Abstract
The goal of this chapter is to review the current state of the emerging field of circadian rhythms and exercise metabolism. Since this area of research is fairly new, we have broken up this chapter into the following sections. (1) The role of circadian rhythms and in maintaining homeostasis; (2) introduction to the circadian clock mechanism and clock output; (3) the role of the circadian clock in regulation of resting fat and carbohydrate metabolism in the skeletal muscle; and (4) interactions between exercise and circadian rhythms. We hope that this chapter can serve as a reference and/or entry point for scientists wanting to integrate circadian concepts in their understanding or research design of exercise metabolism.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
16.1 Circadian Rhythms Underlie Predictive Homeostasis in Physiology
One of the most common questions we are asked about circadian rhythms and physiology is about how or whether this concept of daily variation has implications for our understanding of exercise and metabolism. Our simple answer is that yes, because circadian rhythms are a fundamental and essential mechanism of cell homeostasis. To help frame the role of circadian clocks in maintaining homeostasis, we provide a historical perspective of the interaction between circadian biology, physiology, and homeostasis. Walter Cannon defined homeostasis as the property of a system in which a variable, such as blood glucose, is actively regulated to remain very nearly constant (Moore-Ede 1986). For many years, the majority of mechanisms in place to support homeostasis were reactive processes. For example, after a meal increased blood glucose is sensed, the pancreas releases insulin which functions to facilitate glucose uptake into tissues and bringing blood glucose levels back down. However, in 1986, Dr. Martin Moore-Ede presented the Bowditch Lecture at the American Physiological Society meeting and discussed the concept of “predictive homeostasis” based on several physiological parameters, such as heart rate and cortisol, that were known to change prior to a predictable event such as awakening (Moore-Ede 1986). Dr. Moore-Ede proposed that our systems evolved in an environment with very predictable environmental changes, such as light-dark, and as such there were intrinsic mechanisms in place that anticipate these changes. These predictive homeostatic mechanisms allowed for more rapid adjustments with benefits for survival. It is now clear that the circadian timing mechanism is found in virtually all cells within the body which are critical for predictive homeostasis. Consistent with the physiological benefits of this predictive system, it is now clear that disruption to the circadian timing mechanism, or predictive homeostasis, leads to diminished ability of the system to maintain homeostasis with negative health outcomes. Thus, it is critical to understand that circadian rhythms and the clocks responsible are a fundamental part of daily cell and systems physiology. Additionally, the fact that circadian rhythms have been demonstrated at the subcellular level (e.g., chromatin availability) at the cell and tissue levels (e.g., insulin sensitivity) and systems level (e.g., heart rate and behavior) serves as a reminder that our resting cell, tissue, and system physiology is constantly oscillating. Thus, when we introduce exercise or exercise training, we do so on a moving physiology and metabolic baseline.
16.1.1 A Brief History of Circadian Biology and Exercise Physiology
Exercise physiology and circadian biology are well-established fields with rich histories. The history of exercise physiology is often traced to the musings of ancient Greek physicians (Tipton 2003), while circadian biology can trace roots back around 400 B.C. when Androsthenes, a scribe for Alexander the Great, noted that the leaves of certain plants displayed opening and closing patterns coincident with sunrise and sunset (Persson and Bondke Persson 2019). Despite these histories, integration of these fundamental concepts has been relatively recent, seemingly starting in the late 1940s, but expanding rapidly in the last 10 years.
One of the first circadian and exercise studies looked at time of day as a variable for human physical work capacity (reviewed in: Kleitman 1949). Subsequent studies examined athletic performance at a few times of day (Wahlberg and Astrand 1973; Conroy and O’Brien 1974), while the first experiments to explore circadian variation in exercise tolerance were completed in the late 1960s (Voigt et al. 1968; Crockford and Davies 1969). Contrary to our current understanding, the initial work did not identify circadian variation in exercise performance, though the authors point out this could have been due to the repeated bouts of exercise across the day or large interindividual variability. However, similar to the initial work in chronobiology, the initial integration of circadian concepts into exercise physiology were more systemic (e.g., exercise capacity or heart rate) and not on the molecular scale. Only with the discovery of the molecular components of the circadian timing mechanism has the interest in how exercise and circadian biology work together been identified and explored. For those interested in additional and/or more detailed training in circadian biology, there are numerous online resources, including a digital course directed by Drs. Martha Merrow and Till Roenneberg (https://www.coursera.org/learn/circadian-clocks).
16.2 The Core Molecular Clock Mechanism
Daily rhythms in physiology and metabolism can be considered in a framework of predictive homeostasis, and it is now clear that these daily rhythms are driven by the circadian clock mechanism. In this section we provide an overview of the clock to provide critical framework for integration into exercise metabolism studies. The core molecular clock is a timing mechanism that exists in virtually all cells in the body and is highly conserved across mammals. The basis of the clock is a transcription/translation feedback loop that maintains an approximately 24 h period independent of external inputs (time cues: zeitgebers). The clock is comprised of a set of molecules that form the positive limb (considered transcriptional activators) and a different set of molecules that form the negative limb (considered transcriptional repressors). The components and function of the molecular clock have been presented in significant detail, and reviews of the state of knowledge of the clock mechanism can be found here (Mohawk et al. 2012; Takahashi 2017).
