Abstract
Metagenomics sequencing enables the direct study of microbial communities revealing important information such as taxonomy and relative abundance of species. Metagenomics binning facilitates the separation of these genetic materials into different taxonomic groups. Moving from second-generation sequencing to third-generation sequencing techniques enables the binning of reads before assembly thanks to the increased read lengths. The limited number of long-read binning tools that exist, still suffer from unreliable coverage estimation for individual long reads and face challenges in recovering low-abundance species. In this paper, we present a novel binning approach to bin long reads using the read-overlap graph. The read-overlap graph (1) enables a fast and reliable estimation of the coverage of individual long reads; (2) allows to incorporate the overlapping information between reads into the binning process; (3) facilitates a more uniform sampling of long reads across species of varying abundances. Experimental results show that our new binning approach produces better binning results of long reads and results in better assemblies especially for recovering low abundant species. The source code and a functional Google Colab Notebook are available at https://www.github.com/anuradhawick/oblr.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Recent advancements in sequencing technologies have accelerated microbiome research significantly. Broadly, metagenomics analysis supports the direct study of microbial genetic material from the host environments [3, 32]. One fundamental problem that dominates across a wide range of research is the identification and characterization of microbial genetic material. Metagenomics binning specifically determines the species present in a given sample and further supports the downstream functional analysis of identified microorganisms. There exist two main paradigms for metagenomics binning; (1) reference-based binning (e.g. Kraken2 [36], Centrifuge [11], MeganLR [7], Kaiju [20], etc.) and (2) reference-free binning (e.g. MaxBin 2 [37], MetaBAT 2 [10], VAMB [25], etc.). Reference-free approaches are preferred when unknown species are present or the reference databases are incomplete. Typically, short reads from the second-generation sequencing technologies (e.g. Illumina, etc.) are assembled into much longer contigs to be binned as longer contigs usually carry more pronounced genomic signals, e.g., the coverage and composition information of contigs. The coverage of an assembled contig is estimated by the aligned reads on this contig whereas the composition information is computed from the normalized ologonucleotide frequencies.
Long-read technologies from the third-generation sequencing is continuously gaining popularity [17], especially with the recent introduction of PacBio HiFi and Nanopore Q20+ technologies. As long reads are getting similar to contigs (assembled from short reads) in terms of length and accuracy, it is worth investigating whether long reads themselves can be binned directly before assembly. Note that the contigs binning tools cannot be directly applied to bin accurate long reads due to the absence of a coverage value for each read. MetaBCC-LR [35] and LRBinner [34] are two recent attempts to bin long reads in a reference-free manner. MetaBCC-LR and LRBinner both use k-mer coverage histograms for coverage and trinucleotide frequency vectors for composition. While MetaBCC-LR uses coverage and composition features in two subsequent steps, LRBinner combine coverage and composition features via an auto-encoder. Although these two k-mer based approaches show some promising results in binning long reads, they are highly likely to suffer from unreliable coverage estimation for individual long reads and poor sensitivity for low-abundance species due to imbalance clusters (refer to Fig. 3).
In this paper, we propose a novel binning approach (OBLR) to bin long reads using the read-overlap graph. In contrast with MetaBCC-LR and LRBinner, we adopt a novel coverage estimation strategy and a sampling strategy to form uniform clusters assisted by the read-overlap graph. We show that read-overlap graph assists in better estimation of read coverage and enables us to sample reads more uniformly across species with varying coverages. Moreover, the connectivity information in the read-overlap graph facilitates more accurate binning via inductive learning. Experimental results show that our new binning approach produces better binning results of long reads while reducing the extensive resources otherwise required for the assembly process.
