Abstract
Cancer cell invasion and progression to metastasis have been extensively studied. There is not one all-inclusive model that encompasses the complete picture of the different conditions and pathways operating in human tumors. Application of gene expression signatures is one way of mining the complex tumor landscape and has been proposed to represent a robust method to reflect the many signaling systems.
This chapter gives an update on gene expression signature studies related to breast cancer progress, with particular focus on the supporting stroma. Several signature studies indicate that a combination of extracellular remodeling, activated vascular biology, immune responses, and metabolic reprogramming, in part adipocyte-related, takes place during breast cancer progression. Stromal alterations are likely to be exploited for novel biomarkers and companion treatment targets. Some basic methodological aspects and recent developments are highlighted.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- Breast cancer
- Tumor progress
- Clinical outcome
- Tumor microenvironment
- Gene expression signatures
- Tumor-associated stroma
- Cancer-associated fibroblasts
- Vascular biology signatures
- Immune-related signatures
- Cancer-associated adipocytes
-
Global gene expression data may better reflect the complexity of cancer biology as compared to the detection of single gene or protein alterations, potentially being a powerful resource for identifying markers of the complex biological processes taking place in cancer.
-
Interactions between vascular processes, immune responses, cancer-associated adipocytes, and extracellular remodeling appear to be critically important features of tumor subtypes and their associated outcomes, as reflected in composite signature biomarkers.
-
Identifying genes and proteins with known functions, differentially expressed between subgroups, may provide improved understanding of the biological differences between tumor phenotypes.
-
Gene networks analyses, by means of gene set enrichment analyses and mRNA data paralleling protein–protein interaction analyses, provide increased biological understanding of complex large-scale data.
Introduction
And above all, watch with glittering eyes the whole world around you because the greatest secrets are always hidden in the most unlikely places. Those who don’t believe in magic will never find it.—Roald Dahl (1916–1990)
Tumor cell invasion and metastasis are multistep processes that are detrimental to the organ in which they occur. The route to cancer dissemination is suggested by distinct steps; local infiltration, intravasation, and transport of cancer cells in the lymphatic or hematogenous systems, followed by extravasation of tumor cells from the vessels into the tissue parenchyma (or niche) of the new site where micrometastases may form and grow to macroscopic lesions [1, 2]. The English surgeon Stephen Paget postulated “the seed and soil hypothesis” in 1889, suggesting that tumor cells (denoted “seeds”) have affinity for specific tissue environments (denoted “soil”) in certain organs [3]. Literally, Paget sedded a hypothesis followed by many researchers studying cancer invasion and the metastatic process over the next century.
Before setting off on the invasion-metastasis cascade, it is crucial that tumor cells fulfill prerequisites such as the ability to detach and move from the original colony, with unlimited proliferative potential, and a capacity to evade from destruction [4]. The underlying effectors in the invasion-metastasis cascade are suggested to be classified as metastasis initiating , metastasis progressing and metastasis virulent [5]. Metastasis-initiating genes generate a supportive environment that facilitates tumor infiltration to surrounding tissue. Expression of such genes, in the epithelial cells or the microenvironmental compartments, may promote angiogenesis, vascular invasion, epithelial-mesenchymal transition (EMT), and evasion from immune destruction with important implications to the processes involved in cancer metastasis.
The microenvironment is regarded to play a crucial role both in embryonic organ development and in cancer invasion, two processes with several similar features [6]. Cellular and molecular interactions between the epithelial cells and the microenvironment and between elements within the microenvironment also take place in functional differentiation of the normal mammary tissue. Exploiting the normal microenvironment programs, by a form of “hacking” these pathways, is suggested as potential ways of promoting cancer invasion [7]. This may be reversely exploited when targeting the metastatic processes in the therapy setting, exemplified by a study on a xenograft model of breast cancer, identifying neutrophils within the lung microenvironment supporting metastatic initiation and as drivers of establishing lung metastases [8]. Thus, inhibiting the enzyme Alox5 abolished the pro-metastatic neutrophil activity in the lung microenvironment and reduced the occurrence and growth of lung metastases.
Genes supporting metastasis progression promote extravasation and survival of the cancer cells outside of their original environment [5]. Cancer cells that have entered the circulation may subsequently extravasate and infiltrate distant organs. When entering such new environment, cancer cells are required to adapt rapidly for colonization to occur, where the disseminated cancer cells reside in their new microenvironment and grow into macro-metastases. Specific cancer cell gene expression has been implicated to direct organ-specific tropism. One example is the expression of IL-11, which facilitates breast cancer metastases to the bone [9]. The establishment of a “receptive” environment at the future metastatic location before the colonization of tumor cells (the pre-metastatic niche) is suggested as a mechanistic model explaining metastatic organotropism [10]. Cancer-specific factors released from the primary tumor promote changes in the future metastatic microenvironment before the tumor cells arrive to this location. Also, bone marrow cells may migrate to the pre-metastatic niche in response to the systemically released factors, facilitating the environment for the cancer cells to “thrive” [11, 12].
The tumor microenvironment is increasingly focused in cancer research, both in pre-invasive lesions, primary tumors, pre-metastatic niches, as well as in the metastatic lesions. The tumor microenvironment components have been regarded as genetically more stable than the tumor cells. This is an important factor that renders the stromal components a strategic focus when searching targets for cancer therapy.
Since the discovery of cell signaling, researchers have debated how to best reflect alterations of pathways and levels of pathway activation in different model systems. One major trend in cancer research has been to undertake relatively simple approaches (e.g., measuring one protein or one specific mutation) when searching for markers of deregulated pathways as prognostic and predictive markers. A simplified approach to the complex and unstable cancer biology likely contributes to the lack of biomarker and treatment effects when translating the research findings to the clinical setting.
Global gene expression data may have a stronger potential to reflect the complexity of cancer biology as compared to the detection of single-gene alterations and may be a powerful platform for identifying markers for the complex biological processes taking place in the tumors. When taking the global expression profiles into account, we somehow compensate for the lack of knowledge regarding “the complete picture” of specific signaling pathways and their phenotypic consequences, including potential compensatory mechanisms derived from their deregulation.
From the beginning of this century, gene expression arrays have been increasingly applied in translational cancer research. Some of the first array studies demonstrated that gene expression data could identify known and novel cancer subclasses, with similarities in terms of biological behavior [13, 14]. In addition to identifying molecular phenotypes in various cancer types [15,16,17,18], transcriptional alterations have proven to be powerful tools for creating classifiers predicting cancer recurrences [19,20,21,22], and to identify alterations in functional pathways, suggesting relevant targets for therapy [23].
Improved Understanding of Cancer Biologic Processes
Oncogenic and non-oncogenic alterations underlie and support the cancer biological processes leading to cancer progress and metastatic disease. High-throughput techniques such as DNA microarrays and RNA sequencing measure the expression of a multitude of genes in one single experiment. This enables multi-faceted views on the phenotypes being studied and provides information about associations between complex gene expression alterations and the phenotypes.
In the era of global gene expression studies, two landmark reports in the field introduced the potential of exploring biological function via studies of gene expression alterations [24, 25]. By studying how the gene expression pattern changed when altering the conditions from fermentation to aerobic metabolism in the yeast Saccharomyces cerevisiae, deRisi and colleagues characterized this metabolic reprogramming at a functional genetic and biochemical level [24], and they were amongst the pioneers in applying large-scale gene expression data to biological questions. deRisi also demonstrated how gene expression patterns change according to deletion or overexpression of specific transcription factors, proposing application of DNA gene expression microarrays for examination of the “signature pattern” accompanying such molecular alterations. deRisi stated: “Perhaps the greatest challenge now is to develop efficient methods for organizing, distributing, interpreting, and extracting insights from the large volumes of data these experiments will provide” [24]. And he was right: although such “global analyses” have assisted in some of the major progresses made in translational cancer research, the issues deRisi raised are still major challenges when translating “omics” data and analysis output into biological relevant information.
Hughes and colleagues published one of the earliest reports considering the signaling complexity when relating gene expression data to genetic and phenotypic alterations [25]. The functions of uncharacterized genes were identified through mapping of gene expression alterations induced by specific gene deletions to transcriptional profiles of known perturbations. A few years later, Huang and colleagues demonstrated that the expression pattern from several genes, included in a “metagene,” characterized and predicted the neoplasm classes under study [26]. These studies were amongst the “precursor studies” to the many reports on gene expression signatures that followed the next decade. Several studies have since been conducted, supporting the assertion that multigene markers better reflect the complexity in the signaling of multiple pathways [27,28,29], as demonstrated by Huang et al. for MYC and HRAS pathways [26].
Gene Expression Signatures as Biomarkers
The Biomarkers Definition Working Group defines a biomarker as: “A characteristic that is objectively measured and evaluated as an indicator of normal biologic processes, pathogenic processes, or pharmacologic responses to a therapeutic intervention.” [30]. D. Hanahan and R.A. Weinberg describe in their “Hallmarks of Cancer” reviews, the tumor biologic processes and enabling characteristics that are essential for tumor initiation and progression to take place [31, 32]. Gene expression signatures might reflect such hallmark characteristics of a tumor, including specific biological processes—and may as such function as biomarkers. Gene expression signature studies have assisted in identifying targets for therapy, suggested as prognostic and predictive markers for specific cancer therapies.
Bild and colleagues integrated early on gene expression profiles and oncogenic alterations, identifying metagenes—gene signatures—reflecting activation levels of, for example, MYC and RAS signaling pathways [27]. The identified signatures are is associated with patient outcomes, demonstrating a prognostic effect of these metagenes. Bild suggested that oncogenic signatures may reflect the oncogenic phenotype and point to tumor biological processes underlying the phenotypic alterations. Moreover, measures of pathway deregulation in this study were linked to therapy response for drugs targeting components of the specific pathways. In this manner, Bild suggested a potential for gene expression signatures also as markers guiding therapy selection.
Lamb and colleagues defined in one of their Connectivity Map papers “the ultimate objective of biomedical research:” To connect human diseases with the genes that underlie them and drugs that treat them [33]. He regarded this “a daunting task” but aimed for a solution. The Connectivity Map tool was developed, aiming to reveal functional connections in diseases and linking these to genetic perturbations and drug actions [33]. As part of this endeavor, a reference bank of gene expression signatures derived from the effects on cultured human cells treated with small molecules (e.g., approved drugs and other bioactive compounds) was established. Bioinformatic analyses were integrated into a publicly available (online) tool, making it possible to match any other signature (also the “homemade” ones) to the drug signatures, thereby enabling researchers to pattern-match the specific gene expression profiles under study with gene expression profiles reflecting effects of the small molecules tested as part of the Connectivity Map database [33, 34]. In the primary publication of the Connectivity Map, the authors demonstrated this tool as a powerful resource to link gene expression patterns to functional effects, bio-physiological processes, and targets for therapy in various diseases. However, the Connectivity Map is suggested as a hypothesis-generating tool, and the importance of validating the findings in other model systems is stressed by the authors. In 2017, Subramanian and coworkers published a 1000-fold expanded Connectivity Map version, assessing each perturbation by RNA sequencing of selected 1000 genes, providing 1.3 million perturbation-linked gene expression profiles freely available for the scientific community (https://clue.io) [35].
Gene Expression Signatures in Breast Cancer
Since the gene expression microarray methodology entered the cancer research field, many breast cancer gene expression signatures have been published. Perou, Sørlie and colleagues explored global gene expression data in breast cancer and identified molecular classes [16]. These were further demonstrated with clinical relevance in follow-up studies [17, 36]. In the same decade, van’t Veer described a “poor prognosis gene expression signature” [20]. Subsequently, gene expression signatures like MammaPrint, OncotypeDx, PAM50, and the Genomic Grade Index (GGI) have been approved by the FDA, and have demonstrated prognostic value for breast cancer patients—overall and within subgroups (e.g., stage I/II, ER-positive breast cancer in postmenopausal women, for the Oncotype DX).
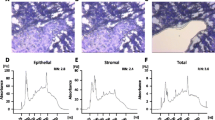
These early signatures were primarily derived as part of a whole section tissue approach, extracting RNA from bulk tumor tissue, with epithelial and stromal cells intermixed. Enrichment of the epithelial component of the tissue samples was promoted, implying lower expression signal from the stromal cells as compared to the epithelial component [37]. For prognostication, adding data about the specific stromal components could add important information. Increased knowledge about the microenvironment, its heterogeneity between tumor subtypes and the epithelial-microenvironmental interactions will likely assist in improving personalized diagnostics and treatment strategies (Fig. 23.1).
Gene Expression Signatures Reflecting the Tumor Microenvironment
As the role of the tumor stroma became a “hot topic” in discussions of the mechanisms for cancer progression, researchers advanced to some extent from bulk tissue gene expression approaches to focusing on specific tumor compartments (Fig. 23.2). Gene expression changes related to the tumor microenvironment (TME) in cancer have been increasingly studied in many types of cancer. Cell-specific alterations (e.g., gene expression changes in immune cells, endothelial cells, cancer-associated fibroblasts, adipocytes) have been described and gene signatures generated, partly as pure prognostic metagenes, partly reflecting cell subsets and cancer biologic processes in the tumor compartments. Deconvolution of bulk gene expression data, a form of “computational dissection,” into information about cell type or compartments, and accompanying counts or expression profiles, came into play a few years ago [39, 40], adding to the methodological approaches applied when deciphering TME data from whole tissue gene expression information.