16.2.1 The Components of the Molecular Clock
The Positive Arm of the Molecular Clock
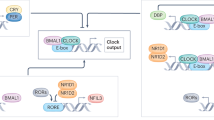
In mammalian tissues, the positive limb of the molecular clock is driven by two transcription factors. Brain and muscle ARNT-like 1 (BMAL1, also known as ARNTL) and CLOCK, these are basic helix-loop-helix Per-Arnt-Sim (bHLH-PAS) transcription factors. These transcription factors form a heterodimer and bind to E-box sequences (5’-CANNTG-3′, and 5’-CACGTG-3′ most often) across the genome to activate transcription. Importantly, BMAL1/CLOCK bind the promoters of the core circadian genes Period (Pers) and cryptochrome (Crys), transcriptionally activating expression of these negative arm genes (Fig. 16.1). Seminal work from the Takahashi lab revealed more than 5000 binding sites for both CLOCK and BMAL1 across the liver genome over a 24 h period and over 75% of those targeted genes were transcriptionally active (Koike et al. 2012). These findings highlight two key points in molecular clock function. First, that BMAL1 and CLOCK directly regulate the Per/Cry genes as a key part of the molecular clock. Second, BMAL1 and CLOCK bind in a time of day-dependent manner to 1000s of sites across the genome, demonstrating that in addition to keeping a 24 h cycle of the Per/Cry families, the core molecular clock is contributing, in a time of day manner, to a diverse transcriptional network within the cell (Takahashi 2017).
A simplified core circadian clock. A simplified schematic that depicts the core circadian transcription factors BMAL1 and CLOCK which heterodimerize and transcribe the circadian repressors Period (Pers) and Cryptochrome (Crys) families of genes. The Pers and Crys mRNAs are translated in the cytoplasm where they form a multimers with casein kinase (CK delta and epsilon). This repressor complex translocates into the nucleus to repress the transcriptional activity of the BMAL1:CLOCK heterodimer. This process takes roughly 24 hours to complete a cycle
The Negative Arm of the Molecular Clock
The negative arm of the core clock is comprised of the period (Per1, Per2, and Per3) and cryptochrome (Cry1 and Cry2) gene families. Studies have shown that, the Per and Cry families of genes are transcribed by BMAL1 and CLOCK in the middle of the rest phase (e.g., afternoon for nocturnal rodents), leading to their protein expression levels peaking in the late rest phase. Upon translation, the PER and CRY proteins interact with each other, as well as with kinases, such as casein kinase 1δ and 1ε (CK1 δ/ε). These protein complexes translocate to the nucleus where they inhibit BMAL1/CLOCK transcriptional activity and thereby decrease their own transcription. As PER/CRY/CK1 represses BMAL1/CLOCK transcription, the abundance of repressive transcripts declines, as does the relative content of the repressor complex proteins, as they have short half-lives due to proteasomal degradation. Once the transcriptional repression subsides, BMAL1/CLOCK transcription increases, beginning the next circadian transcriptional cycle.
Similar to the widespread genomic localization observed for BMAL/CLOCK, the PER/CRY proteins also bind at 1000s of sites across the genome (Koike et al. 2012). While some of the genomic sites are overlapping between the positive and negative arm, indicative of the expected transcription repression by the negative arm on the positive arm, several hundred sites are not shared between positive and negative arm proteins. The specific role of the PER/CRY families binding to other sites of the genome is currently underexplored, though some data suggest the negative arm proteins serve as mediators of metabolic input into circadian timing (Lamia et al. 2009; Schmutz et al. 2010). Additional work exploring the potential functions of the negative arm proteins independent of the circadian time keeping mechanism is warranted. Together, however, the core molecular clock mechanism is responsible for the daily transcriptional program contributing to the cellular capacity to maintain homeostasis.
16.2.2 Clock Output: The Daily Program of Transcription Underlying the “Moving Baseline” in Cell Physiology
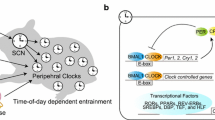
The daily pattern of gene expression outside of the core clock is referred to as the circadian clock output (Fig. 16.2). The most common approach to define the circadian clock output for a tissue is to assess the total number of oscillating mRNAs over a 24 h–48 h time course. To date, numerous studies have explored the circadian clock output in muscle as well as other tissues and found the number of oscillating genes represent up to 50% of known protein coding genes (Miller et al. 2007; McCarthy et al. 2007; Pizarro et al. 2012; Zhang et al. 2014; Mure et al. 2018). Restated, the circadian clock output comprises a significant portion of the daily transcriptional program within a tissue, including contributing to daily oscillations in mRNAs associated with metabolism, and general cellular functions. Importantly, the daily variation of metabolic gene expression is not random, but serves to temporally align cellular metabolic gene expression with both rest and active behavior as well as patterns of food availability and intake. In a simple sense, the clock mechanism in muscle functions to temporally segregate periods of fuel oxidation vs. storage. For the purpose of this chapter, the metabolic clusters of genes regulated by the circadian clock in muscle provide the framework for our understanding of intersection of circadian biology with exercise metabolism.
Output from the core circadian clock. The BMAL1:CLOCK heterodimer also contributes to a daily program of gene expression in all cells, and this is called the circadian clock output. The circadian clock output is specific to each tissue and is important for a number of different cellular functions, including transcriptional regulation, metabolism, and homeostasis
16.2.3 Environmental Inputs Adjust the Timing of the Clock and Clock Output
A fundamental principle of the circadian clock is that the period length is set at ~24 h, but cues from the environment can modify or alter the timing or phase of the clock. The ability to move the phase of the clock allows the timing system and its output to adapt to changes in light/dark cycles. The ability to modulate phase means that the clock and clock output can adjust to the environmental changes and supports the function of predictive homeostasis. To date, a majority of the work exploring the mechanisms through which the molecular clock senses and responds to environmental inputs have focused on how the circadian system responds to the timing of light cues. Research in this area has demonstrated that the clock mechanism is differentially sensitive to the environmental time cue (Aschoff 1965; Pittendrigh and Daan 1976). This means that the exact same cue can induce one of three predictable outcomes, (1) a phase advance, (2) a phase delay, and (3) no change; the outcomes depend on the status of the clock mechanism at the time at which the zeitgeber is received/sensed. The importance of this aspect of circadian biology in relationship to exercise and metabolism is that time of exercise can result in changes in phase of peripheral tissue clocks which then modifies the metabolic factors that are part of clock output.
