Abstract
Breast cancer is a highly diverse disease. With the state-of-the-art methods of molecular studies, novel subgroups of breast cancer can be revealed. The proper identification of subtypes is crucial for treatment choice. Hence, further investigation of breast cancer subtypes is promising in terms of therapy tailoring. We applied various machine learning approaches to the set of protein level measurements to detect subpopulations of breast cancer patients. Those methods involved various dimensionality reduction techniques combined with clustering. The outcomes of those approaches depended on the algorithms involved and on their parameters. Hence, we proposed the methodology to compare the results of clustering algorithms when the proper number of groups is unknown. The used metrices based on the effect size measurements and allowed for the selection of the best machine learning approach. The values of the proposed pooled d measure varied from 1.6847 for the worst method to 2.0568 for the best one. The highest value was obtained for the custom DiviK approach. Potentially, the metrices can also serve for the proteomic characterization of differences between subtypes and the identification of novel biomarkers.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Breast cancer is a diverse disease with highly heterogenous molecular characterization. Its subtypes vary in prognosis and therapy response. Proper diagnosis and subtype identification are crucial for treatment choice and planning.
In the early 2000s, Sørlie et al. [1] proposed a division of breast cancers into five intrinsic molecular subtypes: Luminal A, Luminal B, HER2-enriched, Basal, and Normal-like. This study led to the development of the PAM50 classifier [2], which allowed labeling a tumor with its intrinsic molecular subtype based on the gene expression microarray measurements. However, with the arrival of new technologies for molecular profiling, it became possible to further investigate, extend, and modify well-established breast cancer subtype categorization.
Machine learning provides a variety of methods for clustering and feature extraction or selection. Those techniques can be successfully applied for large genomic or proteomic datasets to investigate the heterogenic and diverse structure of breast cancer. However, results of subtypes identification often distinctly differ between algorithms in terms of both patient assignment to clusters and the final number of clusters detected. Moreover, the clustering outcome strongly depends on the parameters used. Thus, a method to compare and select different grouping approaches and parameters is needed. However, this task seems to be challenging as the method should deal with an unbalanced number of cases among subpopulations, an unknown target number of subtypes, a huge number of features in comparison with observations, and various dissimilarity degrees between resulting clusters. Some of the difficulties result also from the biological background and disease characterization: for instance, basal breast cancers are expected to be far more isolated from other tumors, while luminal family members should tend to group together and then further split into smaller subgroups.
In this study, we aim to test various approaches for clustering evaluation as well as to propose a metrics that would handle the challenges mentioned above.
2 Materials
Data used in this study are the result of the Reverse Phase Protein Arrays (RPPA) experiment. This dataset was created as a part of The Cancer Genome Atlas Breast Invasive Carcinoma (TCGA-BRCA) project [3]. All results were downloaded from the Genomic Data Commons (GDC) Data Portal in the normalized form. Samples used for the RPPA measurements were collected from primary tumors of females suffering from breast cancer. TCGA provided molecular subtype labels obtained with the PAM50 classifier based on the gene expression microarrays [4]. We excluded the samples with missing PAM50 etiquette. Due to the insufficient number of normal-like cases, this group was not considered. We also excluded proteins which levels were missing for some patients due to the requirements of algorithms used in the further analysis. The remaining records were corrected for the batch effect with the ComBat tool [5]. Finally, the dataset consisted of expression levels for 166 proteins and 407 patients. The summary of patients included in the study regarding their PAM50 label is presented in Table 1.
3 Methods
3.1 Subtype Detection
To investigate the dataset composition and identify subpopulations of breast cancer patients, we tested various combinations of clustering algorithms and feature extraction or selection methods. We used the HDBSCAN [6], graph-based Louvain community detection [7], and custom Divisive intelligent k-means (DiviK) [8] algorithms for grouping. Those methods were applied either to the levels of all available proteins or to the reduced feature space. Features were extracted with Principal Components Analysis (PCA) to select top components explaining 90% of the variance in the data and with Uniform Manifold Approximation and Projection (UMAP) [9] performed on the PCA-reduced dataset. For the feature selection, we used the Gaussian Mixture Model (GMM) [10] decomposition of log2-scaled variances of protein levels. All tested combinations were presented in Table 2.