The figure illustrates the components of a breast tumor, including the normal and tumor epithelial cells, structures, and other constituents representing the tumor microenvironment. Their spatial relations stimulate the idea of close interactions between the different cell populations. BC breast cancer, CAA cancer-associated adipocytes, ECM extracellular matrix. With permission, reprinted from C. Zhao et al. (ref. [38]), J Exp Clin Cancer Res 2020. doi: 10.1186/s13046-020-01666-z
Microenvironmental or stromal signatures might help elucidate biological processes critical for the progression of cancer, and may thereby improve the vision of the still quite blurred understanding of tumor progression and development of metastatic disease. In the following sections of this chapter, selected thematic groups of gene expression signatures are elucidated, reflecting biological processes and acting as prognosticators and predictors of therapy response.
Allinen and colleagues were among the first to point to the “bulk tissue approach” as problematic, when exploring stromal tissue features by gene expression analyses [41]. They therefore aimed to elucidate cellular interactions along with paracrine regulatory modules in breast cancer, reporting the transcriptional and genetic alterations in various cell types in invasive breast cancer, ductal carcinoma in situ, and normal breast tissue. All cell types were purified, and the gene expression profiles of the cell types such as the epithelial cells, myoepithelial cells, myofibroblasts, fibroblasts, endothelial cells, and leukocytes were described. The identification of upregulated CXCL14 and CXCL12 specifically in tumor myoepithelial cells and myofibroblasts, causing epithelial cell proliferation and invasion via the binding of these ligands to their cognate receptors on epithelial tumor cells, were among their novel findings [41]. This study uniquely examined cell type-specific gene expression programs and additionally validated the functional consequences of these alterations, proposing novel analysis approaches to study the tumor-stroma interactions.
Gene Expression Signatures Reflecting the Bulk Cancer-Associated Stroma
As the cancer stroma is composed of several cellular components, examination of general stromal gene expression alterations may bring us into challenges of low specificity regarding which cell type the different expression signals originate from. However, the literature on general stromal signatures demonstrates new information as compared to what was derived from the studies on “whole tissue approaches,” as elucidated in the following section.
Several gene expression signatures derived from the tumor stroma have been published, some of them investigated in relation to disease progress. Two studies explored the differences in the tumor stroma by assessing pre-invasive ductal carcinoma in situ lesions and invasive breast carcinomas. Ma and colleagues assessed the global expression alterations specifically in the stromal and epithelial compartments [42], and demonstrated comprehensive gene expression changes in the tumor-associated stroma during progression from normal to the pre-invasive and invasive states. A gene expression signature reflecting histologic tumor grade was identified in the stromal compartment. This study embraced the hypothesis that tumor-stromal-related changes contribute to tumor progression, specifically in the step from pre-invasive to invasive disease.
In a similar manner, Roman-Peréz and colleagues compared the expression pattern of tumor-adjacent tissue from invasive carcinomas and ductal carcinoma in situ, identifying breast cancer subtypes defined by extratumoral expression patterns [43]. Two distinct “microenvironmental subtypes” were identified, denoted as “Active” and “Inactive” types. Tumors with “active signature” shared features of claudin-low breast cancer and were associated with TGF-β induced activation score. The “active signature” also correlated with tumor aggressiveness and clinical outcome in ER-positive breast cancer.
In supervised analyses of global gene expression data, gene expression patterns between different pre-defined groups have been examined. What would be the best groups to compare when investigating the microenvironmental alterations that support or drive tumor progression? Normal versus cancer? Normal versus pre-invasive in situ lesions? The pre-invasive cases versus cancer? Or simply, although a more complex analytical approach, the whole sequence from normal through pre-invasive and eventually invasive carcinomas? In the following section, studies approaching this challenge in different ways are summarized.
Troester and colleagues compared global expression patterns of normal breast tissue from reduction mammoplasty resections and normal breast tissue adjacent to tumor tissue. A 155-gene “cancer adjacent normal tissue” signature was derived [44]. Genes reflecting constituents of the extracellular matrix, and remodeling of this, as well as genes of inflammation were enriched in this signature. Further, some of the signature genes were known to be involved in cell adhesion, angiogenesis, and re-epithelialization such as keratins. Interpreting these transcriptional findings in a functional manner, similarities to wound healing were seen, and the signature was regarded to reflect an in vivo “wound response.” Further, the signature is strongly associated with breast cancer survival, indicating that tumor-related microenvironmental responses might be of importance in the progression of breast carcinomas.
Finak and colleagues applied laser capture tissue microdissection to assess the gene expression pattern of tumor stroma in primary breast cancer. Several gene expression signatures identified in this series are associated with disease course. The 26-gene signature denoted “stroma derived prognostic predictor ” pointed to contrasting immune responses and angiogenic and hypoxic responses in different tumors [45]. This signature also predicted prognosis, as validated in multiple breast cancer data sets. Based on clustering of the 26-gene signature, the authors suggested stroma-dependent breast cancer subtypes. Also, the stroma signature by Finak predicted clinical outcome independent of other signatures, which were also associated with prognosis, indicating that their stroma-derived prognosticator mirrors specific biological processes taking part in directing the clinical disease course. In this study, Finak and colleagues demonstrated an independent stromal impact within the tumor, showing that genes of their stroma-derived prognostic marker did not predict prognosis when assessed in the epithelial component. The prediction of metastatic disease improved when combining the stroma signatures by Finak with other signature scores of prognostic value, indicating an improved reflection of the stroma-related processes when merging signatures developed by different analytical approaches.
How do the stromal and epithelial cells communicate? Are we able to reflect the interplay between these two compartments by the use of gene expression data? To address these questions, Casey and colleagues examined the transcriptomic pattern of epithelial and stromal cells, both in normal breast tissue and in invasive breast cancer [46]. Cell type-specific interactions were also assessed. A “motile phenotype” was identified in the epithelial compartment, and a “reactive phenotype” in the stromal compartment, with genes reflecting remodeling of the extracellular matrix in a proteolytic manner in the invasive cancer. Also, genes promoting epithelial-mesenchymal interaction (EMT), such as FAP (fibroblast activated protein alpha) were identified. This study interestingly supports a molecular crosstalk between the epithelial and stromal cell compartments, suggesting that alterations facilitating invasion are one of the features of cancer-associated stroma.
By examining global gene expression alterations relating to specific tumor microenvironment elements that are microscopically assessable, it might be possible to identify underlying alterations of the histopathologic phenotype. Van den Eynden examined fibrotic tumor foci and associated gene expression patterns [47], and demonstrated Ras signaling and HIF1A-pathway activation along with other hypoxia- and angiogenesis-related genes in the large fibrotic foci. Also, fibrotic foci correlated with an activated wound healing signature and with earlier development of distant metastases.
What would be the model system best fit to capture ongoing microenvironmental processes promoting tumor progression? Marchini and colleagues examined the transcriptomic alterations in A17 mouse mammary carcinoma cells [48]. Three gene expression signatures reflecting stroma-related features and processes were identified: One “stemness signature,” one “angiogenesis signature,” and one “signal transduction signature.” These signatures are associated with mesenchymal stem cell signatures, ER-negative breast cancer, a basal-like phenotype and breast cancer bone metastases. In post-treatment assessment of breast cancer xenograft models, the A17 angiogenesis- and signal transduction signatures were more highly expressed after hormonal therapy. This study indicates a linkage between mesenchymal features, tumor progression and therapy resistance, directing an interpretation of these findings towards EMT, as regarded having critical importance in tumor progression. Recent studies indicate that epithelial-mesenchymal plasticity contributes to stem-like tumor features and generates cancer stem cells [49,50,51].
Are the tumor microenvironmental changes in cancer progression common or specific across tumor types? Planche and colleagues examined this question by laser microdissecting stromal cells of invasive breast and prostate carcinoma. These two tumor types displayed distinctly different stromal gene expression patterns [52]. Gene expression alterations of the cancer type-specific stromal genes clustered both breast and prostate cancer samples into groups with different disease courses. Of note, genes of extracellular matrix constituents and proteolytic enzymes were upregulated in the invasive breast cancer stroma, in line with the observations done on the tumor histology sections.
In most mRNA expression studies, RNA is extracted from tissue that is snap frozen in the surgical theatre, as the RNA is best preserved for quantitative analyses in this manner, as compared to when extracted from formalin fixed paraffin embedded (FFPE) tissue. However, as of the writing of this chapter, FFPE patient-derived tissue is widely available, as this is stored in pathology archives worldwide. Winslow and colleagues made a critical step forward in this field when they succeeded in studying gene expression alterations from laser dissected tumor epithelial and stromal compartments from FFPE invasive breast cancer samples. This study showed that stroma-specific gene expression signatures were segregated into three major thematic groups; (1) extracellular matrix and fibroblast-related genes; (2) vascular-related genes; and (3) immune cell-related genes. Strikingly, the immune-related signature is associated with basal-like breast cancer subtype [53]. As the results from this study were in line with other similarly designed studies on fresh frozen tissue, the study gave new hope for RNA studies on FFPE tissue.
A few studies have related global gene expression data to specific molecular microenvironmental alterations. Specifically, a relationship between CD10+ stromal cell expression and breast cancer progression was previously reported [41], and Desmedt and colleagues followed up on this by exploring gene expression alterations related to CD10+ stromal cells [54]. A “CD10+ stroma signature ” of 12 genes was generated by comparing the gene expression patterns of CD10+ cells isolated from breast carcinomas and normal breast tissue. In co-culture experiments, the CD10+ cells were characterized as specific cell populations: fibroblasts, myoepithelial, and mesenchymal stem cells. As seen in many of the stroma- and CAF-derived signatures, the CD10+ signature was composed of genes related to matrix remodeling. Interestingly, genes related to osteoblast differentiation (e.g., osteopontin) were also upregulated in the CD10+ signature. All the different CD10+ cell types contributed to this stroma-related signature, however, the highest CD10+ stroma signature score was found in mesenchymal stem cells. Of clinical value, the signature was able to differentiate in situ and invasive breast cancer lesions. Also, the CD10+ signature demonstrated a potential to predict response to chemotherapy, and high CD10+ stroma score was associated with reduced survival in HER2 positive breast cancer cases. This study is a good example of how to combine in vivo and in vitro studies, specifically with respect to validating the functionality of a gene expression signature.
In another study describing gene expression alterations reflecting specific molecular alterations, Rajski and colleagues identified a signature associated with IGF-I stimulated stromal cells [55]. Amongst the IGF-I signature genes, there was enrichment of proliferation-associated genes. This signature clustered the cancer samples in two major groups: those with upregulated IGF-1 and those without. Cases in the cluster with genes upregulated by IGF-I experienced shorter survival. An example of a signature related to specific histopathologic tumor features is one necrosis-related signature derived from gene expression alterations between endometrial carcinomas with and without tumor necrosis [56]. In this case, tumor necrosis was found to be associated with gene expression programs of hypoxia, angiogenesis, and inflammatory responses.
The cancer biology underlying phenotypic features of various cancer types may be cancer specific, but also share commonalities with other diseases and non-cancerous conditions. West and colleagues exploited the potential of approaching the research question from a different angle, when postulating that fibroblasts present with different activation states. In their approach to this question, they distinguished fibroblast populations in non-cancerous samples [57], demonstrating that solitary fibrous tumors and desmoid-type fibromatosis exhibited different expression patterns. In particular, the expression of growth factors and extracellular matrix genes were differentially expressed. When assessing the gene signature separating solitary fibrous tumor from desmoid-type fibromatosis in a series of invasive breast cancer, two groups of breast carcinomas were identified and associated with different survival. The cases with an expression pattern similar to the desmoid-type fibromatosis showed more favorable outcome, while the other group was observed with poorer prognosis. These findings supported the hypothesis that tumor stromal response varies among carcinomas of different aggressiveness.
After the first breast cancer subtype classification by Perou et al. [16], further subgroups of the subtypes are identified based on molecular alterations, like the Lehman subgrouping of triple-negative breast cancer into basal-like, immunomodulatory, mesenchymal, mesenchymal-like, and luminal androgen receptor subtypes—some of the subtypes associated with distinct survival patterns [58]. A recent study by Bareche and coworkers aimed to characterize the tumor microenvironment of triple-negative breast cancer subtypes, elucidating how the microenvironment heterogeneity may contribute to the different clinical pictures seen in triple-negative subsets [59]. A broad signature approach was applied, incorporating gene sets reflecting immune activation, angiogenesis, hypoxia, cancer-associated fibroblast, and metabolism (e.g., glycolysis, lipid metabolism) in the analyses. Distinct TME profiles and specific immune cell composition and localization were associated with the different triple-negative subgroups—and associated differently with clinical outcomes. Next, 16 signatures reflecting innate and adaptive immune responses [60] were mapped to the triple-negative subtypes, demonstrating enrichment of adaptive immune response in the immune modulatory subtype, and enriched innate response in the mesenchymal-like subtype. The mesenchymal and basal-like subtypes showed poor immune responses, both innate and adaptive.
