possible for the programmer to specify a different partitioning scheme by using the already existing interface provided by Spark. For the vertices, we would default to the partitioning scheme supplied by the user or, if no scheme was provided, default to a uniform partitioning strategy such as the one based on the hash of each vertex. In cases where the graph becomes unbalanced, the user can repartition the underlying vertex and edge RDDs to either increase or decrease the number of partitions, using Spark’s repartition function. Increasing the number of partitions implies shuffling, which will incur a significant overhead due to network communication between workers. However, when decreasing the number of partitions, it is possible to avoid a shuffling phase by using Spark’s coalesce function.
The GraphX platform already offers several partition strategies such as: EdgePartition2D, this is the strategy described earlier and implements a strategy that divides the adjacency matrix of the graph into several blocks, as well as shuffling the vertices of the graph to provide a more balanced work distribution; EdgePartition1D, which groups together edges with the same source vertex; RandomVertexCut, which distributes the edges based on the hash code of both the source and destination vertex identifiers; CanonicalRandomVertexCut, the same strategy as the RandomVertexCut but also taking in consideration the direction of the edge when performing the hash. Our solution also introduces a new partition strategy, represented by the PKGridPartitionStrategy class. This strategy is similar to EdgePartition2D of GraphX. The main difference between the strategies is that the vertices will not be shuffled, in order not to change the data locality of the edges, thus providing a more space-efficient representation of the entire graph in some cases, at the cost of worse workload distribution in the cluster.
4 Evaluation
To evaluate the implementation of our solution, we performed various benchmarks in a cluster of computing nodes, each node corresponding to a Spark worker that keeps part of the total graph in main memory. We submitted several graph processing jobs to the cluster, executing some basic graph operations and some of the more popular graph algorithms, using relevant graph datasets and analyzing the gains (penalties) our solution has in terms of compression storage improvements and processing performance.
The cluster was prepared using the AWS EMR service [13], which enables the easy setup of a cluster of Spark workers. The cluster uses a single master node and various worker nodes.
The actual number of employed workers varies throughout each test. Each machine in the cluster has a 4-core processor with 16 GB of available main memory, in order to represent typical cost-efficient cloud-provider servers. Note that in edge cloud scenarios, servers would normally include more resource-constrained machines [40] that would make memory efficiency a much more pressing issue.
The Spark jobs are submitted from a driver program in a remote machine and the datasets are retrieved from AWS S3 buckets to be used in the jobs executed in the cluster.
Datasets. The datasets used in the evaluation of our implementation are from the Network Repository [35] and the Stanford Large Network Dataset Collection (SNAP) [23]. The datasets chosen for the benchmarks are the following:
-
YouTube Growth (3M vertices, 12.2M edges)
-
EU (2005) (863K vertices, 19M edges)
-
Indochina (2004) (7M vertices, 194M edges)
-
UK (2002) (18M vertices, 298M edges)
Memory Overhead. Our benchmarks show that the memory overhead of the data structure of the graph remains the same independently of the number of processors. This is due to the fact that the number of used partitions chosen by Spark, based on the size of the file where the dataset was read from, remains the same.
Figure 4 shows the results of the memory usage of the entire graph for all datasets. The results show that our solution has significantly less memory overhead than the GraphX implementation. When testing the memory usage of the entire graph, comparing to the GraphX implementation, results range from a reduction of 30% to 50% (roughly 1.50 to two-fold more memory efficient) of 60% to 70% (roughly three-fold more memory efficient). This is in part due to the partitioning of the graph and its nature. The best performance is observed on the web graphs, since these have much higher edge clustering when compared to other types of graphs. Furthermore, the number of processors has no significant impact on the graph size in memory. Regarding memory efficiency of edges specifically, it improves three fold (30%, of initially used).
Workload Latencies. The workloads presented in this subsection compare the latency between PK-Graph and GraphX for different graph algorithms. Latency is defined as the total time the system takes to execute graph processing jobs.