In the HDBSCAN algorithm, there was a need to assign classes to the cases which were left unclassified. We tested several methods for this prediction, based on:
-
1.
HUMAP-C1: Proximity in 2-dimensional UMAP
-
2.
HUMAP-C2: Proximity in the dataset with all protein levels (complete)
-
3.
HUMAP-C3: Proximity in the set of top principal components explaining 90% of the variance.
3.2 Comparison of Clustering Approaches
To evaluate clustering results and investigate proteomic profiles of identified subpopulations, we compared levels of each protein between the clusters with a one-way ANOVA procedure followed by the Tukey-Kramer post hoc tests. ANOVA results served for calculations of η2 effect size for each protein. The higher the η2 value, the better the cluster separation. The η2 metrics considers all clusters together, so its values do not provide insight into whether all clusters are well-separated, or just some of them are highly isolated.
Moreover, we calculated the values of modification of Cohen’s d effect size to compare each obtained cluster versus all remaining ones considered jointly [11]. This measure was calculated based on the following equation:
Hence, for each protein, we obtained as many d values, as many subtypes were detected with a particular approach. As a result, for each method, we achieved a list of protein η2 values, and several lists of d values corresponding to subtypes.
To integrate η2 per method, we computed mean, median, and 3rd quartile of protein η2 values. To obtain a pooled value of d metrics per method, we proposed to assign the 3rd quartile of protein d absolute values to each subtype. Then, we projected the 3rd quartiles as a point in the k-dimensional space, where k was the number of subtypes detected. Finally, we calculated the pooled d value as a distance between the created point and the beginning of the coordinate system.
Moreover, we assessed the similarity between detected subtypes and PAM50 labels with the Dice coefficient. To further investigate the differences in outcomes of various method combinations, we referred the corresponding clusters to each other for the approaches with the lowest and the highest values of the pooled d metrics. We compared the values of d per protein for each subtype.
3.3 Biological Investigation
To biologically characterize each resulting cluster and evaluate the differences between the worst and the best approaches according to the pooled d metrics, we identified the proteins with significantly increased or decreased levels in each subtype compared to all remaining ones. Hence, we selected proteins with at least large or very large effect, so those with absolute values of d equal at least 0.8 or 1.2, respectively [11, 12]. We matched those proteins to the Kyoto Encyclopedia of Genes and Genomes (KEGG) database pathways in which they are involved [13] (accessed April 13, 2022).
4 Results
All HDBSCAN approaches without GMM filtration provided five clusters corresponding to Basal, HER2-enriched, Luminal A, and Luminal B subtypes. Luminal A cases were divided into two subgroups. All the remaining combinations of methods (HDBSCAN with GMM, Louvain, and DiviK algorithms) gave six clusters. The clusters in all combinations corresponded to Basal, HER2-enriched, Luminal B, and three Luminal A subpopulations.
The distributions of η2 values per method are presented in Fig. 1A. The exemplary distributions of absolute d values for the DiviK method with built-in variance-based GMM filtration per subtype are presented in Fig. 1B.
The distributions of metrices values with quartiles, median, and mean values marked with vertical lines. Panel A density plots showing distributions of η2 values per method. Panel B density plots showing distributions of absolute d values per subtype for the DiviK method with variance-based GMM filtration.
Obtained values of η2 quartiles and mean, pooled d, and Dice coefficient are presented in Table 3. Dice coefficient results are compared with pooled d and the 3rd quartile of η2 in Fig. 2.
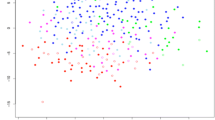
The results of the worst (HUMAP-C3) and the best (DiviK) approaches according to the pooled d values are also marked and compared to original PAM50 labels at the UMAP visualization in Fig. 3.
The primary difference between those two methods is that the DiviK algorithm provides an additional Luminal A3 cluster, containing cases included mainly in HUMAP-C3 Luminal B and Luminal A1 subtypes.
Those two contrasting approaches are further compared in Fig. 4. The protein values of d are referred to each other for corresponding Luminal subtypes: A1 versus A1, A2 versus A2, B versus B (respectively: Panels A, B, and C). Moreover, we compared the HUMAP-C3 Luminal B subtype with an additional Luminal A3 subtype given by DiviK (Panel D).