Qian and coworkers described in 2020 a “pan-cancer blueprint of the heterogeneous tumor microenvironment”—adding substantially to our knowledge about heterogenous microenvironment by analyzing single-cell RNA and proteins [61]. By profiling 233,591 single cells from lung, colorectal, ovary and breast tumors and corresponding tumor-free tissue, profiles of 68 stromal cell populations were identified, 22 unique and 46 shared between cancer types. The stromal cell populations were characterized phenotypically by marker genes, metabolic activities, and tissue-specific expression differences. Applying the analysis approaches to an independent subset of melanoma tumors treated with checkpoint immune inhibitors, a naïve CD4+ T-cell phenotype predictive of response to checkpoint immunotherapy was identified. By applying single cell and signature analyses approaches, this study replies in interesting ways to whether and how the tumor microenvironment heterogeneity is present across cancer types, and generates, as the authors state, “the first panoramic view on the shared complexity of stromal cells in different cancers”—with potential for identification of strong prognostic and predictive cancer biomarkers. interactions.
Gene Expression Signatures Reflecting Cancer-Associated Fibroblasts
Most of the studies above have investigated bulk tissue stroma and have thereby potentially reflected expression contribution from the combination of different stromal cell types. Many of the stromal signatures correlate with clinicopathologic features and disease course, potentially reflecting underlying stroma biology. Still, it is tempting to ask: What is the contribution to the signatures from each of the specific stromal cell types?
Chang and colleagues were among the first to generate a pure fibroblast gene expression signature, where the expression alterations were generated by fibroblasts being exposed to serum [62]. The signature was denoted a “core serum response .” Functional analyses revealed involvement of the signature genes in myofibroblast activation, matrix remodeling, and cell motility. All these processes contribute to wound healing. Based on the expression of this wound healing-related signature, breast cancer samples are segregated into two groups. The group with activated signature pattern was associated with increased risk of metastatic disease and death from breast cancer. Further, the signature pattern was consistent in paired samples of locally advanced breast carcinomas, biopsied before and after chemotherapy, indicating stability of the biological program reflected in this signature. Interestingly, the basal-like molecular breast cancer subtype is significantly associated with the expression pattern of the wound healing-related signature, suggesting that the signature points to intrinsic properties of the basal-like phenotype. The signature was also examined in gene expression data sets of various tumor types, and the findings were striking: The expression pattern of the signature separated the cases into two groups, with significantly increased risk of metastatic disease in the group with the activated signature pattern. Harold F. Dvorak suggested in a review in 1986 that the wound is an analog to the stromal processes observed in tumors [63]. The gene expression signature by Chang might have captured some of the alterations observed by Dvorak.
Tchou and colleagues added information about subtype-specific stromal gene expression patterns in breast cancer [64]. Their analyses demonstrated distinctly different expression profiles in CAFs from breast cancer samples of the HER2 positive subtype, triple-negative cases and ER-positive cases. In particular pathways linked to the cytoskeleton and integrin signaling were differentially enriched in the different CAF groups. The results from this study add to the arguments of specific stroma-related breast cancer subtypes, supporting the hypothesis that fibroblasts participate to the disease biology underlying clinically relevant breast cancer subtypes.
Two projects exploring transcriptional alterations in tumor-associated fibroblasts compared to normal mammary fibroblasts, demonstrated an increased expression of genes involved in tumor progression in the CAFs. Cytokines, genes related to remodeling of the extracellular matrix, and genes reflecting paracrine or intracellular signaling, as well as cell-matrix interactions, were upregulated in the tumor-associated fibroblasts [65, 66]. In the study by Singer, it was noted that these gene expression alterations also take place in the isolated cell culture state, in the absence of adjacent malignant epithelium [66]. In the study by Bauer, the CAF-associated genes were incorporated into a 31-gene signature that was validated by qPCR. Some of the genes upregulated in CAFs were validated at protein levels by immunohistochemistry, with respect to location and quantitation [65]. Taken together, the findings from these two studies indicate fibroblastic subpopulations of the tumor stroma, facilitating tumor progress.
By comparing global gene expression patterns of platelet-derived growth factor (PDGF)-stimulated human fibroblasts and resting fibroblasts, Frings and colleagues identified a 113-gene expression signature reflecting PDGF-activated fibroblasts [67]. This signature had the potential to identify breast cancers with a stroma of PDGF-stimulated fibroblasts. The signature correlated with high expression of the PDGF receptor β (PDGFRB) and its ligands and was enriched for genes related to angiogenesis and regulation of the extracellular matrix. Signature analyses in several breast cancer data sets demonstrated associations between the PDGF signature score and clinicopathologic features reflecting aggressive tumors, such as large tumor size, high histologic grade, HER2 positive, and ER-negative tumors. Moreover, signature activation is correlated with the HER2 positive, basal-like and Luminal B subtypes of breast cancer. In line with these observations, the signature demonstrated a robust association with survival; a high signature score was associated with reduced survival, also in multivariate analyses, when adjusted for other stroma signatures and a proliferation signature.
Sonnenblick et al. developed a stromal gene expression signature based on reactive breast cancer stroma in HER2 positive cases, containing increased amounts of reactive myofibroblasts surrounding the tumor cell nests [68]. This “reactive stroma signature” was associated with trastuzumab resistance in estrogen receptor (ER)-negative tumors, but not in ER-positive tumors, suggesting the reactive stroma and its accompanying signature as a potential predictive marker for Trastuzumab in subsets of breast cancer.
Siletz and colleagues assessed transcription factor signatures and activity specific for mammary CAFs versus normal mammary fibroblasts [69]. A transcription factor activity signature included activation of reporters for ELK1, GATA1, retinoic acid receptor, serum response factor, and vitamin D receptor. An increased activation of reporters for HIF1 and several STAT and proliferation-related transcription factors was seen after induction of fibroblasts by conditioned medium from breast cancer cell lines. These transcription factor activity profiles indicate CAF subtype-specific signaling promoting tumor progression through a pro-invasive stroma.
In recent years, single-cell RNA sequencing has allowed for identification and deep characterization of CAF subpopulations. By defining CAF subpopulations by single-cell RNA sequencing of transcriptomes of mesenchymal cells from a genetically engineered mouse model of breast cancer, Bartoschek and colleagues added knowledge about CAF heterogeneity with functional and clinical implications [70]. Gene signatures reflecting angiogenesis and vascular development, matrix-related genes, cell cycle activation, and development and differentiation were enriched in the CAF groups that accordingly were annotated vascular, matrix, cycling, and developmental CAFs (vCAFs, mCAFs, cCAFs, dCAFs). The vCAFs and mCAFs signatures were validated by RNA sequencing of bulk breast cancer tissue, demonstrating biological and clinical relevance (Fig. 23.3).
Unbiased clustering of fibroblast single-cell transcriptomic data reveals four populations. (a) Schematic representation of negative selection strategy. (b) gating strategy and quantification of flow cytometry for single-cell sequencing. FSC forward scatter, SCC side scatter. (c)Violin plot of detected genes in 784 sorted fibroblasts. (d) t-SNE layout of CAFs (n = 716) by RPKM-normalized transcriptomic data. (e) Expression plots on t-SNE layout. Log2(RPKM+1) levels of CAFs marker genes in individual cells. (f) Cell size and granularity as determined by forward-scattered light (FSC) and side-scattered light (SSC) of different CAF populations. With permission, reprinted from M. Bartoschek et al. (ref. [70]), Nat Commun 2018. doi: 10.1038/s41467-018-07582-3
Wu and colleagues aimed to elucidate stromal heterogeneity in triple-negative breast cancer [71]. Two CAF and two perivascular-like (PVL) subpopulations were identified in the stroma, with distinct spatial relationships and functional properties. The gene signatures reflecting inflammatory CAFs and differentiated PVL revealed associations with cytotoxic T-cell dysfunction in independent cohorts of triple-negative breast cancer, pointing to potential candidate biomarkers for new therapeutic strategies in the treatment of triple-negative breast cancer.
Woelfle and colleagues derived a signature of 86 genes differentially expressed between primary tumors with and without bone marrow metastases [72]. Although the tumor microenvironment was not the focus of this study, most of the signature genes were related to extracellular matrix remodeling, cytoskeleton plasticity and cell adhesion. Also, RAS- and HIF1A signaling were enriched in tumors with bone marrow metastases. The many similarities between this signature and the stroma- and CAF-related signatures described above lead to an intriguing perspective on this signature. In addition to facilitate invasive growth and tumor progression, perhaps the tumor stroma is heavily involved in directing tumor metastases to different locations? Another interesting perspective of this signature was that 77 of the 86 signature genes were downregulated in primary tumors with bone marrow metastases, indicating transcriptional repression as part of the picture in tumor progressive processes.
A few studies have examined transcriptional alterations related specifically to the extracellular matrix in breast cancer. Bergamaschi and colleagues set out to classify breast carcinomas based on constituents of the extracellular matrix (ECM), selecting 278 ECM-related from the literature [73]. The ECM-related genes segregated the breast cancer samples into four ECM classes with different clinical courses. The ECM group associated with best survival showed upregulation of protease inhibitors of the serpin family. The ECM group associated with poorest survival presented with overexpression of integrins and metallopeptidases, and low expression of laminin chains. In a follow-up study, Triulzi and colleagues demonstrated that one of the ECM groups consistently predicted one cluster in several independent breast cancer data sets [74]. The 58-gene signature of this ECM subset contained 43 genes encoding structural ECM proteins. Investigation of gene expression data sets on separate cancer epithelial and stromal cells demonstrated that genes of this ECM signature were expressed both by the epithelial and stromal compartments. In vitro experiments showed induction of signature genes, in particular in fibroblasts and in ER-negative breast cancer cells. Single genes and gene sets reflecting EMT were significantly associated with this ECM signature. In another study validating the functionality of the identified CAF subsets, Bartoschek and colleagues (see above) characterized transcription of genes encoding ECM proteins included in the matrisome. Each of the CAF populations demonstrated distinct ECM transcriptional signatures, supporting their different biological functions [70].
Gene Expression Signatures Reflecting Vascular Biology
Various measures of histologically verified tumor vasculature (e.g., mean vessel density, vascular proliferation) are related to tumor progress and metastatic disease in solid cancer types. The vasculature is viewed as a target for therapy, as exploited in therapeutic programs in several tumor types. Studies on genomic programs measuring the transcriptional alterations have a potential to reveal novel aspects of vascular biology in malignant tumors. With this in mind, Wallgard and colleagues sought to elucidate the transcriptome and molecular processes specific to endothelial cells [75]. Fifty-eight genes specifically linked to microvascular expression were identified, many of them not previously described in relation to functions of endothelial cells. Wallgard suggested several genes and related proteins to be further explored in relation to drugs targeting the microvasculature, like Eltd1, Gpr116, Ramp2, Rasip1. In a recent study, Cleuren and colleagues further characterized the endothelial cell biology by facilitating isolation of endothelial cell (EC) ribosome-associated transcripts, also known as the translatome [76]. By combined endothelial-specific translating ribosome affinity purification (EC-TRAP) and high-throughput RNA sequencing analyses, known and new pan EC-enriched gene signatures and tissue-specific EC transcripts were identified, also demonstrating endothelial cell heterogeneity across tissue types and disease states (Fig. 23.4). Results from this study indicated that the transcriptome of a tissue lysate can serve as a proxy for the corresponding tissue translatome, supporting relevance of the mRNA expression analysis approaches applied in previous studies on vascular biology in cancer.
Identification of enriched transcripts after EC-TRAP. (a) GO analysis of the 500 top-ranked genes with the highest enrichment scores after EC-TRAP (FDR < 10%) shows overrepresentation of transcripts involved in vascular-related processes. (b) Unsupervised hierarchical clustering of EC-enriched genes shows distinct, highly heterogenous vascular bed-specific EC expression patterns. (c) Comparison of the 500 most enriched genes per tissue identifies a group of pan-endothelial and subsets of tissue-specific EC-enriched genes. With permission, reprinted from A.C.A. Cleuren et al. [76], Proc Natl Acad Sci USA 2019. doi: 10.1073/pnas.1912409116
The vasculature is regarded as the main route for breast cancer metastases. Hu and colleagues compared the global transcription pattern of primary tumors and distant metastases, identifying an in vivo hypoxia signature reflecting VEGF activation and predicting poor clinical outcome in breast cancer and other tumor types [77]. This 13-gene signature was composed of several angiogenesis-related genes. Eight of the 13 signature genes contained binding sites for the hypoxia-related transcription factor HIF1α and had been demonstrated to be regulated by HIF1α.