We now know that both feeding and exercise provide environmental cues that impact the phase settings of clocks in peripheral tissues. In particular, current data have demonstrated that one acute exercise bout is sufficient to alter the phase of peripheral circadian clocks in human (Youngstedt et al. 2019) as well as rodent (Gannon and Rea 1995; Schroeder et al. 2012; Kemler et al. 2020) models. Most recently, studies have shown that exercise, like light, can induce a predictable phase response. This means that if one exercises earlier than when they are normally active the phase of the muscle clock will advance and in contrast, if one exercises later than when they are normally active, the phase of the muscle clock will delay. If one exercises around their normal active period we predict that there would be no shift in the phase of the muscle clock. These predictions are primarily based on the effects of an acute bout of exercise. And as a reminder, if the clock shifts in phase, then this will also shift the clock output (Youngstedt et al. 2019; Kemler et al. 2020). Therefore, it is not surprising that there are time of day specific effects of exercise on transcriptomic and metabolomic responses to the exercise bout (Ezagouri et al. 2019; Sato et al. 2019). The known mechanisms through which exercise influences the timing of the molecular clock output are discussed in Sect. 16.4.1. Since this field has only matured in the last 2 years, the profound diversity of exercise interventions, such as resistance vs. endurance, as well as exercise bout duration and intensity represent a wide range of important new areas of research.
16.3 The Role of the Molecular Clock in Daily Patterns of Muscle Metabolism
The ultimate focus of this chapter is on the intersection of circadian rhythms and exercise metabolism. However to extract exercise-specific effects, one first has to have an understanding of the daily changes in metabolism that occur downstream of the circadian clock at rest. Thus, this section reviews the current understanding of substrate metabolism changes downstream of the circadian clock in the skeletal muscle. In particular, we highlight the interplay between circadian control, glucose, and lipid metabolism during steady-state conditions.
16.3.1 Circadian Control of Glucose Metabolism
Skeletal muscle is the largest insulin sensitive organ in the body, and it plays an essential role in whole-body glucose homeostasis being responsible for ~85% of insulin-stimulated glucose uptake (DeFronzo and Tripathy 2009). Insulin sensitivity in the skeletal muscle also displays robust circadian variations in mice (Dyar et al. 2014; Aras et al. 2019) and humans (de Goede et al. 2018). The circadian variation in skeletal muscle insulin sensitivity, peaking during the beginning of the active/feeding phase, stimulates a rise in tissue glucose uptake and oxidation in anticipation of increased locomotor activity. In contrast, during the resting/fasting phase, the muscle displays reduced insulin sensitivity leading to a decrease in glucose uptake and glycolytic flux in muscle cells. Disruption of the intrinsic muscle clock causes muscle insulin resistance and altered muscle glucose metabolism, showing insulin-dependent glucose uptake in skeletal muscle reduced by muscle-specific inactivation of Bmal1 (Dyar et al. 2014). One potential explanation for the daily change in muscle insulin sensitivity is the daily oscillation of the muscle glucose transporter GLUT4 displaying highest expression during active/feeding phase (Harfmann et al. 2016). The circadian control of GLUT4 has been clearly demonstrated as loss of Bmal1 in skeletal muscle leads to a significant reduction (~45%) in GLUT4 protein levels across the diurnal cycle compared to control (Dyar et al. 2014; Harfmann et al. 2016).
In addition to the circadian variation in insulin sensitivity and glucose uptake, studies have shown that the muscle clock contributes to variation in pyruvate dehydrogenase, PDH activity, as well as the two rate limiting enzymes in glucose metabolism: hexokinase and phosphofructokinase (Dyar et al. 2014). Pizarro and colleagues (2013) report hexokinase-2 expression displays a circadian rhythm that peaks in at the beginning of the active/feeding phase, as well as the genes responsible for phosphofructokinase expression (Pfkfb 1/3/4) which also display circadian rhythmicity and peak in expression during the mid-late resting/fasting phase in humans (Pizarro et al. 2012). However, in muscle in which the circadian clock is stopped (i.e., muscle-specific Bmal1 knockout mice), the expression of these enzymes (i.e., PDH, Hk2, Pfkfb) is dampened resulting in a reduced protein expression and diminished enzyme activity, indicating impaired glycolytic flux (Dyar et al. 2014; Hodge et al. 2015; Harfmann et al. 2016). Together these data illustrate that the muscle clock modulates both glucose uptake and utilization over time of day in skeletal muscle.
Excess carbohydrate is stored as glycogen in skeletal muscle which accounts for approximately 70–80% of whole-body stores (Ivy et al. 1988; Jensen et al. 2011). Unlike the liver, skeletal muscle glycogen content is not responsible for maintaining blood glucose concentrations, rather it is a rapidly accessible energy store utilized during active contractions (Jensen et al. 2011), with glycogenesis regulated by the enzymatic activity of both glucose-6-phosphate and glycogen synthase (Viijlar-Palasí and Guinovart 1997) and glycogen breakdown regulated by phosphorylase (Howlett et al. 1998; Jensen and Richter 2012). Previous studies have reported that there is a diurnal rhythm of glycogen content in skeletal muscle, displaying highest values during the mid-active/feeding phase (Leighton et al. 1988). Interestingly, muscle-specific knockout of the clock gene, Bmal1, is associated with significant increases in muscle glycogen content. Whether this clock-dependent change in muscle glycogen is due to diminished glycolytic flux or alteration in glycogenolysis is not yet know (Harfmann et al. 2016).