Basic Iteration. This workload iterates all edges of the graph and applies a user function to each edge (for evaluation, this function simply multiplies the edge’s integer value by a constant). The obtained results are shown in Figs. 5a, 5b, 5c and 5d. As observed with the previous tests, as the number of processors increases, the iteration latency decreases. As the GraphX implementation is more efficient at traversing all edges in an edge partition, it achieves a lower latency compared to PK-Graph, even when using a k value that optimizes processing performance. In terms of iteration latency, overall our implementation is between 15% to 40% slower than the GraphX implementation, depending on the type of graph, obtaining better results for web graphs when compared to social network graphs.
PageRank. For the PageRank algorithm, we observe similar patterns to the basic iteration test, with PK-Graph ’s latency approaching that of GraphX with higher vCPU counts on the Indochina (2004) and UK (2002) datasets. The latency results for PageRank are depicted in Figs. 6a, 6b, 6c and 6d. For larger graphs, as the number of available processors increases, the latency of the graph operation decreases.
Triangle Count. This workload executes an algorithm to count triangles, which is typically used in social network analysis to detect communities and measure clustering coefficients. It is an algorithm which has less latency than PageRank. Triangle Count latency results are presented in Figs. 7a, 7b, 7c and 7d. For this algorithm, the relationship between latency and number of vCPUs exhibited behavior similar to PageRank, with datasets Indochina (2004) and UK (2002) seeing a smaller latency gap between PK-Graph and GraphX when executing with a higher number of vCPUs.
CPU Usage Results. They are presented in Figs. 8a, 8b, 8c and 8d. For this metric, we compare the total run time of Spark executors to their total CPU time for each dataset, showing the percentage of the total run time spent on the processor. PK-Graph achieves a higher CPU usage as the iteration algorithms used by our solution are heavier.
Edge Partition Statistics. The latency of building an edge partition from a list of edges is shown in Fig. 9. As the number of edges in a partition increase, the incurred latency while building the partition also grows. In Fig. 10 we show the behavior of iteration latency as the number of edges increases. The higher the value of parameter k in the \(k^2\)-tree, the better the iterator performance due to the smaller height of the tree.
Analysis and Discussion. Overall, while considering the detailed evaluation of our implementation, our solution provides a significant reduction in memory usage, i.e. between 40% and 50% depending on the k value used for the \(k^2\)-tree, the type of graph and the partitioning strategy employed. As we are using a \(k^2\)-tree as the compressed data structure, the sparser the adjacency matrix of the graph is, the better the compression achieved. This enables the employment of comparatively less capable devices, such as in those deployed in community micro clouds (i.e., in edge cloud and IoT-like scenarios), as well as those low-cost made available by cloud providers (i.e., spot instances and virtual machines tailored to micro-services and serverless computing).
Our implementation also provides competitive processing performance when compared to the GraphX implementation, specially considering that this current GraphX approach focuses mainly on having the best possible processing performance by keeping all edges in an array with no application of compression techniques. The performance penalty of PK-Graph decreases in inverse relation with the complexity of workload algorithm and the size of the dataset. Nonetheless, while requiring less memory (the resource harder to share across time between workloads), results show that at times PK-Graph incurs a higher CPU usage than GraphX, due to the increased graph processing complexity over the compact data structure, as our iteration algorithms are more demanding on it.
5 Conclusion
We improve upon GraphX’s implementation, using a \(k^2\)-tree, a data structure that efficiently represents binary relations between two vertices. GraphX’s implementation uses two arrays to store the local source and destination vertex identifiers and a hash map to keep track of all the direct neighbors of each vertex. Our solution replaces this with a \(k^2\)-tree that can efficiently compute the direct and reverse neighbors of any local vertex.
We focused on reducing the memory usage of graphs while still maintaining a competitive processing performance. We designed, developed and evaluated an extension to the storage component of the GraphX distributed graph processing library of Spark so that the processed graph is made more space-efficient by using the \(k^2\)-tree lossless compressed representation, while also aiming to achieve similar performance to the uncompressed version. We evaluated the performance of PK-Graph in a cluster of Spark workers, using various datasets to showcase the effectiveness of our solution in both web and non-web graphs, as well as how our solution scales as the size of the graph and the number of available processors increase. Our experimental results highlight that our solution offers a significant reduction in memory usage of graphs, specially for web graphs, while achieving competitive processing performance when compared to the GraphX implementation. PK-Graph demonstrates an innovative combination of data representation and processing techniques for distributed processing systems while decreasing space complexity, resulting in a middleware which enables execution in resource-constrained scenarios, with application on less powerful machines and spot-type virtual instances. For the different iteration workloads, the latency difference between GraphX and PK-Graph tended to decrease with bigger datasets and higher number of vCPUs.