2 Methods
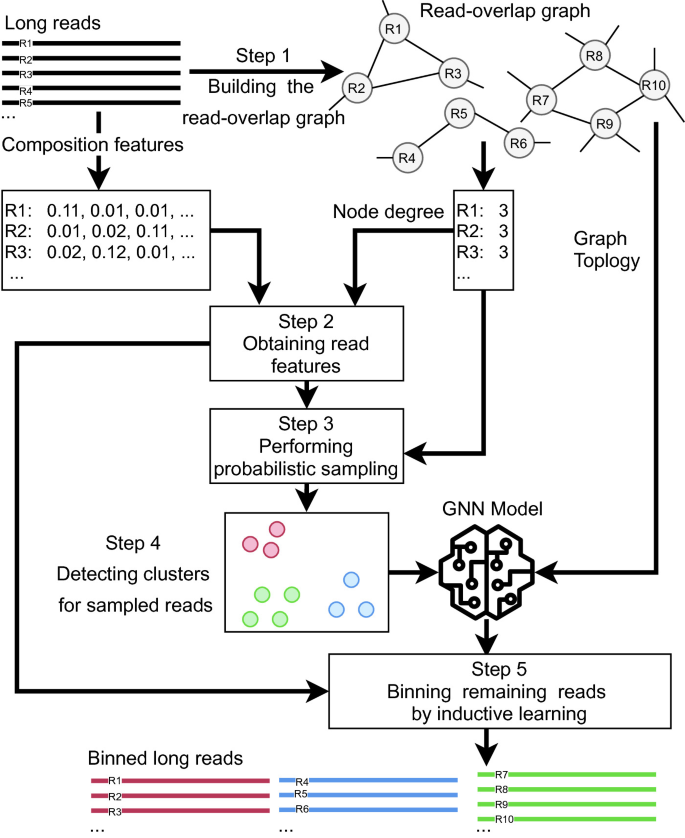
Our pipeline consists of 5 steps performing the tasks, (1) building the read-overlap graph, (2) obtaining read features, (3) performing probabilistic sampling, (4) detecting clusters for sampled reads and (5) binning remaining reads by inductive learning. Figure 1 illustrates the overall pipeline of OBLR. The following sections explain each step in detail.
2.1 Step 1: Constructing Read-Overlap Graph
As the first step of the pipeline, we construct the read-overlap graph. The read-overlap graph is introduced to utilize the overlapping information between raw reads. Earlier works have demonstrated that the topology of read-overlap graph can help binning short reads [2] as well as distinguishing between genomes at the strain level [1]. As for long reads, two reads are overlapping (connected by an edge in the read-overlap graph) if and only if their overlapping length is at least \(L_{overlap}\) and the overhang length is at most \(L_{overhang}\) (computed according to [13]). Note that overhang refers to the region on the sequence that lies along with the aligned sequence, however, does not have matching bases to meet overlap criteria. In our pipeline, we use k-mer bin map (kbm2) program to compute the approximate overlaps between reads. We use the empirically determined values, \(L_{overlap}=2560\) and \(L_{overhang}=512\) as overlap selection criteria in default setting. Note that kbm2 is a sub-routine of the recent assembler wtdbg2 and is extremely fast to detect overlapping reads using k-mer bins without performing pairwise alignment [28]. In the read-overlap graph, each node \(R_i\) represents a read while each edge \((R_i, R_j)\) indicates that \(R_i\) and \(R_j\) are overlapping. We also define \(D(R_i)\) as the degree of \(R_i\) in this read-overlap graph.
2.2 Step 2: Obtaining Read Features
In our pipeline, we intend to derive read features that incorporate both composition and coverage information of long reads.
The composition information of long reads can be computed as their oligonucleotide frequencies which are shown to be conserved within a given species while being reasonably distinct between species [30, 37]. More specifically, we compute a tetra-nucleotide frequency vector for each long read \(R_i\), i.e., \(X(R_i) \in \mathbb {R}^{136}\) as there are 136 distinct tetra-mers when combining reverse complements. This vector is used as the composition feature in our pipeline.
The coverage information of long reads usually refers to the coverage of underlying genomes from which the long reads are drawn. This is also important in metagenomics binning as long reads from the same species tend to have similar coverages [25, 35]. While such coverage information is usually available for contigs assembled from short reads (as a byproduct of assembly), long reads do not come with their coverage information. However, a read from a high-coverage genome is likely to have more overlaps compared to that from a low-coverage genome. Therefore, it is a natural choice to use the node degree in the read-overlap graph to estimate the coverage of the corresponding read. This choice is supported by Fig. 2 which shows a clear correlation between the node degree in the read-overlap graph and the coverage information of the corresponding read.