Pepin and colleagues identified two distinct tumor vasculature types by analyzing global transcription patterns across laser capture microdissected tumor-associated and matched normal vasculature [78]. The two tumor vasculature types demonstrated specific gene expression signatures, whereas one related to anti-angiogenic signaling. Samples enriched for this signature demonstrated lower mean vessel density as compared to the group enriched for the gene signature associated with active vascular remodeling and reduced vascular shear stress. Reduced vascular shear stress is suggested to reflect reduced vessel flow rate and may reflect inappropriate tumor perfusion. Significantly, several therapeutic targets with potential relevance in anti-angiogenic treatment (e.g., MET, PDGFRβ, ITGAV) were differentially expressed between the vasculature subtypes.
When studying alterations in vascular gene expression, different study designs may reveal different layers of the complete picture. Bender and colleagues demonstrated by supervised analyses of angiogenesis-related genes that the gene expression of the VEGF and semaphorin families was altered in pro-angiogenic manners in triple-negative breast cancer [79]. Compiling these genes into a composite biomarker, the gene expression signature is associated with triple-negative breast cancer and reduced survival in non-triple-negative subtypes.
Wallace and colleagues approached angiogenesis-related biology in a more indirect manner. In an analysis of genes and pathways mediating fibroblast contribution in cancer progression [80], the authors studied how Ets2 function varied between mammary stromal fibroblasts and epithelial cells. In HER2 positive breast cancer mouse models, Ets2 inactivation in fibroblasts reduced tumor growth. The same effects were not seen when inhibiting Ets2 in epithelial cells. An Ets2-dependent gene signature was derived, enriched in genes related to remodeling of the extracellular matrix, cell migration, and angiogenesis. Supportive to these functional interpretations, fewer functional blood vessels were found in tumors lacking fibroblast Ets2. The Ets2-dependent gene expression signature was able to segregate human breast cancer stroma and normal stroma and indicated a link between Ets2 and the fibroblast-endothelial crosstalk, pointing to a contribution of Ets2 in the angiogenic process.
Xiao and colleagues [81] developed in vitro models studying breast cancer-specific endothelial cells, identifying multiple subpopulations of tumor-associated endothelial cells, each population with distinct gene expression patterns. A relationship with tumor-associated endothelial cells had not previously been established for several genes. Irx2 and Zfp503 were without previously known relevance to vascular biology, but were found highly upregulated in tumor endothelial cells. These genes are known to regulate neuronal patterning and developmental differentiation [82, 83], and may point to new information on vascular-related mechanisms and co-regulatory circuits in vascular biology.
Mannelqvist and colleagues published an 18-gene expression signature related to vascular invasion in endometrial carcinomas, also relating to features of aggressive disease and disease outcome [84]. In a follow-up study on multiple breast cancer gene expression data sets, the vascular invasion signature was associated with tumor progression and clinical course in breast cancer [85]. Also, a high signature score was associated with the basal-like phenotype and response to neoadjuvant chemotherapy. The signature was composed of genes related to angiogenesis, immune responses, and extracellular matrix biology. Further, the signature was correlated with other gene expression profiles of vascular biology, hypoxia, EMT, immune response, and tumor progression.
The same research group later published a 32-gene signature reflecting tissue-based vascular proliferation. Microvessel proliferation was assessed by dual endothelial immunostaining of Factor-VIII/Ki67, and global gene expression data as well as copy number information were explored in supervised manners [86]. Several genes in the signature had previously been linked to processes such as neovascularization, endothelial cell migration and adhesion, supporting this signature as relevant for tumor angiogenesis. Also, amplification of the region 6p21, potentially harboring VEGF, is associated with high microvessel proliferation.
Tobin et al. elucidated how gene transcripts representative of normal endothelium related to breast cancer progress. A composite microvasculature (MV) score was derived from expression values of 57 mouse microvasculature transcripts [87]. In 993 breast cancer tumors, the MV score did not associate with microvessel density, but indicated decreased risk of metastasis in endocrine-treated patients. Further, the MV score was increased from pre-treatment to post-treatment samples in metastatic breast tumors after treatment with sunitinib and docetaxel, compared to cases with only docetaxel treatment, supporting the concept of vascular normalization following treatment with an angiogenic inhibitor.
Physiological angiogenesis is regarded molecularly different from the pathologic angiogenesis, indicating different mechanisms involved in the two processes. Guarischi-Sousa and colleagues followed up on this idea, identifying a 153-gene signature reflecting pathologic angiogenesis from oxygen-induced retinopathy [88]. Applying a machine learning algorithm, a signature of 11 of the 153 genes was compiled together with information on age and stage into a signature strongly predicting breast cancer survival. The authors propose the signature as a potential marker for pointing to tumors relevant for angiogenesis-targeted therapies.
Harrell and colleagues sought to determine whether tumor-associated vascular properties could identify mechanisms contributing to the different risks of metastatic disease across the intrinsic subtypes of breast cancer [89]. They found that claudin-low and basal-like tumors were enriched for transcriptional programs reflecting vascular quantity, vascular proliferation, and a VEGF/Hypoxia signature. Incorporating several of the vascular gene signatures described above-added information about risk of metastatic disease. Furthermore, experimental studies demonstrated that claudin-low cells exhibited endothelial-like morphology, and claudin-low xenograft tumors were highly perfused through intercellular spaces and non-vascular tumor cell lined channels. This study combined the transcriptional studies with experimental validation in an interesting manner, demonstrating both endothelial-like characteristics of cancer cells, and how the vasculature in conceptually new manners may contribute to breast cancer progression. Also, the gene expression signatures were suggested as predictive markers to anti-angiogenic therapy.
Mendiola and colleagues set out to identify a biomarker predicting response to bevacizumab and paclitaxel in metastatic breast cancer by exploring angiogenesis-related genes and clinical markers [90]. An 11-gene signature predicted improved progression-free and overall survival in patients on bevacizumab–paclitaxel treatment, with added prognostic value when combining the signature with five clinical covariates. The value of these composite biomarkers as predictive markers to bevacizumab in metastatic breast cancer should be tested.
Krüger et al. recently studied predictive markers for combined neoadjuvant bevacizumab and chemotherapy treatment in a randomized trial [91]. Along with tissue-based angiogenesis biomarkers (microvessel density, proliferative microvessel density, glomeruloid microvascular proliferation), the authors explored how an angiogenesis-based mRNA signature previously published from the same group [86], reflected pathologic complete response. High baseline MVD predicted pCR in the bevacizumab-arm, whereas vascular proliferation and a high angiogenesis score were associated with the triple-negative and basal-like phenotypes but did not predict therapy response.
Inflammation is regarded to promote tumor angiogenesis. Pro-inflammatory cytokines work through mediators, enhancing or suppressing angiogenesis. Combinations of these factors may contribute to the tumor’s vascular invasive and metastasizing properties. Pitroda and colleagues explored how vascular inflammation influences cancer prognosis [92]. A gene expression signature reflecting inflammation in tumor-associated endothelial cells was developed. The endothelial-derived 6-gene inflammatory signature predicted reduced overall survival in breast cancer and other tumor types. Also, inflammatory pathways activated in endothelial cells are linked to tumor progression in mice, supporting a vasculo-immunogenic link contributing to tumor progression in breast cancer.
Oshi et al. aimed to explore relations between intratumoral angiogenesis, inflammation, and metastasis in breast cancer [93]. They derived an angiogenesis-related signature score that did not correlate with clinicopathologic variables or survival, nor with breast cancer molecular subtypes. However, a high score is associated with a low fraction of immune cell infiltrations, both favorable and unfavorable. Unfavorable inflammation-related gene sets (IL6, TNFα, TGFβ) and metastasis-related gene sets were enriched in high-score tumors. Further, a high angiogenesis score was significantly associated with metastasis to brain and bone.
Gene Expression Signatures Reflecting Immune-Related Alterations
The immune system is considered to play an important role in cancer initiation and progression and is a promising multi-faceted target in novel therapeutic strategies [94]. Important interactions between the immune cells and other tumor microenvironmental elements are brought to discussion [95]. How immune system alterations contribute to cancer progress is not yet well understood. Studies on breast cancer have demonstrated survival benefit from immunotherapy, mainly in advanced triple-negative and HER2 subtypes [96,97,98,99,100]. European Medicine Agency approved in 2020 the PD-L1 checkpoint inhibitor atezolizumab in combination with chemotherapy (nab-paclitaxel), to patients with PD-L1 positive, unresectable, locally advanced or metastatic triple-negative breast cancer [99, 101]. PD-L1 immunohistochemistry is approved as a predictive biomarker test for this treatment regimen, but study results indicate a need for improved predictive biomarkers for checkpoint inhibitors. Although the field of cancer immunology has been extensively explored and exploited for diagnostic and therapeutic purposes, the words of Winston Churchill still seem valid: “This is not the end. It is not even the beginning of the end. But it is, perhaps, the end of the beginning.”
Perou and colleagues touched upon the transcriptional heterogeneity of ER-negative breast cancer in their early breast cancer classification study [16]. Teschendorff et al. followed up on this, demonstrating transcriptional alterations associated with the clinical course of ER-negative breast cancer [102]. Distinct subclasses among ER-negative tumors were shown based on transcriptional patterns. One of the classes consisted of basal-like tumors with upregulation of genes related to immune response and complement activation. This subset of ER-negative samples demonstrated better survival pattern as compared to the rest of ER-negative tumors. Based on this study, a seven-gene immune response signature was derived. Downregulation of this module is associated with increased risk of advanced disease. In a later study, Rody and colleagues focused on the clinically and prognostically heterogenous triple-negative breast cancer subtype [103]. The basal-like and claudin-low subtypes were described by metagenes reflecting angiogenesis, inflammation, and non-neoplastic cell types like immune cells, adipocytes, and fibroblasts. High immune cell score is associated with improved survival, and high inflammation and angiogenesis scores are correlated with reduced survival. By applying a ratio of the B-cell and IL-8 metagenes, Rody identified a subgroup (32%) of triple-negative cases with high B-cell and low IL-8 scores, experiencing improved outcome. Further, two other breast cancer studies have underpinned the association between an immune response and tumor subsets with milder disease courses [104, 105]. In the study by Alexe and colleagues, a HER2-positive subtype with low recurrence rate was associated with high expression of lymphocyte-associated genes [104]. Also, a prominent lymphocytic infiltration was seen by histologic examination of these tumor cases. In the study by Schmidt and colleagues, a high B-cell metagene score was associated with metastasis-free survival in node-negative cases with high proliferation, as validated both in high-grade cases and in young breast cancer patients [105]. Schmidt and colleagues [106] followed up on this study, aiming to identify one single immune system marker for cancer progression. Immunoglobulin κC (IGKC) demonstrated similarly predictive and prognostic value as the entire B-cell metagene [105]. IGKC gene expression is associated with improved survival across different molecular subtypes in node-negative breast cancer. Also, levels of IGKC measured by immunostaining in a series of FFPE breast cancer tissues correlated with clinical outcomes. Tumor-infiltrating plasma cells were identified as the source of the protein. These findings suggest relevance of further exploration of the humoral immune response and its relevance in the therapeutic setting.
One study pointing in this direction, specifically examined genes related to TH1-mediated adaptive immunity in breast cancer [107], and demonstrated that inflammation and immune suppression predicted tumor subsets with different clinical outcomes. Data sets on various tumor types were analyzed, and Hsu showed that upregulation of the TH1-mediated adaptive immunity genes correlated with good prognosis in young breast cancer patients (<45 years). Two other studies demonstrated better survival in cases of high immune signature score in breast cancer [108, 109]. Bianchini et al. demonstrated association between high expression of a B-cell/plasma cell signature and improved survival in ER-positive cases with high proliferation, also when adjusting for standard prognostic variables and other transcriptional scores [108]. In the study by Nagalla and coworkers, a cluster of cases without distant metastases was associated with genes related to immunological functions. These genes could be clustered into three major “immune metagenes,” one cluster reflecting B-cells and/or plasma cells, another cluster reflecting T-cells and natural killer cells, and a third cluster reflecting monocytes and/or dendritic cells [109]. In tumors of high proliferation, high immune metagene score was associated with reduced risk of metastasis—cases with low immune metagene scores are associated with poorer outcome.
A few studies of immune-related signatures have suggested therapy strategies based on their findings. Ascierto and colleagues elucidated how immune function networks related to tumor-infiltrating immune cells were more highly expressed in cases without recurrent disease [110]. The network genes were related to B-cell development, interferon signaling, autoimmune reactions, and antigen presentation pathways. The results indicated crosstalk between the adaptive and innate immune systems. Five B-cell response genes predicted relapse-free survival (>85% accuracy), also validated by qPCR. The authors thus suggested immunotherapy, in the neoadjuvant setting, to patients with high risk of recurrent disease, potentially by inducing genes of immune function.