Future Work. We envision the integration of the \(k^2\)-tree data structure on other processing systems such as Flink, as well as exploring the possibility of integrating other schemes such as the WebGraph [5] representation. Orthogonal to this, it would be relevant to expand evaluated datasets to include more diverse real-world graphs and to evaluate further ideas with datasets of greater size. Evaluating our contribution with other algorithms and also comparing with other similar works will be relevant.
References
Álvarez-García, S., Brisaboa, N.R., Gómez-Pantoja, C., Marin, M.: Distributed query processing on compressed graphs using K2-trees. In: Kurland, O., Lewenstein, M., Porat, E. (eds.) SPIRE 2013. LNCS, vol. 8214, pp. 298–310. Springer, Cham (2013). https://doi.org/10.1007/978-3-319-02432-5_32
Angles, R.: The Property Graph Database Model (2018). http://ceur-ws.org/Vol-2100/paper26.pdf. Accessed 24 Apr 2020
Besta, M., Fischer, M., Kalavri, V., Kapralov, M., Hoefler, T.: Practice of streaming and dynamic graphs: concepts, models, systems, and parallelism. CoRR abs/1912.12740 (2019). http://arxiv.org/abs/1912.12740
Boldi, P., Vigna, S.: The WebGraph framework II: codes for the World-wide Web. In: 2004 Data Compression Conference (DCC 2004), 23–25 March 2004, Snowbird, UT, USA, p. 528. IEEE Computer Society (2004). https://doi.org/10.1109/DCC.2004.1281504
Boldi, P., Vigna, S.: The WebGraph framework I: Compression techniques. In: Feldman, S.I., Uretsky, M., Najork, M., Wills, C.E. (eds.) Proceedings of the 13th International Conference on World Wide Web, WWW 2004, New York, NY, USA, 17–20 May 2004, pp. 595–602. ACM, New York, NY, USA (2004). https://doi.org/10.1145/988672.988752
Brisaboa, N.R., Ladra, S., Navarro, G.: k2-trees for compact web graph representation. In: Karlgren, J., Tarhio, J., Hyyrö, H. (eds.) SPIRE 2009. LNCS, vol. 5721, pp. 18–30. Springer, Heidelberg (2009). https://doi.org/10.1007/978-3-642-03784-9_3
Busato, F., Green, O., Bombieri, N., Bader, D.A.: Hornet: an efficient data structure for dynamic sparse graphs and matrices on GPUs. In: 2018 IEEE High Performance Extreme Computing Conference (HPEC), pp. 1–7. IEEE (2018)
Chen, R., Shi, J., Chen, Y., Zang, B., Guan, H., Chen, H.: PowerLyra: differentiated graph computation and partitioning on skewed graphs. ACM Trans. Parallel Comput. (TOPC) 5(3), 1–39 (2019)
Ching, A., Edunov, S., Kabiljo, M., Logothetis, D., Muthukrishnan, S.: One trillion edges: graph processing at Facebook-scale. Proc. VLDB Endow. 8(12), 1804–1815 (2015)
Coimbra, M.E., Esteves, S., Francisco, A.P., Veiga, L.: VeilGraph: incremental graph stream processing. J. Big Data 9(1), 1–29 (2022)
Coimbra, M.E., Francisco, A.P., Russo, L.M.S., de Bernardo, G., Ladra, S., Navarro, G.: On dynamic succinct graph representations. In: Data Compression Conference (DCC), p. 10. IEEE, January 2020. https://sigport.org/documents/dynamic-succinct-graph-representations
Coimbra, M.E., et al.: A practical succinct dynamic graph representation. Inf. Comput. 285, 104862 (2021)
Deyhim, P.: Best practices for amazon EMR. Technical report, Amazon Web Services Inc. (2013)
Francis, N., et al.: Cypher: an evolving query language for property graphs. In: Proceedings of the 2018 International Conference on Management of Data, pp. 1433–1445 (2018)
Gabielkov, M., Legout, A.: The complete picture of the twitter social graph. In: Proceedings of the 2012 ACM Conference on CoNEXT Student Workshop, pp. 19–20 (2012)