In summary, the circadian clock in muscle is important for regulating pathways involved in glucose uptake/insulin sensitivity, glucose utilization, and glucose storage. Thus, independent of exercise there are time of day differences in these important muscle metabolic parameters. A key job of the muscle clock is to prepare the tissue for the activity onset following a fast upon awakening and for changes in substrate availability associated with feeding. These temporal patterns likely have consequences for exercise outcome measures. For example, if the phase of the muscle clock is disrupted, such as during shift work or jet lag, it would be predicted that this would result in impaired exercise performance, in part, through a reduction in glucose uptake and expression of key glycolytic enzymes.
16.3.2 Circadian Control of Lipid Metabolism
The muscle clock also has been shown to contribute to the regulation of skeletal muscle lipid oxidation over time of day. Genes that regulate each step of lipid metabolism, from fatty acid transport to oxidation and back to storage, are known to exhibit circadian rhythms (Zhang et al. 2014; Hodge et al. 2015; Dyar et al. 2018a). Fatty acid transfer into the inner mitochondrial matrix occurs in a time of day manner, peaking in the early to mid-active phase, to support oxidative metabolism with energy availability. Analysis of microarray datasets from circadian time course studies reveals that the gene encoding for acyl-carnitine translocase, Slc25a20, reaches peak expression in the middle of the resting/fasting period (Indiveri et al. 2011; Zhang et al. 2014). Further, two genes that encode for lipid transport are fatty acid binding proteins Fabp3 and 4, which are also expressed in a circadian manner, showing highest mRNA expression levels in the early and mid-resting/fasting periods (Syamsunarno et al. 2013; Hodge et al. 2015; Dyar et al. 2018a). Consistent with these data, multiple genes that encode for β-oxidation, such as enoyl CoA hydratase, tri-functional enzyme subunits Hadh α/β, and the acetyl-CoA acyltransferase, have also been identified to be circadian, reaching peak expression around the mid-resting/fasting phase (Zhang et al. 2014). These observations are consistent with predictive homeostasis with important enzymes for oxidative metabolism increasing prior to waking and activity onset. In addition, these observations illustrate that the muscle clock can regulate with malonyl-CoA, known to promote β-oxidation reaching peak expression during similar rest period as that of the circadian β-oxidation genes, aligning lipid metabolism to fit a circadian profile in skeletal muscle (Schmidt and Herpin 1998; Saggerson 2008). In contrast to increases in fat oxidation patterns during rest, the lipogenic genes Acly, Acaca, and Fasn all peak expression at the end of the active/feeding phase (Funai and Semenkovich 2011; Ameer et al. 2014) Thus, the muscle clock contributes to a baseline pattern that temporally segregates fat oxidation versus fat storage over time of day.
Analysis of the circadian time course from metabolomic and lipidomic data from mice in which the core clock gene, Bmal1, is knocked out was performed in 2018 by Dyar et al., The major findings were that loss of muscle clock function led to significant perturbations in fatty acid, triglyceride, and phospholipid metabolism. For example, Bmal1 KO mice displayed reduced levels of muscle triglycerides, and this was associated with significant downregulation of Dgat2 gene expression. Dgat2 is implicated in fat storage and known to encode for the enzyme responsible for the conversion of diacylglycerols to triglycerides. Interestingly, this finding was concomitant with the upregulation of genes known to be involved in fatty acid synthesis (Dyar et al. 2018b), thus serving to effectively flip the temporal periods of lipid storage and utilization in animals without a functional muscle clock. Since these mice maintain a normal central clock and their activity and feeding patterns are normal, this flipping of substrate storage and utilization in Bmal1 KO mice illustrates the significant contribution that the circadian clock makes to substrate metabolism under resting conditions.
16.4 Exercise, Circadian Rhythms, and Metabolism
The temporal gating of substrate metabolism by circadian clocks is an important component of homeostasis, highlighted by studies in which circadian rhythms are disrupted being linked to a variety of metabolic diseases (Masri and Sassone-Corsi 2018). The majority of daily physiological processes, including many aspects of metabolism, undergo changes about the relative time of day. Many of these metabolic variations, which we refer to as the “moving baseline,” are downstream of the circadian clock within each tissue, including skeletal muscle. In this section, we discuss two topics; (1) what is understood about the integration of exercise metabolism and circadian rhythms in skeletal muscle and (2) what happens when we exercise at different times of the day.
It has been recognized for over 10 years that exercise at different times of the day leads to distinct performance outcomes. For example, daily variations in resistance and endurance exercise peak performance have been reported to fluctuate when studied during the active/feeding phase in humans (Souissi et al. 2004; Ab Malik et al. 2020; Mirizio et al. 2020) and rodents (Ezagouri et al. 2019). As such, consistently, studies across species have revealed that variables such as skeletal muscle strength and oxidative capacity demonstrate significant differences in performance outcomes over time of day (Atkinson and Reilly 1996; van Moorsel et al. 2016; de Goede et al. 2018). For example, numerous studies have demonstrated increased maximal isometric strength in the later afternoon versus morning (Douglas et al. 2021), while oxidative capacity peaks in the late evening (Reilly and Waterhouse 2009; van Moorsel et al. 2016). Thus, these exercise performance outcomes provide evidence that the circadian timing of our metabolism throughout the day (i.e., morning vs. afternoon) likely impacts exercise response and in turn performance outcomes such as endurance capacity and maximal strength (Ezagouri et al. 2019; Douglas et al. 2021).