Iglesia and colleagues aimed to elucidate transcriptional alterations related to the cancer immune response of breast and ovarian cancers with high lymphocyte infiltration and improved survival [111]. RNAseq data and a microarray dataset were applied to identify signatures reflecting the adaptive immune response. The B-cell signatures predicted improved survival in the basal-like and HER2 subtypes. Further, analyses of B-cell receptor (BCR) sequences were assessed through RNAseq data. It was previously shown that a clonal expansion of the B-cells and somatic hypermutations in B-cell tumor-infiltrating lymphocytes in breast tissue represent an antigen-directed response [112,113,114], and the response of antigen-specific B-cell populations actively demonstrate features of clonal expansion. A part of the basal-like and HER2-enriched cases with shorter survival showed upregulation of BCR gene segments with low diversity, indicating lack of B-cell clonal expansion, and were also indicative of an ineffective antigen-directed response in these cases, potentially contributing to their poorer prognosis. More and varied BCR segments with increased expression are associated with improved prognosis. The results indicate a limited B-cell antitumor response in a subset of basal-like breast cancer. Also, immunomodulatory therapies were suggested, and supporting B-cell responses may be one relevant approach in B-cell infiltrated carcinomas.
Perez and coworkers developed a transcriptional signature of immune-related genes predicting clinical benefit in a clinical trial of adjuvant Trastuzumab in combination with chemotherapy in HER2 positive breast cancer [115]. Signature enrichment is associated with increased recurrence-free survival only in the study arms receiving Trastuzumab. Cases in the Trastuzumab study arms without immune signature enrichment did not benefit from Trastuzumab, suggesting interactions between immune-related genes and therapy response. Immune-related signatures associate with improved survival in several studies. However, when it comes to immune responses, the picture is not black and white. Rody and colleagues elucidated how the transcriptional changes of immune metagenes related to clinical outcomes [116]. An IgG metagene, which was found to be a marker for B-cells, did not associate with prognosis. However, high expression of a T-cell/lymphocyte-specific kinase signature is associated with survival in ER-negative cases and cases of concurrently ER and HER2 positivity. This study also suggests inhibition of the IL-8 pathway as a potential therapeutic strategy in breast cancer. Adding to the complexity, a link between the EMT program and immune evasion seen in cancer has been suggested [117,118,119,120].
By unsupervised analysis, identifying co-expressed breast cancer transcripts in global gene expression data, Yang and coworkers identified two co-expressed gene clusters with significant enrichment of gene sets reflecting immune responses and cell cycle activity. A condensed 17-gene signature was derived, correlating well with overall levels of tumor-infiltrating lymphocytes in triple-negative breast cancer [121]. The immune cell signature demonstrated prognostic value in subtype- and immunity-adjusted risk of distant metastasis (iRDM) analysis, as validated by independent cohorts.
The immune microenvironment in triple-negative breast cancer has been in the spotlight over the last years. The study by Zhang and colleagues contributes to our understanding of the mechanisms that promote cancer progress in triple-negative breast cancer, exploring the gene expression profiles of tumor-infiltrating CD4+T cells, elucidating how they contribute to modulating immune cell functions in triple-negative breast cancer [122]. The contribution of CD4 + T cells to the tumor-promoting biology was examined by assessing differentially expressed genes between tumor and peripheral blood CD4+T cells from patients with triple-negative breast cancer. Expression patterns associated with increased levels of T regulatory (Treg) cells and exhausted lymphocytes, and decreased effector/memory and cytotoxic T-cells were demonstrated in tumor samples. Additionally, genes overexpressed in CD4+ TILs contributed to exhaustion of lymphocytes and regulation of chemotaxis.
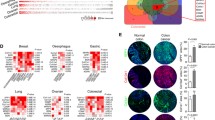
Adding information about spatial gene- and protein relations is recently increasingly focused on translational cancer studies. Considering the risk of losing compartment-specific information by studies on bulk tumor, Gruosso et al. integrated spatial tissue immune response information and gene expression profiling data from matched stromal and epithelial tumor compartments, identifying distinct tumor immune microenvironment (TIME) profiles in the triple-negative subgroup (Fig. 23.5) [123].
(a) The analysis pipeline from Gruosso et al. [123], demonstrating discovery and validation of gene expression signatures identifying spatially context-dependent immune cell profiles. The authors visualize correlations between the immune cell profiles and pathway signaling (b), how signature combinations into meta-signatures stratify clinical outcome (c), and propose identification of new tumor phenotypes (d). With permission, reprinted from Gruosso et al. [123], J Clin Invest 2019. doi: 10.1172/JCI96313
Biological processes identified by analyses of gene expression data from laser capture microdissected tissue from matched stromal and epithelial tumor compartments, pointed to distinct TIME subtypes, believed to support the development of TIME-dependent targeted therapeutic approaches to treat triple-negative breast cancer. Based on the high versus low tumor core or margin CD8+ T-cell infiltration, with information on stromal and epithelial T-cell infiltration, the triple-negative subset was grouped into “margin-restricted” (MR), “immune desert” (ID), “fully inflamed” (FI), and “stroma-restricted.” The tumor characterization based on CD8+ T-cell localization identified mRNA signatures identify distinct biological processes in the different tumor compartments, like enrichment of cholesterol biosynthesis and IL17-related immunosuppression in restricted stromal tumors. The TIME subtype-specific mRNA signatures provided new survival knowledge within the group of triple-negative breast cancer.
Adipocytes and Glycolysis-Related Gene Signatures
At the invasive front of breast cancer, we frequently find the adipocytes, more specifically the breast cancer-associated adipocytes (CAAs). The crosstalk between adipocytes and cancer cells results in phenotypical and functional changes for both cell types [38, 124]. Ultimately, the interplay between CAAs and tumor cells shapes the tumor microenvironment towards an oncogene-driven state favoring proliferation, angiogenesis, invasion, and metastasis.
CAAs are proposed as key players in breast cancer progression, like in the study by Wu and colleagues [124], where the authors investigated the adipocyte-cancer cell crosstalk to gain insight into tumor biology and uncover novel therapeutic targets. Adipocytes are regarded as a giant energy storage upon interaction with BC cells, providing high energy metabolites. With this in mind, theories discuss that tumors may induce reprogramming of metabolic cooperation in adipocytes, adjusting to intracellular metabolic processes supporting proliferation through interplay and interactions between CAAs and breast cancer cells [125, 126]. Importantly, dividing cells demand extreme amounts of energy, and to meet these requirements, alterations in the metabolism of all macromolecules take place in cancer cells. Metabolic changes are well known as a hallmark of cancer [32], and glucose utilization and uptake are heavily increased in order to fuel cell growth and division in multiple cancer types. Interestingly, glycolysis is preferred in malignant tumors rather than oxidative phosphorylation in mitochondria [38, 127].
Targeting tumor metabolism has become a promising therapeutic strategy in cancer treatment. Investigating tumor glycolysis was the main objective for Tang and colleagues [128], who presented a glycolysis-related gene expression signature aiming to predict the prognosis of breast cancer patients. A total of 878 patients were included in the analyses, revealing 129 glycolysis-related genes significantly associated with breast cancer prognosis. From these, a robust four-gene signature was established in a prognostic model, separating breast cancer patients into high- and low-risk groups. Survival analysis demonstrated significantly better prognosis in the low-risk group. Moreover, the glycolysis-related gene signature showed excellent prognostic accuracy, also when stratified by clinicopathological risk factors. To validate the prognostic value of the signature, external validation sets were applied, demonstrating both statistically significant and clinical relevance of the signature.
Accompanying the hunt for glycolysis-related gene signatures in breast cancer, Li and colleagues [129] identified a prognosis-associated signature related to energy metabolism in triple-negative breast cancer (TNBC). Herein, 1097 cases were studied. An 8-gene signature associated with energy metabolism were identified, distinguishing patients’ outcome into low-risk and high-risk groups. The 8-gene signature is distinctively associated with the patients’ clinical characteristics, representing an independent factor in predicting TNBC patient prognosis. Also, the signature could potentially be used as a prognostic marker and for predicting response to therapy targeting the energy metabolism.
With similar study design, Zhang and colleagues [130] identified a glycolysis-related 11-gene signature for prognostic evaluation of breast cancer patients. In contrast to the common workflow applied in most studies working on gene expression and gene signature identification, this study selected genes mainly by performing gene set enrichment analysis (GSEA). The authors argued that as GSEA does not require significant differences in gene thresholds and screens genes based on overall expression levels, the risk of overlooking genes with important biological functions decreases. This study was the first in line to identify glycolysis-related genes with prognostic information in breast cancer. The 11-gene signature was proposed as a promising prognostic marker in breast cancer, and potentially with value as a screening tool to identify persons at high risk of developing breast cancer.
Regarding the topic of evading risk of overlooking important genes, protein–protein interaction network analysis (PPI) has become a popular tool when screening for prognostic factors in cancer, appearing to be a more effective method due to its ability to compare the relationship between candidate genes through network interactions. Moreover, interaction networks allow for visualization, which invites the human perspective along with computed calculations to inspect candidate genes based on their network relations, thus minimizing the risk of overlooking potential genes that may seem unimportant at first. When mining for key genes, protein–protein interaction analyses can be applied to single out genes with dense connections and central roles either in the network as a whole or in specific sub-clusters.
Studying the gene expression differences in distant and tumor-adjacent adipose tissues may reflect distance to the tumor, rather than the presence of tumor cells. By gene expression analyses on distant and tumor-adjacent adipose tissue related to invasive breast cancers and on adipose tissue from non-malignant breasts from postmenopausal women, Sturtz and colleagues aimed to identify genes supporting tumor development and progression [131]. The authors demonstrated that highly expressed genes in tumor-adjacent compared to distant adipose tissue promote tumor growth and progression, due to increased cellular proliferation, invasion, migration, metastasis, and angiogenesis.
Methodological Aspects of Gene Expression Signatures
When exploring biological characteristics of the tumor microenvironment and the ongoing processes underlying cancer development and progression, we may feel like Mr. Jones in the song of Bob Dylan (1941–): “… something is happening here, but you don’t know what it is. Do you, Mr. Jones?” How can we best capture the “something happening here” in the microenvironment surrounding the tumor? When using global gene expression data, is there a “perfect” way of picturing the stromal activities? The statistician George E.P. Box (1919–2013) stated that “All models are wrong, but some are useful,” indicating that not one single model is able to catch the complete picture, and combining different and complementary approaches is probably one way out.
In dealing with gene expression analyses as one model, we most likely assess relevant information about the processes and pathway signaling taking place in the tumor microenvironment. The results from these studies are dependent on the input and analysis strategies. Gene expression analysis approaches can be divided into unsupervised and supervised analyses. The former requires no supplementary information to the expression data and provides great exploratory potential. The latter is driven by sample characteristics, typically in two groups, e.g., “positive” versus “negative” molecular phenotype, or high versus low tumor stage.
Unsupervised Analyses and Class Discovery: Unbiased Exploring
By unsupervised analyses, without guidance by additional data except for the gene expression information itself, the aim is to find patterns in the expression profiles where no pre-defined class is presented. Hierarchical clustering is one example of unsupervised analysis. This method aims to group together objects based on measures of similarity and dissimilarities between them [132]. Hierarchical clustering requires specification of similarity metrics and linkage. The similarity metric describes how similar two samples are, by reflecting the distance between them. Additional information for the distance between clusters is needed, as reflected by the linkage method (single, average, or complete linkage). Complete linkage is demonstrated to be superior for clustering genes, while for clustering of samples, both average and complete linkage is proven useful [133]. Validation of the identified clusters is crucial, including validation of both biological and clinical plausibility, and the level of statistical evidence.
Supervised Analyses: Genes Differentially Expressed Between Groups
Identifying genes with known functions that are differentially expressed between two groups may provide better understanding of biological differences between the pre-defined groups [133]. If the genes identified are of unknown function, the analyses have the potential to provide novel insight into new gene functions. Supervised analyses require supplementary information about the groups, such as clinicopathologic or molecular phenotypic data. An increased risk of false-positive findings due to multiple testing occurs as we run, e.g., 20,000 tests simultaneously on the same data, when searching for genes differentially expressed between classes. There are various methods to adjust for multiple testing, all of them with the aim to provide greater certainty that the genes in our analysis output are truly differentially expressed between the groups we examine, and not listed due to chance. Being very strict in the adjustment of the multiple testing might mask true biological effects. The adjustments will thus be a “trade-off” between too few and too many genes correctly identified as differentially expressed between classes. It is generally accepted that applying filters that results in no false-positive genes in the output is a too stringent approach, with a high risk of losing relevant biological findings in the analysis output. When searching for single genes differentially expressed between classes, the genes identified should nevertheless be further validated, and elimination of false-positive candidate genes or biomarkers occurs at these stages. In the search for the optimal cut-off on the output lists, it is important to remember that statistical significance does not imply biological relevance—and that biological relevance will not always provide statistical significance.
The number of genes differentially expressed between classes might be reduced to a limited number of genes with specific biological and/or prognostic information, and may be presented as gene expression signatures . Such signatures (i.e., gene sets) might be regarded as metagenes with respect to expression value, and a signature score is calculated to evaluate the metagene expression value [26]. Such signature scores have been derived in various ways. One simple approach is to generate a “sum score” or “average score” (the score value of one sample equals the sum or the average of the expression values of the genes in the signature). One potential way of better preserving the biological information in a signature score is an algorithm where each sample is given a score value by subtracting the sum of downregulated genes from the sum of upregulated signature genes. More complex algorithms for derivation of gene expression signatures exists [134]. Which algorithm to select depends on how the signature gene list is derived, and the question you want to reply to by use of the signature.