Guia, J., Soares, V.G., Bernardino, J.: Graph databases: Neo4j analysis. In: ICEIS (1), pp. 351–356 (2017)
Iyer, A.P., Li, L.E., Das, T., Stoica, I.: Time-evolving graph processing at scale. In: Proceedings of the Fourth International Workshop on Graph Data Management Experiences and Systems, pp. 1–6 (2016)
Kaepke, M., Zukunft, O.: A comparative evaluation of big data frameworks for graph processing. In: 2018 4th International Conference on Big Data Innovations and Applications (Innovate-Data), pp. 30–37. IEEE (2018)
Kang, U., Tong, H., Sun, J., Lin, C.Y., Faloutsos, C.: GBASE: an efficient analysis platform for large graphs. VLDB J. 21(5), 637–650 (2012)
Katsifodimos, A., Schelter, S.: Apache Flink: stream analytics at scale. In: 2016 IEEE International Conference on Cloud Engineering Workshop, IC2E Workshops, Berlin, Germany, 4–8 April 2016, p. 193. IEEE Computer Society (2016). https://doi.org/10.1109/IC2EW.2016.56
Ko, J., Kook, Y., Shin, K.: Incremental lossless graph summarization. In: Proceedings of the 26th ACM SIGKDD International Conference on Knowledge Discovery & Data Mining, pp. 317–327 (2020)
Kyrola, A., Blelloch, G., Guestrin, C.: GraphChi: large-scale graph computation on just a \(\{\)PC\(\}\). In: Presented as Part of the 10th \(\{\)USENIX\(\}\) Symposium on Operating Systems Design and Implementation (\(\{\)OSDI\(\}\) 2012), pp. 31–46 (2012)
Leskovec, J., Krevl, A.: SNAP Datasets: Stanford large network dataset collection. http://snap.stanford.edu/data, June 2014
Low, Y., Gonzalez, J.E., Kyrola, A., Bickson, D., Guestrin, C.E., Hellerstein, J.: GraphLab: a new framework for parallel machine learning. arXiv preprint arXiv:1408.2041 (2014)
Maass, S., Min, C., Kashyap, S., Kang, W., Kumar, M., Kim, T.: Mosaic: processing a trillion-edge graph on a single machine. In: Proceedings of the Twelfth European Conference on Computer Systems, pp. 527–543, EuroSys 2017. ACM, New York, NY, USA (2017). https://doi.org/10.1145/3064176.3064191
Malewicz, G., et al.: Pregel: a system for large-scale graph processing. In: Proceedings of the 2010 ACM SIGMOD International Conference on Management of Data, SIGMOD 2010, pp. 135–146. ACM, New York, NY, USA (2010). https://doi.org/10.1145/1807167.1807184
Mariappan, M., Vora, K.: GraphBolt: dependency-driven synchronous processing of streaming graphs. In: Proceedings of the Fourteenth EuroSys Conference 2019, EuroSys 2019, pp. 25:1–25:16. ACM, New York, NY, USA (2019). https://doi.org/10.1145/3302424.3303974
Martínez-Bazan, N., Águila-Lorente, M.Á., Muntés-Mulero, V., Dominguez-Sal, D., Gómez-Villamor, S., Larriba-Pey, J.L.: Efficient graph management based on bitmap indices. In: Proceedings of the 16th International Database Engineering & Applications Sysmposium, pp. 110–119 (2012)
Munro, J.I., Nekrich, Y., Vitter, J.S.: Dynamic data structures for document collections and graphs. In: ACM Symposium on Principles of Database Systems (PODS), pp. 277–289 (2015)
Navarro, G.: Compact Data Structures: A Practical Approach. Cambridge University Press, Cambridge (2016)
Page, L., Brin, S., Motwani, R., Winograd, T.: The PageRank citation ranking: bringing order to the web. Technical report 1999-66, Stanford InfoLab (1999). http://ilpubs.stanford.edu:8090/422/
Palankar, M.R., Iamnitchi, A., Ripeanu, M., Garfinkel, S.: Amazon S3 for science grids: a viable solution? In: Proceedings of the 2008 International Workshop on Data-Aware Distributed Computing, pp. 55–64 (2008)