16.4.1 The Influence of Circadian Rhythms on Acute Exercise Responses
Daily variations in exercise capacity likely stem from differences in metabolic starting points derived from the daily changes in the metabolic status that differs between early and late exercisers. For example, exercise during the early active/feeding phase, when hepatic glycogen content is reduced, rather than exercise at the late active/feeding phase, when hepatic glycogen content is increased, results in the rapid depletion of carbohydrate energy stores in skeletal muscle and a shift toward utilization of fatty acid metabolism thereby favoring endurance type exercise. Ezagouri et al. (2019) investigated this concept by assigning animals to sedentary and exercise groups during either the early (ZT14) or late (ZT22) active period, subjecting both wild-type and double Per1/2 knockout mice to moderate-intensity exercise (reported ~55% VO2max). Interestingly, wild-type mice ran ~67% longer at ZT22 than ZT13, whereas the time of day effect on endurance capacity was abolished in the Per1/2 knockouts. These results were also corroborated in healthy, young humans who were subjected to a submaximal constant-load exercise protocol, equivalent to the moderate-intensity exercise protocol of the mice. Each participant performed the exercise protocol on two occasions (0800 and 1800), separated by appropriate wash out period. Strikingly, maximal oxygen consumption (VO2max) was significantly lower at 1800 than 0800, but the respiratory exchange ratio (RER) was significantly higher at 1800 than 0800, indicating greater use of carbohydrate metabolism in the later exercisers. However, blood glucose levels were reported to be higher at exercise performed at 0800 in comparison to 1800. Previous studies have shown a greater aerobic capacity in the evening compared to the morning hours in humans, with improved capacity associated with a greater reliance on carbohydrates (higher RER) (Drust et al. 2005; Reilly and Waterhouse 2009; Küüsmaa et al. 2016; Thosar et al. 2018). These findings are consistent with the rodent data described above (Ezagouri et al. 2019) and thus suggest that the time of day differences in aerobic capacity are likely orchestrated through the circadian clock impact on substrate metabolism in muscle and likely other metabolic tissues.
Another important question that arises from the “moving baseline concept” is whether the time of exercise elicits a differential molecular response. Analysis of clock output data from rodents and humans provides perspective on the underlying variation of gene expression over time of day (McCarthy et al. 2007; Andrews et al. 2010; Zhang et al. 2014; Perrin et al. 2018). Now, when the exact same bout of exercise is performed at two distinctly different times of day, Sato et al. and Ezagouri et al. (2019) found that the exercise-induced gene expression changes were significantly different. While this may be surprising, when you consider that the exercise intervention is occurring on a moving, and not flat, transcriptomic baseline, this outcome is actually expected (Fig. 16.3). Work from two different labs over the last 2+ years highlights the impact of time of day to an acute response to treadmill exercise. In the study by Ezagouri et al., (2019) they analyzed skeletal muscle gene expression and the metabolite profile from two different exercised groups of mice during the early active vs. late active phase and found that a greater number of significantly regulated transcripts were unique to exercise at the early active phase, ZT14 (343 genes), rather than those unique to the late active phase, ZT22 (125 genes), with a there was a modest overlap of 160 genes (Ezagouri et al. 2019). Functional cluster analysis of the differentially regulated RNAseq data found that genes associated with insulin signaling and glucose metabolism were upregulated when exercise was performed at early active phase, ZT14, but not with later exercise at ZT22. In contrast, genes linked to the FoxO signaling pathway were the more enriched category in the late exercise, ZT22 group. Functional analysis of the genes that changed in common between groups was many nuclear encoded mitochondrial genes.
Distinct time of day gene expression responses to treadmill exercise. Treadmill running exercise at the early inactive phase (ZT3) versus the early active phase (ZT15) or at the early active phase (ZT14) compared to the late active phase (ZT22) produces different transcriptional responses. Examples of functional enrichment pathways are provided to demonstrate differential responses based on time of day. The venn diagrams display the upregulated genes only as a percentage of significantly regulated genes, unique to each exercise period: ZT3 vs. ZT15 (blue and green) and ZT14 vs. ZT22 (green and red). All data adapted from Sato et al. (2019) and Ezagouri et al. (2019). Running mouse images were downloaded from BioRender.com
A similar study was reported in Sato et al. (2019), in which they subjected mice to a 1 h acute bout of treadmill exercise at the early resting/fasting phase (ZT3) or during the early active/feeding phase (ZT15) with respective control groups (Sato et al. 2019). For reference, both the Sato et al. and Ezagouri et al. studies have one group of mice that exercise in the later active phase (ZT14 or ZT15). However, the Sato et al. (2019) paper had their second group of mice run at ZT3 which is running at 3 hours in the light/rest phase. Transcriptomic analysis of gastrocnemius muscle revealed that both times of exercise demonstrated a significant change in gene expression. However, there was only a small overlap between the number of exercise-responsive transcripts that were upregulated (12%) and downregulated (5%) when comparing the exercise response at ZT3 to ZT15, confirming a large time of day specificity. Moreover, gene ontology clustering of exercise-responsive genes at different times of day also highlighted the transcriptional response to exercise is time of day specific. This was consistent with metabolomic analysis, which showed upregulated and downregulated metabolites also have little overlap, 13% and 1%, respectively, between exercise at different time points. While this does not address potential changes with long-term training, it does highlight the interaction of exercise and the circadian clock, providing evidence that clock output is different over time of day even in response to an acute exercise intervention.