In summary, we combine both tetra-nucleotide frequency vector \(X(R_i)\) (for composition) and the node-degree information \(D(R_i)\) (for coverage) to derive the read feature vector as \(XD(R_i)\) = \(X(R_i) \times max(1, lg(D(R_i)))\) for each long read \(R_i\). Note that the max( ) and lg( ) are introduced to dampen rigorous fluctuations in coverage, especially for low-coverage genomes. Henceforth, \(XD(R_i)\) refers to the read features using degree and composition for read \(R_i\).
2.3 Step 3: Performing Probabilistic Sampling
Clustering entire dataset at once can lead to the well-known class-imbalance problem [9] because metagenomics samples consist of species with varying coverages, i.e., imbalance clusters. However, probabilistic down sampling can effectively address this problem [16]. In order to perform such under sampling, we recall the degree information of nodes and use the Eq. 1 to compute the relative probability of sampling \(R_i\). Note that \(D(R_i) = 0\) when \(R_i\) is an isolated node and helps OBLR to discard chimeric reads. The effect of down sampling is illustrated in Fig. 3. It is evident that clusters after down sampling are of similar sizes and with less isolated points.
2.4 Step 4: Detecting Clusters for Sampled Reads
We use UMAP [19] to project the sampled reads into lower dimensions and HDBSCAN [18] then is applied to detect clusters for sampled reads. Note than UMAP is a dimensionality reduction technique that is fast and scalable. HDBSCAN is a variant of DBSCAN, however it is capable of determining clusters without fixed parameters. Thus, HDBSCAN is more robust in scenarios where the cluster densities can vary significantly. To accommodate long-read datasets with different sizes, 25,000, 50,000, 100,000, 200,000 and 400,000 are used as the sampled number of reads to detect clusters. For each instance, the Silhouette score [27] is computed. We use the sample size with the highest Silhouette score as the chosen sample size for the dataset. This enables us to determine the size to sample from a given dataset in an unsupervised manner. At the end of this step; we have a sample of reads with their bin labels i.e., cluster labels.
2.5 Step 5: Binning Remaining Reads by Inductive Learning
As the read-overlap graphs may contain millions of nodes, the classic label propagation approaches face scalability issues to bin the remaining reads [15]. Therefore, OBLR employs GraphSAGE [6] to bin the remaining reads into the identified clusters in the previous step. GraphSAGE is a Graph Neural Network (GNN) architecture and has been designed to perform inductive learning using large-scale graphs [6]. GraphSAGE can be represented as a layer in a GNN that aggregates the neighborhood features to represent the features of a node itself. Formally, the l-th layer can be formulated according to Eqs. 2 and 3 [38].
where \(h_{i}^{(l)}\) is the feature vector of node \(R_i\) at layer l. Note that \(h_{v}^{(0)}\) \(=\) \(XD(R_i)\) and \(\mathcal {N}(R_i)\) represent neighbors of node \(R_i\). While GraphSAGE supports arbitrary aggregate functions, we choose the Mean( ) as the aggregation operator to be tolerant towards noise and false connections in the read-overlap graph due to repeats. Furthermore, we use Concatenation( ) as the layer-wise feature combination strategy to retain features from both the node itself and its neighborhood.
We use two GraphSAGE layers followed by a fully-connected layer with K outputs, where K is the number of bins estimated in Step 4. Two GraphSAGE layers use LeakyRELU activation while the final layer uses log softmax activation resulting in the output probabilities for K bins. We train the GNN using 200 epochs using sampled reads binned in Step 4 and use negative log-likelihood (cross-entropy) as the loss function. During the training phase, we use a neighbor sampler that samples up to 20 neighbours in GraphSAGE layers. We use Adam optimizer for gradient descent. The trained GNN on sampled reads provides assignment probabilities for remaining reads to the bins derived in Step 4. The remaining reads are thus assigned to the bins with the highest probabilities.