Total numbers of proteins with significantly higher or lower level for a certain subtype (with at least large or very large effects) are presented in Table 4 per subtype for the worst and the best approach. This table also contains the numbers of corresponding KEGG pathways.
UMAP visualization with results of two clustering approaches referred to the original PAM50 subtype labels. Panel A corresponds to the worst approach according to the pooled d values (HDBSCAN algorithm with the proximity in the set of top principal components explaining 90% of the variance for prediction, preceded by UMAP dimension reduction - HUMAP-C3). Panel B corresponds to the best approach according to the pooled d values (DiviK algorithm with variance-based GMM filtration).
Protein d values for the best (DiviK algorithm with variance-based GMM filtration) and the worst (HDBSCAN algorithm with the proximity in the set of top principal components explaining 90% of the variance for prediction, preceded by UMAP dimension reduction - HUMAP-C3) approach according to the pooled d metrics. Comparison of d values for the corresponding: Luminal A1 subtypes (Panel A), Luminal A2 subtypes (Panel B), Luminal B subtypes (Panel C), and DiviK Luminal A3 versus HUMAP-C3 Luminal B subtypes (Panel D). Dashed lines mark the threshold values for the large effect size, equal to −0.8 and 0.8 [11]. Values for proteins with small or medium effect according to both approaches are marked in grey.
5 Discussion
Obtained results suggest the dataset should be divided into five or six clusters, with one cluster corresponding to each of the Basal, HER2-enriched, and Luminal B subtypes, and two or three subgroups for Luminal A cases.
Based on the η2 and d distributions we concluded that the 3rd quartile is an appropriate representation of metrics values for all proteins. It sufficiently reflects the impact of proteins which expression levels significantly vary between clusters. Still, it remains resistant to outliers.
The DiviK method obtained maximal values of all metrices based on η2 and d. However, in terms of Dice similarity coefficients, all methods that gave six clusters performed better. However, the aim was not to maximize the similarity to the original PAM50 labels but to obtain as distant clusters as possible. All effect size metrices were higher when six clusters were obtained instead of five. GMM filtration improved the values of the 3rd quartile of η2 for both HDBSCAN and Louvain algorithms and pooled d for HDBSCAN. This can be especially noticed for the 3rd quartile of η2 in Fig. 2B, in which results of the Louvain approach with and without filtration are more separated. Hence, it is beneficial to compare the pooled d metrics with other criteria, including the Dice similarity index.
The methods with the highest (DiviK algorithm) and the lowest (HUMAP-C3) values of the pooled d metrics differ mainly regarding Luminal cases handling. HUMAP-C3 gave only two Luminal A subgroups and one bigger Luminal B subtype. DiviK, on the other hand, distinguished one more Luminal A subgroup that consists of patients clustered as Luminal A1 or B by the HUMAP-C3 approach. Moreover, the HER2-enriched subtype is more numerous for the DiviK algorithm, as it also contains a part of patients grouped as Luminal B with the HUMAP-C3 approach.
Division obtained with the DiviK algorithm greatly increased the number of proteins with an effect at least large (with decreased or increased levels in a subtype) for Luminal A1 and B subtypes. In the case of the Luminal A1 cluster, the number of proteins with at least a very large effect is also distinctly higher. Consequently, the number of associated KEGG pathways increased. Luminal A2 clusters do not vary much between the methods. However, the number of proteins and KEGG signaling pathways identified for the HER2-enriched subtype is smaller for the DiviK algorithm than for the HUMAP-C3 approach.
6 Conclusions
We performed breast cancer subtype identification with various combinations of machine learning methods for clustering and data dimensionality reduction. The outcomes were evaluated with several metrices, including the Dice coefficient and η2 effect size. We also proposed a custom effect size-based measure that represents the differences between each cluster and all remaining ones. The results of all metrices were consistent in terms of the best machine learning approach for breast cancer subpopulation detection. However, we believe it is beneficial to consider at least two different criteria for the comparison of various clustering algorithms and their parameters. Moreover, the metrices we used can serve for the characterization of proteomic profiles of breast cancer groups and the identification of novel biomarkers.