Exercise outcomes within important signaling pathways for metabolism have also been reported to be affected by circadian timing in a time of day-specific manner. For example, the mechanistic target of rapamycin complex 1 (mTORC1) and peroxisome proliferator activated receptor gamma coactivator 1 alpha (PGC1α) are two pathways that have been widely reported in the exercise physiology literature. However, these exercise stimulated signaling pathways have also been identified as being downstream of the molecular clock, thus providing a mechanism by which circadian timing may modulate exercise responses (Um et al. 2011; Ramanathan et al. 2018). Further, there is evidence to suggest that the core components of the molecular clock can interact with some of these pathways. For example, PER2, which peaks at the end of the resting/fasting phase, reduces mTORC1 activity in the liver (Wu et al. 2019). However, it is not yet clear whether the interaction of clock components and such pathways are conserved across tissues, e.g., skeletal muscle. Nevertheless, the molecular responses of exercise in skeletal muscle exhibit a clear time-dependent effect for acute exercise. For example, resistance exercise performed in humans at the early active/feeding phase leads to acute mTORC1 activation in skeletal muscle, whereas the same exercise in the late active/feeding phase has been documented to produce a more blunted signaling response (Sedliak et al. 2009). Despite this influence of acute resistance exercise bout performed at different times on hypertrophic signaling, training studies have found that when the resistance exercise is performed at different times of day, there is no difference in the magnitude of skeletal muscle hypertrophy (Küüsmaa et al. 2016; Sedliak et al. 2018). However, it is important to note that training (regularly repeated exercise bouts) at either morning or afternoon/evening will lead to shifting of the muscle clock and clock output in phase. Thus, these findings to not rule out that time of exercise does not matter for muscle growth, but rather they reinforce that maintaining a regular exercise training schedule is important. This is a very new area of research, and thus, there is still very much to be learned.
16.4.2 Exercise Can Target the Molecular Clock and Modify the Moving Baseline
Exercise and scheduled physical activity have been established to be environmental time cues, or zeitgeber, that can modify the phase of the muscle clock. The evidence in support of this has been accumulating since the late 1980s and early 1990s when novel wheel access at different times of day was found to be sufficient to shift the phase of circadian behavioral rhythms in mice and hamsters (Edgar et al. 1991; Edgar and Dement 1991). In follow-up work to these original studies, forced treadmill exercise training in rodents (Wolff and Esser 2012; Schroeder et al. 2012) and humans (Youngstedt et al. 2019) further confirmed exercise serves as a zeitgeber. Specifically, depending on the time of exercise, the muscle clock, and its output, will shift in phase in a predictable manner.
The mechanisms through which exercise modifies the phase of the molecular clock and subsequently the clock output are incompletely understood, but have been the target of numerous recent investigations. Emerging data demonstrate that muscle contractions, as a component of exercise, can directly modulate the expression of core clock components in a time of day-dependent manner. Kemler et al. (et al. 2020) subjected PER2::LUC circadian reporter mice to an acute bout of 60 min treadmill exercise at three different times of day (Kemler et al. 2020). Exercise at ZT5 induced a phase advance of the clock, whereas exercise at ZT11 induced a phase delay of the clock. However, exercise in the middle of the active phase, at ZT17, did not alter the muscle clock phase. These time of day-specific responses mirror the anticipated phase response curves. Ex vivo muscle contraction studies showed that the expression of Per2 is modified acutely, and this results in an altered phase (Small et al. 2020). Small et al. (2020) provided more mechanistic data finding that calcium influx stimulated by contraction leads to the binding of the phosphorylated form of cAMP response element-binding protein (CREB) to the Per2 promoter. Together, these experiments point to CREB activation as a key mechanism, whereby exercise can alter the phase of the circadian clock in muscle.
While exercise seems to directly affect the expression levels of the core clock genes, well-known exercise-induced kinases have been also shown to modify the phase of the circadian clock. While still early, these associations suggest the potential for these kinases to also contribute to the muscle clock and clock output. For the purpose of this chapter, we will discuss four well-known exercise-induced factors that have been reported to interact with the clock mechanism (Fig. 16.4). Specifically, exercise induces activation of the cAMP response element binding protein (CREB), 5’ AMP-activated protein kinase (AMPK), hypoxia inducible factor 1 alpha (HIF-1α), and sirtuin 1 (SIRT1). We review the current understanding of these four exercise and circadian factors in the following paragraphs.
Exercise as an input to the core circadian clock. Exercise is an effector of the core skeletal muscle circadian clock. Bouts of acute exercise increase the phosphorylation of CRE binding protein 1 (CREB) and AMP-activated protein kinase (AMPK) and upregulate hypoxia-inducible factor 1 alpha (HIF-1α) and the NAD-dependent deacetylase sirtuin 1 (SIRT1). Individually, each of these exercise responsive targets has been linked to changes in core clock gene expression or protein levels. The anticipated effect of these changes in core clock gene/protein expression is changes in the timing of the circadian system. Solid lines represent direct experimental evidence, while dotted lines represent anticipated exercise-induced effects. Running mouse images were downloaded from BioRender.com
Exercise increases CREB phosphorylation, and CREB plays a role in changing circadian phase in other tissues. Recent data in skeletal muscle following acute exercise or electrical pulse stimulated contractions indicate that there are also CREB-induced increases in Per2 expression (Small et al. 2020). CREB is activated by increases in cAMP levels and protein kinase A signaling and calcium-specific signaling, as well as through activation of the MAP kinase pathways. It is currently unclear if exercise activates CREB through one or a combination of all of these pathways. Additionally, the time of day-specific effects of exercise on core clock gene expression (Ezagouri et al. 2019; Sato et al. 2019; Kemler et al. 2020) suggest that either (1) exercise-induced CREB activity is different over time of day or (2) the effect of CREB of core clock gene expression varies over time of day. Further work is needed to explore these potential differences and inform the mechanism of CREB-induced changes in core clock gene expression and the subsequent changes in the timing of the circadian phase and output.