Perez, Y., et al.: Ringo: interactive graph analytics on big-memory machines. In: Proceedings of the 2015 ACM SIGMOD International Conference on Management of Data, SIGMOD 2015. ACM, New York, NY, USA (2015). https://doi.org/10.1145/2723372.2735369
ur Rehman, S., Nawaz, A., Ali, T., Amin, N.: g-Sum: a graph summarization approach for a single large social network (2021)
Rossi, R.A., Ahmed, N.K.: The network data repository with interactive graph analytics and visualization. In: AAAI (2015). http://networkrepository.com
Roy, A., Bindschaedler, L., Malicevic, J., Zwaenepoel, W.: Chaos: scale-out graph processing from secondary storage. In: Proceedings of the 25th Symposium on Operating Systems Principles, SOSP 2015, pp. 410–424. ACM, New York, NY, USA (2015). https://doi.org/10.1145/2815400.2815408
Roy, A., Mihailovic, I., Zwaenepoel, W.: X-Stream: edge-centric graph processing using streaming partitions. In: Proceedings of the Twenty-Fourth ACM Symposium on Operating Systems Principles, SOSP 2013, , pp. 472–488. ACM, New York, NY, USA (2013). https://doi.org/10.1145/2517349.2522740
Sakr, S., et al.: The future is big graphs: a community view on graph processing systems. Commun. ACM 64(9), 62–71 (2021). https://doi.org/10.1145/3434642
Salihoglu, S., Widom, J.: GPS: a graph processing system. In: Proceedings of the 25th International Conference on Scientific and Statistical Database Management, pp. 1–12 (2013)
Selimi, M., Cerdà Alabern, L., Freitag, F., Veiga, L., Sathiaseelan, A., Crowcroft, J.: A lightweight service placement approach for community network micro-clouds. J. Grid Comput. 17(1), 169–189 (2019)
Shvachko, K., Kuang, H., Radia, S., Chansler, R.: The Hadoop distributed file system. In: 2010 IEEE 26th Symposium on Mass Storage Systems and Technologies (MSST), pp. 1–10, May 2010. https://doi.org/10.1109/MSST.2010.5496972
Tian, Y., Balmin, A., Corsten, S.A., Tatikonda, S., McPherson, J.: From “Think Like a Vertex” to “Think Like a Graph”. Proc. VLDB Endow. 7(3), 193–204 (2013). https://doi.org/10.14778/2732232.2732238
Wheatman, B., Xu, H.: Packed compressed sparse row: a dynamic graph representation. In: 2018 IEEE High Performance Extreme Computing Conference (HPEC), pp. 1–7. IEEE (2018)
Xin, R.S., Gonzalez, J.E., Franklin, M.J., Stoica, I.: GraphX: a resilient distributed graph system on spark. In: First International Workshop on Graph Data Management Experiences and Systems, GRADES 2013, pp. 2:1–2:6. ACM, New York, NY, USA (2013). https://doi.org/10.1145/2484425.2484427
Zaharia, M., et al.: Apache Spark: a unified engine for big data processing. Commun. ACM 59(11), 56–65 (2016). https://doi.org/10.1145/2934664
Acknowledgements
This work was supported by national funds through FCT, Fundação para a Ciência e a Tecnologia, under projects UIDB/50021/2020 and PTDC/EEI-COM/30644/2017.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Morais, B., Coimbra, M.E., Veiga, L. (2022). PK-Graph: Partitioned \(k^2\)-Trees to Enable Compact and Dynamic Graphs in Spark GraphX. In: Sellami, M., Ceravolo, P., Reijers, H.A., Gaaloul, W., Panetto, H. (eds) Cooperative Information Systems. CoopIS 2022. Lecture Notes in Computer Science, vol 13591. Springer, Cham. https://doi.org/10.1007/978-3-031-17834-4_9
Download citation
DOI: https://doi.org/10.1007/978-3-031-17834-4_9
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-17833-7
Online ISBN: 978-3-031-17834-4
eBook Packages: Computer ScienceComputer Science (R0)