The approach which outperformed all the others was the custom Divisive intelligent k-means (DiviK) algorithm with the feature filtration based on the decomposition of the Gaussian Mixture Model of the log2-scaled protein level variance. For the other clustering methods, the GMM-based filtration also improved all or some metrices, depending on the algorithm.
We detected subgroups of the Luminal A breast cancer subtype: three with best performing approaches and two with the worst ones. We also identified the proteins with significantly increased or decreased levels in particular subgroups and related them to the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways. The selection of the additional third Luminal A subgroup increased the number of proteins with elevated or decreased levels characteristic for Luminal clusters as well as the number of the associated KEGG pathways, especially for the Luminal B subtype.
References
Sørlie, T., et al.: Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl. Acad. Sci. 98(19), 10869–10874 (2001)
Parker, J.S., et al.: Supervised risk predictor of breast cancer based on intrinsic subtypes. J. Clin. Oncol. 27(8), 1160 (2009)
Berger, A.C., et al.: A comprehensive pan-cancer molecular study of gynecologic and breast cancers. Cancer Cell 33(4), 690–705 (2018)
Koboldt, D.C.F.R., et al.: Comprehensive molecular portraits of human breast tumours. Nature 490(7418), 61–70 (2012)
Leek, J.T., et al.: sva: Surrogate Variable Analysis. R package version 3.38.0. (2020)
Campello, R.J.G.B., Moulavi, D., Sander, J.: Density-based clustering based on hierarchical density estimates. In: Pei, J., Tseng, V.S., Cao, L., Motoda, H., Xu, G. (eds.) PAKDD 2013. LNCS (LNAI), vol. 7819, pp. 160–172. Springer, Heidelberg (2013). https://doi.org/10.1007/978-3-642-37456-2_14
Blondel, V.D., Guillaume, J.L., Lambiotte, R., Lefebvre, E.: Fast unfolding of communities in large networks. J. Stat. Mech: Theory Exp. 2008(10), P10008 (2008)
Mrukwa, G., Polanska, J.: DiviK: divisive intelligent K-means for hands-free unsupervised clustering in biological big data. arXiv preprint arXiv:2009.10706 (2020)
McInnes, L., Healy, J., Melville, J.: Umap: uniform manifold approximation and projection for dimension reduction. arXiv preprint arXiv:1802.03426 (2018)
Marczyk, M., Jaksik, R., Polanski, A., Polanska, J.: Gamred—Adaptive filtering of high-throughput biological data. IEEE/ACM Trans. Comput. Biol. Bioinf. 17(1), 149–157 (2018)
Cohen, J.: Statistical Power Analysis for the Behavioral Sciences. Lawrence Earlbaum Associates, New York (1988)
Sawilowsky, S.S.: New effect size rules of thumb. J. Mod. Appl. Stat. Methods 8(2), 26 (2009)
Kanehisa, M., Furumichi, M., Tanabe, M., Sato, Y., Morishima, K.: KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 45(D1), D353–D361 (2017)
Acknowledgment
This study is supported by European Social Fund grant no. POWR.03.02.00-00-I029 [JT] and Silesian University of Technology grant no. 02/070/BK_22/0033 for Support and Development of Research Potential [JP]. The results published here are based upon data generated by the TCGA Research Network: https://www.cancer.gov/tcga.
Author information
Authors and Affiliations
Corresponding authors
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 Springer Nature Switzerland AG
About this paper
Cite this paper
Tobiasz, J., Polanska, J. (2022). How to Compare Various Clustering Outcomes? Metrices to Investigate Breast Cancer Patient Subpopulations Based on Proteomic Profiles. In: Rojas, I., Valenzuela, O., Rojas, F., Herrera, L.J., Ortuño, F. (eds) Bioinformatics and Biomedical Engineering. IWBBIO 2022. Lecture Notes in Computer Science(), vol 13347. Springer, Cham. https://doi.org/10.1007/978-3-031-07802-6_26
Download citation
DOI: https://doi.org/10.1007/978-3-031-07802-6_26
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-07801-9
Online ISBN: 978-3-031-07802-6
eBook Packages: Computer ScienceComputer Science (R0)