AMPK is a well-known exercise-responsive protein (Richter and Ruderman 2009). AMPK is often studied in its context as an energy sensor and thereby metabolic regulator. Therefore the interaction between AMPK and the molecular clock potentially represents the primary interface linking changes in metabolism to change in circadian timing. Current data indicate that AMPK affects the timing of the circadian clock through altering the stability of CRY1 proteins through phosphorylation and subsequent targeted degradation of CRY1 by the proteasome (Lamia et al. 2009). While the endurance exercise-induced activation of AMPK was not different at different times of day (Ezagouri et al. 2019), the AMPK-induced reduction in CRY1 content is beneficial for the upregulation of fatty acid oxidation genes (Jordan et al. 2017). Further, the CRY proteins have a strong repressive effect on these lipid metabolism genes, reiterating the daily variation in metabolic gene expression is influenced directly by the core clock machinery.
Endurance exercise also increases the expression of HIF-1α, a protein with a structure similar to Bmal1 (Wu et al. 2017). And HIF-1α may contribute to the exercise-induced transcription of core clock genes (Adamovich et al. 2014; Peek et al. 2017). Importantly, loss of HIF-1α was required for the molecular clock adaptation to an experimental jet-lag paradigm, suggesting a role of HIF-1α in phase changes, which is specifically remodeling the timing of the molecular clock output. Moreover, there was a time of day-specific effect of an acute exercise bout on both the induction of HIF-1α and the corresponding increase in HIF-1α gene targets (Peek et al. 2017). Recent data also indicate that HIF-1α binds to the Per2 promoter and increases its expression, potentially explaining its role in changing circadian phase (Wu et al. 2017).
Finally, current data suggest that SIRT1, the NAD+ dependent deacetylase, can target both the positive and negative arms of the molecular clock. SIRT1 not only binds to BMAL1:CLOCK to increase amplitude of circadian transcription, but it also deacetylates PER2, increasing its degradation (i.e., repressing the repressor) (Asher et al. 2008; Foteinou et al. 2018). The SIRT1-induced increase in circadian output amplitude may represent a mechanism through which exercise training enhances the function of the circadian clock, though data are needed to support this postulation. Additionally, because SIRT1 contributes to changes in histone acetylation, SIRT1-induced changes in chromatin structure may influence the exercise-induced circadian output, but more data are still needed.
While we have highlighted the interaction between exercise-inducible factors and the molecular clock, additional targets likely exist that will further explain how exercise modifies the timing of the circadian clock and its output. Moreover these data have focused on skeletal muscle, and the exercise effect on the molecular clock in other peripheral tissues remains largely unexplored. Regardless of the mechanism through which exercise is modifying the timing of the core circadian clock, there are clear exercise-induced changes in the timing of the core circadian clock with implications for the timing of its output (e.g., lipid oxidation genes). These exercise-induced changes in circadian clock output timing therefore likely result in differences in daily substrate metabolism, insulin sensitivity, and other processes that contribute to maintaining homeostasis within tissues and across the organism. Finally, most of the data discussed in the context of exercise and the circadian clock are from acute exercise studies. The effect of exercise training on the circadian clock and clock output remains significantly understudied and represents an exciting future direction of the circadian exercise metabolism field.
16.5 Conclusions
In summary, circadian rhythms and the circadian clock are fundamental parts of our cell and systems biology. The circadian clocks function to support homeostatis through their role regulating gene expression in anticipation of predictable changes in the environment and behavior. It is well established that the circadian clock in muscle and other tissues function to regulate gene expression and the tissue metabolome in a time of day manner. A significant component of clock output is genes that are important for substrate metabolism including pathways for fuel oxidation as well as storage. These findings are supported by studies in both humans and rodents and highlight that the metabolic response to an acute bout of exercise will differ based on time of day. While these findings are quite clear, there are still many large gaps in the field. For example, the majority of exercise studies are done with more endurance exercise with very limited data on resistance exercise. We also know very little about how exercise intensity or duration impacts aspects of circadian clock output. The issue of the impact of training, and not just acute exercise, is also not well studied. These large gaps represent exciting new opportunities for both basic exercise science and applied and clinical interventions. In particular there is growing interest in whether time of day exercise strategies could be helpful with preventing or delaying the development of metabolic diseases. Lastly, we want to note that moving forward, future exercise studies in both human and rodent interventions must take care to provide transparent reporting of circadian conditions (e.g., light/dark cycles, feeding status, and habitual activity), as well as robust time of day sampling rates with appropriate controls in order to draw meaningful comparisons and interpretations from the data.