3 Experimental Setup
We evaluate our pipeline using two simulated and three publicly available real datasets. Note that, all the experiments are conducted and evaluated using the same set of parameters. Detailed information about the datasets are available in Appendix A.
3.1 Simulated Datasets
We simulate four PacBio datasets using SimLoRD [29] containing 8, 20, 50 and 100 [35] species with average read length 5,000 bp and the default PacBio error profiles in SimLoRD (insertion = 0.11, deletion = 0.04 and substitution = 0.01). These two datasets are named as Sim-8, Sim-20, Sim-50 and Sim-100 respectively.
3.2 Real Datasets
Three real datasets with known reference genomes are also used to evaluate read-level binning performance. Long reads from these datasets were aligned to references using Minimap2 [14] to obtain the ground truth.
-
ZymoEVEN: Oxford Nanopore reads sequenced from GridION device from NCBI Accession Number ERR3152364 [24]. The dataset consists of 10 species with average read length 4,119.
-
SRR9202034: PacBio CCS reads of the ATCC MSA-1003 Mock Microbial Community from NCBI BioProject number PRJNA546278 Accession Number SRR9202034. The dataset contains 15 species with more than 0.1% relative abundance and average read length 8,263.
-
SRX9569057: PacBio-HiFi reads of the NCBI BioSample SAMN16885726 Accession Number SRX9569057. This dataset contains 13 species (18 strains) with more than 0.1% relative abundance and average read length 9,093.
Table 1 summarizes the information about the datasets including the number of species, dataset sizes and the number of nodes (reads) and edges of the read-overlap graphs.
3.3 Baselines and Evaluation Criteria
We benchmark our approach against two recent long-read binners, MetaBCC-LR [35] and LRBiner [34]. For a binning result of K bins against the ground truth of S species, we populate a matrix a of size \(K\times S\), where \(a_{ks}\) denotes the number of reads assigned to bin k from the species s of the sample. The binning results are evaluated using precision Eq. 4, recall Eq. 5 and F1-score Eq. 6 [37]. We used AMBER [21] to compute completeness and purity, genome fractions using MetaQUAST [22], assembly CPU-time and memory usage to evaluate performance.
4 Results and Discussion
We evaluate binning results at the read-level using precision, recall, F1 score, the number of bins produced. We further evaluate each bin using per-bin F1-scores using AMBER [21]. Moreover, we conducted a quantitative evaluation of assemblies using MetaQuast [22] before and after binning.
4.1 Binning Results
We benchmark OBLR against MetaBCC-LR [35] and LRBiner [34] which are two recent long-read binning tools as presented in Table 2. We observed that OBLR results in the highest F1-scores across all the datasets with the overall best performance. OBLR also produces more accurate estimates of the number of bins in most datasets.
These observations are further supported by the AMBER [21] evaluation summarized in Fig. 4 and 5 where OBLR produces the best per bin F1-scores among all three long-read binners. Per bin F1-score evaluates each bin separately using their purity and completeness while penalizing false bin splits and merged bins. Note that MetaBCC-LR and LRBinner suffer from fragmented binning results (i.e., overestimated number of bins) because bins with low completeness are significantly penalized by AMBER. This observation is explained further in Appendix B.
Per bin F1-score comparison between MetaBCC-LR, LRBinner and OBLR computed by AMBER [21] for simulated datasets.
Per bin F1-score comparison between MetaBCC-LR, LRBinner and OBLR computed by AMBER [21] for real datasets.
4.2 Assembly Results
We perform assembly using the long-read metagenomic assembler metaFlye [12]. Table 3 demonstrates the genome fraction and resource usage for assembling raw reads (termed Raw) and assembling reads binned by OBLR (termed Binned), respectively. Gains in genome fraction are relatively higher for simulated datasets with more species (e.g., Sim-50 and Sim-100) while the most significant benefit being the drastic reduction of the resources consumed. Binning long reads from real datasets before assembly in general maintains the genome fraction (94.13% to 94.27% in SRX9569057, 86.51% to 86.67% in ZymoEVEN and 90.30% to 90.39% in SRR9202034) while significantly saving on the resources. The saving on peak memory varies from 40% to 80%. However, CPU hours consumed remains comparable due to re-indexing of reads and the near-linear time complexity of the assembly process.