Gene Networks Differentially Enriched Between Classes
Gaining further insight into biological mechanisms involved in a given process is a major challenge when working on high-throughput gene expression data. Subramanian et al. pointed to a few of the obstacles in how to interpret the single-gene lists into new and/or relevant biological information [35]: We may miss information about pathway alterations by single-gene analyses, as the interpretation of these is heavily dependent on the researcher’s pre-existing knowledge of the field. Pathway signaling may involve large gene networks and thus should not be too focused on “large enough” fold changes of single genes in the search for biological information in our data output. Minor changes in all genes known to be involved in a signaling pathway may be of higher importance than large fold changes of a few genes.
Gene Set Enrichment Analysis (GSEA) is a method that determines whether an a priori defined set of genes shows statistically significant differences between two classes (e.g., phenotypes). GSEA is an open access tool (www.broadinstitute.org/gsea), incorporating The Molecular Signatures Database (MSigDB), a publicly available collection of seven major classes of annotated gene sets (www.broadinstitute.org/gsea/msigdb). The gene expression signatures applied in GSEA/MSigDB are generated in various ways, and caution needs to be drawn when interpreting the results. To draw conclusions on gene set analyses, it is crucial to understand how the gene sets and signatures in question are generated, evaluating whether the specific gene sets are relevant for the current study. As for all large-scale analyses, considering the adjustment for multiple testing is required before interpreting the analysis output.
Linking gene expression alterations to network patterns of experimentally verified protein–protein interactions (PPI) provides improved understanding of the transcriptional patterns underlying the tumor and microenvironmental phenotypic characteristics [135, 136]. When analyzing the microenvironmental alterations and the interplay between the epithelial and microenvironmental compartments in tumor progression, integrating multiple levels of data will likely add information [137]. Large breast cancer studies have aimed at such integrative analyses, although a similar “all-level approach” not yet has been done with the microenvironment in focus.
Some Future Perspectives
Over the last decade, RNA sequencing has increasingly replaced the gene expression microarray method for global gene expression profiling. Analysis approaches have advanced, although a lot of the same analysis approaches are applied. Assessment of large-scale DNA, RNA, and protein single-cell information is one of the later methodological adds. Adding spatial information on top of the single-cell profiling seems like a promising approach for better elucidating network biology, intratumor heterogeneity, and the accompanying biological and clinical consequences. Imaging mass cytometry (IMC) assesses multiple protein-based markers with high-dimensional spatial resolution at the single-cell level—a promising tool for developing complex models of cellular interactions with particular relevance for the tumor microenvironment [138]. The method provides possibilities for multiplexed detection of up to 40 metal-bound proteins, showing promise regarding improved understanding of cancer biology, with accompanying development of functional biomarkers [139,140,141]. By applying this method, Jackson et al. recently described novel microenvironment subgroups splitting the classic molecular subtypes, informing clinical outcome [139]. More recent developments of the IMC method have provided a possibility to concurrently detect multiple mRNA and proteins at single-cell level, preserving the spatial information. A recent study describing this technology demonstrated strong correlation between HER2 mRNA and proteins at the cell population level in breast cancer (Fig. 23.6) [142]. Also, other platforms provide the possibility to detect concurrent single-cell co-expression of multiple transcripts and proteins [143, 144]. Combining mRNA and protein information into multi-level signatures, pointing to new subclasses in cancer, will likely be an approach exploited in future biomarker research.
To build comprehensive models of cellular states and interactions in normal and diseased tissue, genetic and proteomic information must be extracted with single-cell and spatial resolution. Schulz et al. extended imaging mass cytometry to enable multiplexed detection of mRNA and proteins in tissues. Three mRNA target species were detected by RNAscope-based metal in situ hybridization with simultaneous antibody detection of 16 proteins. Analysis of 170 breast cancer samples showed that HER2 and CK19 mRNA and protein levels are moderately correlated on the single-cell level, but only HER2, and not CK19, has strong mRNA-to-protein correlation on the cell population level. The chemoattractant CXCL10 was expressed in stromal cell clusters, and the frequency of CXCL-expressing cells correlated with T-cell presence. With permission, reprinted from D. Schulz et al. [142], Cell Syst 2018. doi: 10.1016/j.cels.2017.12.001
To understand how metastases are initiated and how they progress, Lawson and colleagues aimed to elucidate the properties of metastasis-initiating cells in human breast cancer. By single-cell analyses from early-stage metastatic lesions, Lawson demonstrated that cells from these lesions are characterized by a gene expression signature reflecting stemness [145]. Strikingly, the gene expression signature patterns in metastatic cells from tissues in early and advanced stage metastatic disease (patient-derived xenograft models) were distinctly different. The early-stage metastatic cells demonstrated increased expression of stem cell markers, epithelial-to-mesenchymal transition, as well as pro-survival and dormancy-associated genes. The metastatic cells from the advanced stage were more heterogeneous and displayed an expression pattern like the matched primary tumor. This study adds important information about the role of stem-like cells to the picture of the early stages of the metastasis process.
Two elegantly designed studies, linking information about tumor-stroma interactions, pointed at integrin signaling as being of major importance in tumor progression and in the organotropism of the metastatic lesions. Reuter and colleagues profiled gene expression data of both epithelium and stroma at specific time points during tumor progression in an experimental 3D tumor model [146]. A “core cancer progression signature ” was identified, and data indicated extracellular matrix-interacting network hubs as essential in tumor progression. Blocking the β1-integrin hub, inhibited tumor development. A study on the role of exosomes in the metastatic process demonstrated that tumor-derived exosomes prepare the pre-metastatic niche in organ-specific cells [147]. Lung and liver metastases were associated with specific integrin expression patterns. Targeting these integrins decreased the exosome uptake as well as lung and liver metastases, and Hoshino suggested that exosomal integrins have a potential role in directing metastatic cells in organotropic manners.
Deconvolution methods, a computational dissection of bulk gene expression data, providing cell compartment or cell type-specific counts or expression profiles, is a novel approach, potentially assisting in decoding complex data, with improved understanding of the tumor compartments [40, 148,149,150]. In a recent study by Zhu et al., gene expression data (by RNAseq) was analyzed from 50 primary breast tumors and their matched metastatic tumors [151]. Based on gene expression data, deconvolution methods demonstrated lower abundance of immune cells in the metastatic lesions, except for M2 macrophages, that occurred with higher levels in the metastatic lesions compared to primary tumors. Validation by immunohistochemistry analyses of tumor tissue confirmed the mRNA results, proposing immune escape as a potential mechanism for the lower infiltration of immune cells in metastases.
Concluding Remarks/Summary
Do signature approaches, as outlined in this chapter, seem promising when searching to understand the microenvironment biology in cancer? The summarized studies indicate that signature analyses are valuable tools in cancer research. Capturing gene expression alterations in multigene signatures may better reflect the complex biological programming both driving and supporting tumor development and progression. Stroma-related alterations are probably exploitable with respect to treatment identification. As underlined from many of the studies on transcriptional alterations of the tumor-associated microenvironment, interplay between extracellular remodeling, vascular biology and immune-related signaling appears to be critically important features of tumor subtypes and their associated patient outcomes. How to best reflect the functional interactions between the compartments is a daunting task. Integrating, interpreting, and validating results from global gene expression analyses are still major challenges, as deRisi stated in the very beginning of the “omics” era [24]. Developing new technology and analysis approaches, and steadily increasing the detection possibilities and the level of molecular complexity outlined, including molecular networks across molecular levels, provide new knowledge potentially impacting how we understand tumor biology and clinical diseases. Adding context-depending spatial information to large-scale single-cell data is proposed as a promising way forward—potentially further improving our understanding of microenvironment heterogeneity, and its biological and clinical consequences. How we embrace this methodology, should likely go along the line suggested by the mathematician Richard W. Hamming (1915–1998): If you believe too much, you will never notice the flaws; if you doubt too much you won’t get started. It requires a lovely balance.”
References
Fidler IJ. The pathogenesis of cancer metastasis: the ‘seed and soil’ hypothesis revisited. Nat Rev Cancer. 2003;3(6):453–8.
Talmadge JE, Fidler IJ. AACR centennial series: the biology of cancer metastasis: historical perspective. Cancer Res. 2010;70(14):5649–69.
Paget S. The distribution of secondary growths in cancer of the breast. Lancet. 1889;1:571–3.
Nguyen DX, Massague J. Genetic determinants of cancer metastasis. Nat Rev Genet. 2007;8(5):341–52.
Nguyen DX, Bos PD, Massague J. Metastasis: from dissemination to organ-specific colonization. Nat Rev Cancer. 2009;9(4):274–84.
Bhat R, Bissell MJ. Of plasticity and specificity: dialectics of the microenvironment and macroenvironment and the organ phenotype. Wiley Interdiscip Rev Dev Biol. 2014;3(2):147–63.
Boudreau A, van't Veer LJ, Bissell MJ. An “elite hacker”: breast tumors exploit the normal microenvironment program to instruct their progression and biological diversity. Cell Adhes Migr. 2012;6(3):236–48.
Wculek SK, Malanchi I. Neutrophils support lung colonization of metastasis-initiating breast cancer cells. Nature. 2015;528(7582):413–7.
Valastyan S, Weinberg RA. Tumor metastasis: molecular insights and evolving paradigms. Cell. 2011;147(2):275–92.
Oppenheimer SB. Cellular basis of cancer metastasis: a review of fundamentals and new advances. Acta Histochem. 2006;108(5):327–34.
Kaplan RN, Rafii S, Lyden D. Preparing the “soil”: the premetastatic niche. Cancer Res. 2006;66(23):11089–93.
Kaplan RN, Riba RD, Zacharoulis S, Bramley AH, Vincent L, Costa C, et al. VEGFR1-positive haematopoietic bone marrow progenitors initiate the pre-metastatic niche. Nature. 2005;438(7069):820–7.
Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403(6769):503–11.
Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP, et al. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286(5439):531–7.
Kim H, Nam SW, Rhee H, Shan Li L, Ju Kang H, Hye Koh K, et al. Different gene expression profiles between microsatellite instability-high and microsatellite stable colorectal carcinomas. Oncogene. 2004;23(37):6218–25.
Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–52.
Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98(19):10869–74.
Sotiriou C, Neo SY, McShane LM, Korn EL, Long PM, Jazaeri A, et al. Breast cancer classification and prognosis based on gene expression profiles from a population-based study. Proc Natl Acad Sci U S A. 2003;100(18):10393–8.
Li W, Wang R, Yan Z, Bai L, Sun Z. High accordance in prognosis prediction of colorectal cancer across independent datasets by multi-gene module expression profiles. PLoS One. 2012;7(3):e33653.
van ’t Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415(6871):530–6.
Wang Y, Klijn JG, Zhang Y, Sieuwerts AM, Look MP, Yang F, et al. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet. 2005;365(9460):671–9.
van de Vijver MJ, He YD, van’t Veer LJ, Dai H, Hart AA, Voskuil DW, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347(25):1999–2009.
Rouzier R, Rajan R, Wagner P, Hess KR, Gold DL, Stec J, et al. Microtubule-associated protein tau: a marker of paclitaxel sensitivity in breast cancer. Proc Natl Acad Sci U S A. 2005;102(23):8315–20.
DeRisi JL, Iyer VR, Brown PO. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278(5338):680–6.
Hughes TR, Marton MJ, Jones AR, Roberts CJ, Stoughton R, Armour CD, et al. Functional discovery via a compendium of expression profiles. Cell. 2000;102(1):109–26.
Huang E, Ishida S, Pittman J, Dressman H, Bild A, Kloos M, et al. Gene expression phenotypic models that predict the activity of oncogenic pathways. Nat Genet. 2003;34(2):226–30.
Bild AH, Yao G, Chang JT, Wang Q, Potti A, Chasse D, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439(7074):353–7.
Black EP, Huang E, Dressman H, Rempel R, Laakso N, Asa SL, et al. Distinct gene expression phenotypes of cells lacking Rb and Rb family members. Cancer Res. 2003;63(13):3716–23.
Lamb J, Ramaswamy S, Ford HL, Contreras B, Martinez RV, Kittrell FS, et al. A mechanism of cyclin D1 action encoded in the patterns of gene expression in human cancer. Cell. 2003;114(3):323–34.
Biomarkers Definitions Working G. Biomarkers and surrogate endpoints: preferred definitions and conceptual framework. Clin Pharmacol Ther. 2001;69(3):89–95.
Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100(1):57–70.
Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–74.
Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, Wrobel MJ, et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313(5795):1929–35.
Lamb J. The Connectivity Map: a new tool for biomedical research. Nat Rev Cancer. 2007;7(1):54–60.
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–50.
Sorlie T, Tibshirani R, Parker J, Hastie T, Marron JS, Nobel A, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci U S A. 2003;100(14):8418–23.
Manjili MH, Najarian K, Wang XY. Signatures of tumor-immune interactions as biomarkers for breast cancer prognosis. Future Oncol. 2012;8(6):703–11.
Zhao C, Wu M, Zeng N, Xiong M, Hu W, Lv W, et al. Cancer-associated adipocytes: emerging supporters in breast cancer. J Exp Clin Cancer Res. 2020;39(1):156.
Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12(5):453–7.
Peng XL, Moffitt RA, Torphy RJ, Volmar KE, Yeh JJ. De novo compartment deconvolution and weight estimation of tumor samples using DECODER. Nat Commun. 2019;10(1):4729.
Allinen M, Beroukhim R, Cai L, Brennan C, Lahti-Domenici J, Huang H, et al. Molecular characterization of the tumor microenvironment in breast cancer. Cancer Cell. 2004;6(1):17–32.
Ma XJ, Dahiya S, Richardson E, Erlander M, Sgroi DC. Gene expression profiling of the tumor microenvironment during breast cancer progression. Breast Cancer Res. 2009;11(1):R7.
Roman-Perez E, Casbas-Hernandez P, Pirone JR, Rein J, Carey LA, Lubet RA, et al. Gene expression in extratumoral microenvironment predicts clinical outcome in breast cancer patients. Breast Cancer Res. 2012;14(2):R51.
Troester MA, Lee MH, Carter M, Fan C, Cowan DW, Perez ER, et al. Activation of host wound responses in breast cancer microenvironment. Clin Cancer Res. 2009;15(22):7020–8.
Finak G, Bertos N, Pepin F, Sadekova S, Souleimanova M, Zhao H, et al. Stromal gene expression predicts clinical outcome in breast cancer. Nat Med. 2008;14(5):518–27.
Casey T, Bond J, Tighe S, Hunter T, Lintault L, Patel O, et al. Molecular signatures suggest a major role for stromal cells in development of invasive breast cancer. Breast Cancer Res Treat. 2009;114(1):47–62.
Van den Eynden GG, Smid M, Van Laere SJ, Colpaert CG, Van der Auwera I, Bich TX, et al. Gene expression profiles associated with the presence of a fibrotic focus and the growth pattern in lymph node-negative breast cancer. Clin Cancer Res. 2008;14(10):2944–52.
Marchini C, Montani M, Konstantinidou G, Orru R, Mannucci S, Ramadori G, et al. Mesenchymal/stromal gene expression signature relates to basal-like breast cancers, identifies bone metastasis and predicts resistance to therapies. PLoS One. 2010;5(11):e14131.
Guo W, Keckesova Z, Donaher JL, Shibue T, Tischler V, Reinhardt F, et al. Slug and Sox9 cooperatively determine the mammary stem cell state. Cell. 2012;148(5):1015–28.
Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133(4):704–15.
Morel AP, Lievre M, Thomas C, Hinkal G, Ansieau S, Puisieux A. Generation of breast cancer stem cells through epithelial-mesenchymal transition. PLoS One. 2008;3(8):e2888.
Planche A, Bacac M, Provero P, Fusco C, Delorenzi M, Stehle JC, et al. Identification of prognostic molecular features in the reactive stroma of human breast and prostate cancer. PLoS One. 2011;6(5):e18640.
Winslow S, Leandersson K, Edsjo A, Larsson C. Prognostic stromal gene signatures in breast cancer. Breast Cancer Res. 2015;17:23.
Desmedt C, Majjaj S, Kheddoumi N, Singhal SK, Haibe-Kains B, El Ouriaghli F, et al. Characterization and clinical evaluation of CD10+ stroma cells in the breast cancer microenvironment. Clin Cancer Res. 2012;18(4):1004–14.
Rajski M, Zanetti-Dallenbach R, Vogel B, Herrmann R, Rochlitz C, Buess M. IGF-I induced genes in stromal fibroblasts predict the clinical outcome of breast and lung cancer patients. BMC Med. 2010;8:1.
Bredholt G, Mannelqvist M, Stefansson IM, Birkeland E, Bo TH, Oyan AM, et al. Tumor necrosis is an important hallmark of aggressive endometrial cancer and associates with hypoxia, angiogenesis and inflammation responses. Oncotarget. 2015;6(37):39676–91.
West RB, Nuyten DS, Subramanian S, Nielsen TO, Corless CL, Rubin BP, et al. Determination of stromal signatures in breast carcinoma. PLoS Biol. 2005;3(6):e187.
Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y, et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest. 2011;121(7):2750–67.
Bareche Y, Buisseret L, Gruosso T, Girard E, Venet D, Dupont F, et al. Unraveling triple-negative breast cancer tumor microenvironment heterogeneity: towards an optimized treatment approach. J Natl Cancer Inst. 2020;112(7):708–19.
Tamborero D, Rubio-Perez C, Muinos F, Sabarinathan R, Piulats JM, Muntasell A, et al. A pan-cancer landscape of interactions between solid tumors and infiltrating immune cell populations. Clin Cancer Res. 2018;24(15):3717–28.
Qian J, Olbrecht S, Boeckx B, Vos H, Laoui D, Etlioglu E, et al. A pan-cancer blueprint of the heterogeneous tumor microenvironment revealed by single-cell profiling. Cell Res. 2020;30(9):745–62.
Chang HY, Sneddon JB, Alizadeh AA, Sood R, West RB, Montgomery K, et al. Gene expression signature of fibroblast serum response predicts human cancer progression: similarities between tumors and wounds. PLoS Biol. 2004;2(2):E7.
Dvorak HF. Tumors: wounds that do not heal. Similarities between tumor stroma generation and wound healing. N Engl J Med. 1986;315(26):1650–9.
Tchou J, Kossenkov AV, Chang L, Satija C, Herlyn M, Showe LC, et al. Human breast cancer associated fibroblasts exhibit subtype specific gene expression profiles. BMC Med Genet. 2012;5:39.
Bauer M, Su G, Casper C, He R, Rehrauer W, Friedl A. Heterogeneity of gene expression in stromal fibroblasts of human breast carcinomas and normal breast. Oncogene. 2010;29(12):1732–40.
Singer CF, Gschwantler-Kaulich D, Fink-Retter A, Haas C, Hudelist G, Czerwenka K, et al. Differential gene expression profile in breast cancer-derived stromal fibroblasts. Breast Cancer Res Treat. 2008;110(2):273–81.
Frings O, Augsten M, Tobin NP, Carlson J, Paulsson J, Pena C, et al. Prognostic significance in breast cancer of a gene signature capturing stromal PDGF signaling. Am J Pathol. 2013;182(6):2037–47.
Sonnenblick A, Salmon-Divon M, Salgado R, Dvash E, Ponde N, Zahavi T, et al. Reactive stroma and trastuzumab resistance in HER2-positive early breast cancer. Int J Cancer. 2020;147(1):266–76.
Siletz A, Kniazeva E, Jeruss JS, Shea LD. Transcription factor networks in invasion-promoting breast carcinoma-associated fibroblasts. Cancer Microenviron. 2013;6(1):91–107.
Bartoschek M, Oskolkov N, Bocci M, Lovrot J, Larsson C, Sommarin M, et al. Spatially and functionally distinct subclasses of breast cancer-associated fibroblasts revealed by single cell RNA sequencing. Nat Commun. 2018;9(1):5150.
Wu SZ, Roden DL, Wang C, Holliday H, Harvey K, Cazet AS, et al. Stromal cell diversity associated with immune evasion in human triple-negative breast cancer. EMBO J. 2020;39(19):e104063.
Woelfle U, Cloos J, Sauter G, Riethdorf L, Janicke F, van Diest P, et al. Molecular signature associated with bone marrow micrometastasis in human breast cancer. Cancer Res. 2003;63(18):5679–84.
Bergamaschi A, Tagliabue E, Sorlie T, Naume B, Triulzi T, Orlandi R, et al. Extracellular matrix signature identifies breast cancer subgroups with different clinical outcome. J Pathol. 2008;214(3):357–67.
Triulzi T, Casalini P, Sandri M, Ratti M, Carcangiu ML, Colombo MP, et al. Neoplastic and stromal cells contribute to an extracellular matrix gene expression profile defining a breast cancer subtype likely to progress. PLoS One. 2013;8(2):e56761.
Wallgard E, Larsson E, He L, Hellstrom M, Armulik A, Nisancioglu MH, et al. Identification of a core set of 58 gene transcripts with broad and specific expression in the microvasculature. Arterioscler Thromb Vasc Biol. 2008;28(8):1469–76.
Cleuren ACA, van der Ent MA, Jiang H, Hunker KL, Yee A, Siemieniak DR, et al. The in vivo endothelial cell translatome is highly heterogeneous across vascular beds. Proc Natl Acad Sci U S A. 2019;116(47):23618–24.
Hu Z, Fan C, Livasy C, He X, Oh DS, Ewend MG, et al. A compact VEGF signature associated with distant metastases and poor outcomes. BMC Med. 2009;7:9.
Pepin F, Bertos N, Laferriere J, Sadekova S, Souleimanova M, Zhao H, et al. Gene-expression profiling of microdissected breast cancer microvasculature identifies distinct tumor vascular subtypes. Breast Cancer Res. 2012;14(4):R120.
Bender RJ, Mac GF. Expression of VEGF and semaphorin genes define subgroups of triple negative breast cancer. PLoS One. 2013;8(5):e61788.
Wallace JA, Li F, Balakrishnan S, Cantemir-Stone CZ, Pecot T, Martin C, et al. Ets2 in tumor fibroblasts promotes angiogenesis in breast cancer. PLoS One. 2013;8(8):e71533.
Xiao L, Harrell JC, Perou CM, Dudley AC. Identification of a stable molecular signature in mammary tumor endothelial cells that persists in vitro. Angiogenesis. 2014;17(3):511–8.
Chang CW, Tsai CW, Wang HF, Tsai HC, Chen HY, Tsai TF, et al. Identification of a developmentally regulated striatum-enriched zinc-finger gene, Nolz-1, in the mammalian brain. Proc Natl Acad Sci U S A. 2004;101(8):2613–8.
Matsumoto K, Nishihara S, Kamimura M, Shiraishi T, Otoguro T, Uehara M, et al. The prepattern transcription factor Irx2, a target of the FGF8/MAP kinase cascade, is involved in cerebellum formation. Nat Neurosci. 2004;7(6):605–12.
Mannelqvist M, Stefansson IM, Bredholt G, Hellem Bo T, Oyan AM, Jonassen I, et al. Gene expression patterns related to vascular invasion and aggressive features in endometrial cancer. Am J Pathol. 2011;178(2):861–71.
Mannelqvist M, Wik E, Stefansson IM, Akslen LA. An 18-gene signature for vascular invasion is associated with aggressive features and reduced survival in breast cancer. PLoS One. 2014;9(6):e98787.
Stefansson IM, Raeder M, Wik E, Mannelqvist M, Kusonmano K, Knutsvik G, et al. Increased angiogenesis is associated with a 32-gene expression signature and 6p21 amplification in aggressive endometrial cancer. Oncotarget. 2015;6(12):10634–45.
Tobin NP, Wennmalm K, Lindstrom LS, Foukakis T, He L, Genove G, et al. An endothelial gene signature score predicts poor outcome in patients with endocrine-treated, low genomic grade breast tumors. Clin Cancer Res. 2016;22(10):2417–26.
Guarischi-Sousa R, Monteiro JS, Alecrim LC, Michaloski JS, Cardeal LB, Ferreira EN, et al. A transcriptome-based signature of pathological angiogenesis predicts breast cancer patient survival. PLoS Genet. 2019;15(12):e1008482.
Harrell JC, Pfefferle AD, Zalles N, Prat A, Fan C, Khramtsov A, et al. Endothelial-like properties of claudin-low breast cancer cells promote tumor vascular permeability and metastasis. Clin Exp Metastasis. 2014;31(1):33–45.
Mendiola M, Martinez-Marin V, Herranz J, Heredia V, Yebenes L, Zamora P, et al. Predictive value of angiogenesis-related gene profiling in patients with HER2-negative metastatic breast cancer treated with bevacizumab and weekly paclitaxel. Oncotarget. 2016;7(17):24217–27.
Kruger K, Silwal-Pandit L, Wik E, Straume O, Stefansson IM, Borgen E, et al. Baseline microvessel density predicts response to neoadjuvant bevacizumab treatment of locally advanced breast cancer. Sci Rep. 2021;11(1):3388.
Pitroda SP, Zhou T, Sweis RF, Filippo M, Labay E, Beckett MA, et al. Tumor endothelial inflammation predicts clinical outcome in diverse human cancers. PLoS One. 2012;7(10):e46104.
Oshi M, Newman S, Tokumaru Y, Yan L, Matsuyama R, Endo I, et al. Intra-tumoral angiogenesis is associated with inflammation, immune reaction and metastatic recurrence in breast cancer. Int J Mol Sci. 2020;21(18):6708.
Chen DS, Mellman I. Elements of cancer immunity and the cancer-immune set point. Nature. 2017;541(7637):321–30.
Joyce JA, Fearon DT. T cell exclusion, immune privilege, and the tumor microenvironment. Science. 2015;348(6230):74–80.
Adams S, Diamond JR, Hamilton E, Pohlmann PR, Tolaney SM, Chang CW, et al. Atezolizumab plus nab-paclitaxel in the treatment of metastatic triple-negative breast cancer with 2-year survival follow-up: a phase 1b clinical trial. JAMA Oncol. 2019;5(3):334–42.