5 Implementation
We use multiple optimizations in the OBLR pipeline. In Step 1, we chunk the reads in to blocks of 250,000 reads, and each block of reads is mapped with entire dataset resulting in several mapping files. Finally, the mapping files are merged into a single file containing edges between reads and degree. For Step 2 we use seq2vec [33] to compute the composition vectors. In Steps 3 and 4 we use Rapids.AI [31] GPU libraries [26] (on NVIDIA RTX 3090 with 24 GB VRAM) for UMAP and HDBSCAN. Finally, we use PyTorch Geometric [5] in Step 5 for GraphSAGE. We conducted our experiments on an Ubuntu 20.04.3 LTS system running on AMD Ryzen 9 5950X with 16-core Processor with 32 threads and 128 GB of RAM.
Table 4 tabulates the resource usage of each step in OBLR pipeline. In addition we also present the resource utilization of kbm2 [28]. Note that the GPU memory utilization is fixed as we perform clustering and silhouette scores only upto 400,000 data points. However, resource usage for other steps vary depending on the read size distribution on each read block and the number of data points.
6 Conclusion
We presented a novel long-read binner, OBLR, which utilizes the read-overlap graph to bin long reads in metagenomic samples. Recent advances such as the k-mer bins mapping (kbm2) [28] enables extremely fast detection of overlapping reads and construction of the read-overlap graph before assembly. OBLR thus makes use of the read-overlap graph to improve the state-of-the-art long-read binning approaches. The read-overlap graph not only helps to estimate the coverage of single long read, but also allow us to sample the long-reads more uniformly across species of varying abundances. The connectivity information in the read-overlap graph further incorporates the overlapping information between reads into the binning process as overlapped reads are more likely to be in the same species. As a result, OBLR demonstrated promising results in producing more accurate bins for long-read datasets and has the potential to improve on metagenomics assemblies in terms of computing resources and genome fraction, especially for low-abundance species. In the future, we plan to investigate how OBLR can be adapted to take the advantage of the high-accuracy long reads including PacBio HiFi and Nanopore Q20+, how to incorporate the binning process into long-read metagenomics assemblies [4, 12], and how to connect metagenomics binning to comparative genomics and phylogenetic studies [8, 23].
References
Baaijens, J.A., El Aabidine, A.Z., Rivals, E., Schönhuth, A.: De novo assembly of viral quasispecies using overlap graphs. Genome Res. 27(5), 835–848 (2017)
Balvert, M., Luo, X., Hauptfeld, E., Schönhuth, A., Dutilh, B.E.: Ogre: overlap graph-based metagenomic read clustering. Bioinformatics 37(7), 905–912 (2021)
Chen, K., Pachter, L.: Bioinformatics for whole-genome shotgun sequencing of microbial communities. PLOS Comput. Biol. 1(2) (2005)
Feng, X., Cheng, H., Portik, D., Li, H.: Metagenome assembly of high-fidelity long reads with hifiasm-meta. arXiv:2110.08457 (2021)
Fey, M., Lenssen, J.E.: Fast graph representation learning with PyTorch Geometric. In: ICLR Workshop on Representation Learning on Graphs and Manifolds (2019)
Hamilton, W.L., Ying, R., Leskovec, J.: Inductive representation learning on large graphs. In: Proceedings of the 31st International Conference on Neural Information Processing Systems, pp. 1025–1035 (2017)
Huson, D.H., et al.: Megan-LR: new algorithms allow accurate binning and easy interactive exploration of metagenomic long reads and contigs. Biol. Direct 13(1), 1–17 (2018)
Huson, D.H., Richter, D.C., Mitra, S., Auch, A.F., Schuster, S.C.: Methods for comparative metagenomics. BMC Bioinf. 10(1), 1–10 (2009)
Japkowicz, N., Stephen, S.: The class imbalance problem: a systematic study. Intell. Data Analysis 6(5), 429–449 (2002)
Kang, D.D., et a.: Metabat 2: an adaptive binning algorithm for robust and efficient genome reconstruction from metagenome assemblies. PeerJ 7, e7359 (2019)