Loi S, Giobbie-Hurder A, Gombos A, Bachelot T, Hui R, Curigliano G, et al. Pembrolizumab plus trastuzumab in trastuzumab-resistant, advanced, HER2-positive breast cancer (PANACEA): a single-arm, multicentre, phase 1b-2 trial. Lancet Oncol. 2019;20(3):371–82.
Schmid P, Adams S, Rugo HS, Schneeweiss A, Barrios CH, Iwata H, et al. Atezolizumab and nab-paclitaxel in advanced triple-negative breast cancer. N Engl J Med. 2018;379(22):2108–21.
Schmid P, Rugo HS, Adams S, Schneeweiss A, Barrios CH, Iwata H, et al. Atezolizumab plus nab-paclitaxel as first-line treatment for unresectable, locally advanced or metastatic triple-negative breast cancer (IMpassion130): updated efficacy results from a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol. 2020;21(1):44–59.
Solinas C, Gombos A, Latifyan S, Piccart-Gebhart M, Kok M, Buisseret L. Targeting immune checkpoints in breast cancer: an update of early results. ESMO Open. 2017;2(5):e000255.
Emens LA. Immunotherapy in triple-negative breast cancer. Cancer J. 2021;27(1):59–66.
Teschendorff AE, Miremadi A, Pinder SE, Ellis IO, Caldas C. An immune response gene expression module identifies a good prognosis subtype in estrogen receptor negative breast cancer. Genome Biol. 2007;8(8):R157.
Rody A, Karn T, Liedtke C, Pusztai L, Ruckhaeberle E, Hanker L, et al. A clinically relevant gene signature in triple negative and basal-like breast cancer. Breast Cancer Res. 2011;13(5):R97.
Alexe G, Dalgin GS, Scanfeld D, Tamayo P, Mesirov JP, DeLisi C, et al. High expression of lymphocyte-associated genes in node-negative HER2+ breast cancers correlates with lower recurrence rates. Cancer Res. 2007;67(22):10669–76.
Schmidt M, Bohm D, von Torne C, Steiner E, Puhl A, Pilch H, et al. The humoral immune system has a key prognostic impact in node-negative breast cancer. Cancer Res. 2008;68(13):5405–13.
Schmidt M, Hellwig B, Hammad S, Othman A, Lohr M, Chen Z, et al. A comprehensive analysis of human gene expression profiles identifies stromal immunoglobulin kappa C as a compatible prognostic marker in human solid tumors. Clin Cancer Res. 2012;18(9):2695–703.
Hsu DS, Kim MK, Balakumaran BS, Acharya CR, Anders CK, Clay T, et al. Immune signatures predict prognosis in localized cancer. Cancer Investig. 2010;28(7):765–73.
Bianchini G, Qi Y, Alvarez RH, Iwamoto T, Coutant C, Ibrahim NK, et al. Molecular anatomy of breast cancer stroma and its prognostic value in estrogen receptor-positive and -negative cancers. J Clin Oncol. 2010;28(28):4316–23.
Nagalla S, Chou JW, Willingham MC, Ruiz J, Vaughn JP, Dubey P, et al. Interactions between immunity, proliferation and molecular subtype in breast cancer prognosis. Genome Biol. 2013;14(4):R34.
Ascierto ML, Kmieciak M, Idowu MO, Manjili R, Zhao Y, Grimes M, et al. A signature of immune function genes associated with recurrence-free survival in breast cancer patients. Breast Cancer Res Treat. 2012;131(3):871–80.
Iglesia MD, Vincent BG, Parker JS, Hoadley KA, Carey LA, Perou CM, et al. Prognostic B-cell signatures using mRNA-seq in patients with subtype-specific breast and ovarian cancer. Clin Cancer Res. 2014;20(14):3818–29.
Coronella JA, Spier C, Welch M, Trevor KT, Stopeck AT, Villar H, et al. Antigen-driven oligoclonal expansion of tumor-infiltrating B cells in infiltrating ductal carcinoma of the breast. J Immunol. 2002;169(4):1829–36.
Hansen MH, Nielsen H, Ditzel HJ. The tumor-infiltrating B cell response in medullary breast cancer is oligoclonal and directed against the autoantigen actin exposed on the surface of apoptotic cancer cells. Proc Natl Acad Sci U S A. 2001;98(22):12659–64.
Nzula S, Going JJ, Stott DI. Antigen-driven clonal proliferation, somatic hypermutation, and selection of B lymphocytes infiltrating human ductal breast carcinomas. Cancer Res. 2003;63(12):3275–80.
Perez EA, Thompson EA, Ballman KV, Anderson SK, Asmann YW, Kalari KR, et al. Genomic analysis reveals that immune function genes are strongly linked to clinical outcome in the north central cancer treatment group n9831 adjuvant trastuzumab trial. J Clin Oncol. 2015;33(7):701–8.
Rody A, Holtrich U, Pusztai L, Liedtke C, Gaetje R, Ruckhaeberle E, et al. T-cell metagene predicts a favorable prognosis in estrogen receptor-negative and HER2-positive breast cancers. Breast Cancer Res. 2009;11(2):R15.
Akalay I, Janji B, Hasmim M, Noman MZ, Andre F, De Cremoux P, et al. Epithelial-to-mesenchymal transition and autophagy induction in breast carcinoma promote escape from T-cell-mediated lysis. Cancer Res. 2013;73(8):2418–27.
Akalay I, Tan TZ, Kumar P, Janji B, Mami-Chouaib F, Charpy C, et al. Targeting WNT1-inducible signaling pathway protein 2 alters human breast cancer cell susceptibility to specific lysis through regulation of KLF-4 and miR-7 expression. Oncogene. 2015;34(17):2261–71.
Kudo-Saito C, Shirako H, Takeuchi T, Kawakami Y. Cancer metastasis is accelerated through immunosuppression during Snail-induced EMT of cancer cells. Cancer Cell. 2009;15(3):195–206.
Ye X, Weinberg RA. Epithelial-mesenchymal plasticity: a central regulator of cancer progression. Trends Cell Biol. 2015;25(11):675–86.
Yang B, Chou J, Tao Y, Wu D, Wu X, Li X, et al. An assessment of prognostic immunity markers in breast cancer. NPJ Breast Cancer. 2018;4:35.
Zhang H, Qin G, Yu H, Han X, Zhu S. Comprehensive genomic and immunophenotypic analysis of CD4 T cell infiltrating human triple-negative breast cancer. Cancer Immunol Immunother. 2021;70(6):1649–65.
Gruosso T, Gigoux M, Manem VSK, Bertos N, Zuo D, Perlitch I, et al. Spatially distinct tumor immune microenvironments stratify triple-negative breast cancers. J Clin Invest. 2019;129(4):1785–800.
Wu Q, Li B, Li Z, Li J, Sun S, Sun S. Cancer-associated adipocytes: key players in breast cancer progression. J Hematol Oncol. 2019;12(1):95.
Nieman KM, Kenny HA, Penicka CV, Ladanyi A, Buell-Gutbrod R, Zillhardt MR, et al. Adipocytes promote ovarian cancer metastasis and provide energy for rapid tumor growth. Nat Med. 2011;17(11):1498–503.
Wang YY, Attane C, Milhas D, Dirat B, Dauvillier S, Guerard A, et al. Mammary adipocytes stimulate breast cancer invasion through metabolic remodeling of tumor cells. JCI Insight. 2017;2(4):e87489.
Wu Z, Wu J, Zhao Q, Fu S, Jin J. Emerging roles of aerobic glycolysis in breast cancer. Clin Transl Oncol. 2020;22(5):631–46.
Tang J, Luo Y, Wu G. A glycolysis-related gene expression signature in predicting recurrence of breast cancer. Aging (Albany NY). 2020;12(24):24983–94.
Li C, Li X, Li G, Sun L, Zhang W, Jiang J, et al. Identification of a prognosisassociated signature associated with energy metabolism in triplenegative breast cancer. Oncol Rep. 2020;44(3):819–37.
Zhang D, Zheng Y, Yang S, Li Y, Wang M, Yao J, et al. Identification of a novel glycolysis-related gene signature for predicting breast cancer survival. Front Oncol. 2020;10:596087.
Sturtz LA, Deyarmin B, van Laar R, Yarina W, Shriver CD, Ellsworth RE. Gene expression differences in adipose tissue associated with breast tumorigenesis. Adipocytes. 2014;3(2):107–14.
Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95(25):14863–8.
Simon RMKE, McShane LM, Radmacher MD, Wright GW, Zhao Y. Statistics for biology and health: design and analysis of DNA microarray investigations. New York, NY: Springer; 2004.
Kato K. Algorithm for in vitro diagnostic multivariate index assay. Breast Cancer. 2009;16(4):248–51.
Deng JL, Xu YH, Wang G. Identification of potential crucial genes and key pathways in breast cancer using bioinformatic analysis. Front Genet. 2019;10:695.
Tang D, Zhao X, Zhang L, Wang Z, Wang C. Identification of hub genes to regulate breast cancer metastasis to brain by bioinformatics analyses. J Cell Biochem. 2019;120(6):9522–31.
Petersen K, Rajcevic U, Abdul Rahim SA, Jonassen I, Kalland KH, Jimenez CR, et al. Gene set based integrated data analysis reveals phenotypic differences in a brain cancer model. PLoS One. 2013;8(7):e68288.
Giesen C, Wang HA, Schapiro D, Zivanovic N, Jacobs A, Hattendorf B, et al. Highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry. Nat Methods. 2014;11(4):417–22.
Jackson HW, Fischer JR, Zanotelli VRT, Ali HR, Mechera R, Soysal SD, et al. The single-cell pathology landscape of breast cancer. Nature. 2020;578(7796):615–20.
Wagner J, Rapsomaniki MA, Chevrier S, Anzeneder T, Langwieder C, Dykgers A, et al. A single-cell atlas of the tumor and immune ecosystem of human breast cancer. Cell. 2019;177(5):1330–45 e18.
Schapiro D, Jackson HW, Raghuraman S, Fischer JR, Zanotelli VRT, Schulz D, et al. histoCAT: analysis of cell phenotypes and interactions in multiplex image cytometry data. Nat Methods. 2017;14(9):873–6.
Schulz D, Zanotelli VRT, Fischer JR, Schapiro D, Engler S, Lun XK, et al. Simultaneous multiplexed imaging of mRNA and proteins with subcellular resolution in breast cancer tissue samples by mass cytometry. Cell Syst. 2018;6(1):25–36 e5.
Brady L, Kriner M, Coleman I, Morrissey C, Roudier M, True LD, et al. Inter- and intra-tumor heterogeneity of metastatic prostate cancer determined by digital spatial gene expression profiling. Nat Commun. 2021;12(1):1426.
Merritt CR, Ong GT, Church SE, Barker K, Danaher P, Geiss G, et al. Multiplex digital spatial profiling of proteins and RNA in fixed tissue. Nat Biotechnol. 2020;38(5):586–99.
Lawson DA, Bhakta NR, Kessenbrock K, Prummel KD, Yu Y, Takai K, et al. Single-cell analysis reveals a stem-cell program in human metastatic breast cancer cells. Nature. 2015;526(7571):131–5.
Reuter JA, Ortiz-Urda S, Kretz M, Garcia J, Scholl FA, Pasmooij AM, et al. Modeling inducible human tissue neoplasia identifies an extracellular matrix interaction network involved in cancer progression. Cancer Cell. 2009;15(6):477–88.
Hoshino A, Costa-Silva B, Shen TL, Rodrigues G, Hashimoto A, Tesic Mark M, et al. Tumour exosome integrins determine organotropic metastasis. Nature. 2015;527(7578):329–35.
Li B, Li T, Liu JS, Liu XS. Computational deconvolution of tumor-infiltrating immune components with bulk tumor gene expression data. Methods Mol Biol. 2020;2120:249–62.
Racle J, Gfeller D. EPIC: a tool to estimate the proportions of different cell types from bulk gene expression data. Methods Mol Biol. 2020;2120:233–48.
Newman AM, Steen CB, Liu CL, Gentles AJ, Chaudhuri AA, Scherer F, et al. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat Biotechnol. 2019;37(7):773–82.
Zhu L, Narloch JL, Onkar S, Joy M, Broadwater G, Luedke C, et al. Metastatic breast cancers have reduced immune cell recruitment but harbor increased macrophages relative to their matched primary tumors. J Immunother Cancer. 2019;7(1):265.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this chapter
Cite this chapter
Wik, E., Ingebriktsen, L.M., Akslen, L.A. (2022). Gene Expression Signatures of the Tumor Microenvironment: Relation to Tumor Phenotypes and Progress in Breast Cancer. In: Akslen, L.A., Watnick, R.S. (eds) Biomarkers of the Tumor Microenvironment. Springer, Cham. https://doi.org/10.1007/978-3-030-98950-7_23
Download citation
DOI: https://doi.org/10.1007/978-3-030-98950-7_23
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-98949-1
Online ISBN: 978-3-030-98950-7
eBook Packages: MedicineMedicine (R0)