Kim, D., Song, L., Breitwieser, F.P., Salzberg, S.L.: Centrifuge: rapid and sensitive classification of metagenomic sequences. Genome Res. 26(12), 1721–1729 (2016)
Kolmogorov, M., et al.: metaflye: scalable long-read metagenome assembly using repeat graphs. Nat. Methods 17(11), 1103–1110 (2020)
Li, H.: Minimap and miniasm: fast mapping and de novo assembly for noisy long sequences. Bioinformatics 32(14), 2103–2110 (2016)
Li, H.: Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34(18), 3094–3100 (2018)
Liang, D.M., Li, Y.F.: Lightweight label propagation for large-scale network data. In: IJCAI, pp. 3421–3427 (2018)
Liu, X.Y., Wu, J., Zhou, Z.H.: Exploratory undersampling for class-imbalance learning. IEEE Trans. Syst. Man Cybern. Part B (Cybernetics) 39(2), 539–550 (2009). https://doi.org/10.1109/TSMCB.2008.2007853
Logsdon, G.A., Vollger, M.R., Eichler, E.E.: Long-read human genome sequencing and its applications. Nat. Rev. Genet. 21(10), 597–614 (2020)
McInnes, L., Healy, J., Astels, S.: HDBSCAN: hierarchical density based clustering. J. Open Source Softw. 2(11), 205, e7359 (2017)
McInnes, L., Healy, J., Melville, J.: Umap: Uniform manifold approximation and projection for dimension reduction (2020)
Menzel, P., Ng, K.L., Krogh, A.: Fast and sensitive taxonomic classification for metagenomics with Kaiju. Nat. Commun. 7, 11257 (2016)
Meyer, F., et al.: Amber: assessment of metagenome binners. Gigascience 7(6), giy069 (2018)
Mikheenko, A., Saveliev, V., Gurevich, A.: Metaquast: evaluation of metagenome assemblies. Bioinformatics 32(7), 1088–1090 (2016)
Nayfach, S., Pollard, K.S.: Toward accurate and quantitative comparative metagenomics. Cell 166(5), 1103–1116 (2016)
Nicholls, S.M., Quick, J.C., Tang, S., Loman, N.J.: Ultra-deep, long-read nanopore sequencing of mock microbial community standards. Gigascience 8(5), giz043 (2019)
Nissen, J.N., et al.: Improved metagenome binning and assembly using deep variational autoencoders. Nat. Biotechnol. 39(5), 555–560 (2021)
Nolet, C.J., et al.: Bringing UMAP closer to the speed of light with GPU acceleration (2020)
Rousseeuw, P.J.: Silhouettes: a graphical aid to the interpretation and validation of cluster analysis. J. Comput. Appl. Math. 20, 53–65, e7359 (1987)
Ruan, J., Li, H.: Fast and accurate long-read assembly with WTDBG2. Nat. Methods 17(2), 155–158, e7359 (2020)
Stöcker, B.K., Köster, J., Rahmann, S.: Simlord: simulation of long read data. Bioinformatics 32(17), 2704–2706 (2016)
Strous, M., Kraft, B., Bisdorf, R., Tegetmeyer, H.: The binning of metagenomic contigs for microbial physiology of mixed cultures. Front. Microbiol. 3, 410 (2012)
Team, R.D.: RAPIDS: Collection of Libraries for End to End GPU Data Science (2018). https://rapids.ai
Tyson, G.W., et al.: Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature 428(6978), 37–43 (2004)
Wickramarachchi, A.: anuradhawick/seq2vec: release v1.0 (2021). https://doi.org/10.5281/zenodo.5515743, https://doi.org/10.5281/zenodo.5515743
Wickramarachchi, A., Lin, Y.: Lrbinner: binning long reads in metagenomics datasets. In: 21st International Workshop on Algorithms in Bioinformatics (WABI 2021). Schloss Dagstuhl-Leibniz-Zentrum für Informatik (2021)
Wickramarachchi, A., Mallawaarachchi, V., Rajan, V., Lin, Y.: Metabcc-LR: meta genomics binning by coverage and composition for long reads. Bioinformatics 36(Supplement_1), i3–i11 (2020)
Wood, D.E., Lu, J., Langmead, B.: Improved metagenomic analysis with kraken 2. Genome Biol. 20(1), 1–13 (2019)
Wu, Y.W., Simmons, B.A., Singer, S.W.: Maxbin 2.0: an automated binning algorithm to recover genomes from multiple metagenomic datasets. Bioinformatics 32(4), 605–607 (2016)
Xu, K., Hu, W., Leskovec, J., Jegelka, S.: How powerful are graph neural networks? arXiv:1810.00826 (2018)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Appendices
A Dataset Information
Tables 5 and 7 demonstrate the simulated and real dataset information respectively. Note that Table 5 and 6 tabulate the coverages used for simulation using SimLoRD [29] while Table 7 indicate abundances from the dataset sources.
B Interpretation of AMBER Per-bin F1-Score
The binning evaluations are presented using Precision, Recall and F1 score. Furthermore, stricter evaluations are presented using AMBER [21] for MetaBCC-LR, LRBinner and OBLR. This section explains the evaluation metrics in detail and discuss as to why AMBER evaluations are poor in some cases where number of bins predicted is further away from the actual number of species in the dataset. Note that the bin assignment matrix a can be presented as \(M\times N\), illustrated in Table 8. Note that \(N=5\) and \(M=7\).
Recall is computed for each species, by taking the largest assignment to a bin. Precision is computed per bin taking the largest assignment of the bin to a given species. In contrast, AMBER uses purity and completeness to compute the per-bin F1 score using the following equations, for each bin b.
The true positives are computed using the majority species in a given bin. Because of this, if a bin appears as a result of a false bin split (1% reads), the Completeness of the bin will be very low as the majority of it (approximately 1%) according to AMBER evaluation. In comparison, the recall of the species using Eq. 5 will report 99% since 99% of the reads are in a single bin despite having the false bin split. Similarly, the false split of the bin will report a greater precision as long as the bin has no other species mixed according to Eq. 4. Consider the following running example.
Example 1. Suppose Species 1 has \(a_{11}=99\) and \(a_{16}=1\) with rest of the row having no reads and Bin 1 and 6 has no reads from another species. Purity in this case will be 100% for both bins 1 and 6 while completeness will be 99% and 1% respectively. F1-score will be 99.5% and 1.98% with average being very low at 50.7%. Recall will be 99% for Species 1 with 100% precision on both bins 1 and 6 since there are no impurities in each bin, thus, F1-score is 99.5% for each bin.
Example 2. Suppose Bin 2 has \(a_{22}=100\) and \(a_{32}=20\), with two species 2 and 3, with no other contaminants and species 2 and 3 are fully contained in the bin. Now, the purity of the bin is 83.33% and completeness is 83.33%, hence, F1-score is 83.33%. Recall for species 2 and 3 will be 100% since it is not broken into multiple bins. However, the precision for Bin 2 will be 83.33%, hence a F1 score of 90.91%.
This means, AMBER penalize whenever a species is broken into pieces across bins while not significantly penalizing bin mergers between large bins and smaller bins. This is because, dominant species in the bin will determine the purity and completeness.
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Wickramarachchi, A., Lin, Y. (2022). Metagenomics Binning of Long Reads Using Read-Overlap Graphs. In: Jin, L., Durand, D. (eds) Comparative Genomics. RECOMB-CG 2022. Lecture Notes in Computer Science(), vol 13234. Springer, Cham. https://doi.org/10.1007/978-3-031-06220-9_15
Download citation
DOI: https://doi.org/10.1007/978-3-031-06220-9_15
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-06219-3
Online ISBN: 978-3-031-06220-9
eBook Packages: Computer ScienceComputer Science (R0)