References
Ab Malik Z, Bowden Davies KA, Hall ECR et al (2020) Diurnal differences in human muscle isometric force in vivo are associated with differential phosphorylation of Sarcomeric M-band proteins. Proteomes 8:22
Adamovich Y, Rousso-Noori L, Zwighaft Z et al (2014) Circadian clocks and feeding time regulate the oscillations and levels of hepatic triglycerides. Cell Metab 19:319–330
Ameer F, Scandiuzzi L, Hasnain S et al (2014) De novo lipogenesis in health and disease. Metabolism 63:895–902
Andrews JL, Zhang X, McCarthy JJ et al (2010) CLOCK and BMAL1 regulate MyoD and are necessary for maintenance of skeletal muscle phenotype and function. Proc Natl Acad Sci 107:19090–19095
Aras E, Ramadori G, Kinouchi K et al (2019) Light entrains diurnal changes in insulin sensitivity of skeletal muscle via ventromedial hypothalamic neurons. Cell Rep 27:2385–2398.e3
Aschoff J (1965) CIRCADIAN RHYTHMS IN MAN. Science 148:1427–1432
Asher G, Gatfield D, Stratmann M et al (2008) SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell 134:317–328
Atkinson G, Reilly T (1996) Circadian variation in sports performance. Sport Med 21:292–312
Conroy RT, O’Brien M (1974) Proceedings: diurnal variation in athletic performance. J Physiol 236:51P
Crockford GW, Davies CT (1969) Circadian variations in responses to submaximal exercise on a bicycle ergometer. J Physiol 201:94P–95P
de Goede P, Wefers J, Brombacher EC et al (2018) Circadian rhythms in mitochondrial respiration. J Mol Endocrinol 60:R115–R130
DeFronzo RA, Tripathy D (2009) Skeletal muscle insulin resistance is the primary defect in type 2 diabetes. Diabetes Care 32:S157–S163
Douglas CM, Hesketh SJ, Esser KA (2021) Time of day and muscle strength: a circadian output? Physiology 36:44–51
Drust B, Waterhouse J, Atkinson G et al (2005) Circadian rhythms in sports performance--an update. Chronobiol Int 22:21–44
Dyar KA, Ciciliot S, Wright LE et al (2014) Muscle insulin sensitivity and glucose metabolism are controlled by the intrinsic muscle clock. Mol Metab 3:29–41
Dyar KA, Hubert MJ, Mir AA et al (2018a) Transcriptional programming of lipid and amino acid metabolism by the skeletal muscle circadian clock. PLoS Biol 16:e2005886
Dyar KA, Lutter D, Artati A et al (2018b) Atlas of circadian metabolism reveals system-wide coordination and communication between clocks. Cell 174:1571–1585.e11
Edgar DM, Dement WC (1991) Regularly scheduled voluntary exercise synchronizes the mouse circadian clock. Am J Physiol Integr Comp Physiol 261:R928–R933
Edgar DM, Martin CE, Dement WC (1991) Activity feedback to the mammalian circadian pacemaker: influence on observed measures of rhythm period length. J Biol Rhythm 6:185–199
Ezagouri S, Zwighaft Z, Sobel J et al (2019) Physiological and molecular dissection of daily variance in exercise capacity. Cell Metab 30:78–91.e4
Foteinou PT, Venkataraman A, Francey LJ et al (2018) Computational and experimental insights into the circadian effects of SIRT1. Proc Natl Acad Sci U S A 115:11643–11648
Funai K, Semenkovich CF (2011) Skeletal muscle lipid flux: running water carries no poison. Am J Physiol Metab 301:E245–E251
Gannon RL, Rea MA (1995) Twelve-hour phase shifts of hamster circadian rhythms elicited by voluntary wheel running. J Biol Rhythm 10:196–210
Harfmann BD, Schroder EA, Kachman MT et al (2016) Muscle-specific loss of Bmal1 leads to disrupted tissue glucose metabolism and systemic glucose homeostasis. Skelet Muscle 6:12
Hodge BA, Wen Y, Riley LA et al (2015) The endogenous molecular clock orchestrates the temporal separation of substrate metabolism in skeletal muscle. Skelet Muscle 5:17
Howlett RA, Parolin ML, Dyck DJ et al (1998) Regulation of skeletal muscle glycogen phosphorylase and PDH at varying exercise power outputs. Am J Phys 275:R418–R425
Indiveri C, Iacobazzi V, Tonazzi A et al (2011) The mitochondrial carnitine/acylcarnitine carrier: function, structure and physiopathology. Mol Asp Med 32:223–233
Ivy JL, Katz AL, Cutler CL et al (1988) Muscle glycogen synthesis after exercise: effect of time of carbohydrate ingestion. J Appl Physiol 64:1480–1485
Jensen TE, Richter EA (2012) The journal of physiology regulation of glucose and glycogen metabolism during and after exercise. J Physiol 590:1069–1076
Jensen J, Rustad PI, Kolnes AJ, Lai Y-C (2011) The role of skeletal muscle glycogen breakdown for regulation of insulin sensitivity by exercise. Front Physiol 2:112
Jordan SD, Kriebs A, Vaughan M et al (2017) CRY1/2 selectively repress PPARδ and limit exercise capacity. Cell Metab 26:243–255.e6
Kemler D, Wolff CA, Esser KA (2020) Time-of-day dependent effects of contractile activity on the phase of the skeletal muscle clock. J Physiol 598:3631–3644
Kleitman N (1949) Biological rhythms and cycles. Physiol Rev 29:1–30
Koike N, Yoo S-H, Huang H-C et al (2012) Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science 338:349–354
Küüsmaa M, Schumann M, Sedliak M et al (2016) Effects of morning versus evening combined strength and endurance training on physical performance, muscle hypertrophy, and serum hormone concentrations. Appl Physiol Nutr Metab 41:1285–1294
Lamia KA, Sachdeva UM, DiTacchio L et al (2009) AMPK regulates the circadian clock by cryptochrome phosphorylation and degradation. Science 326:437–440
Leighton B, Kowalchuk JM, Challiss RAJ, Newsholme EA (1988) Circadian rhythm in sensitivity of glucose metabolism to insulin in rat soleus muscle. Am J Physiol Metab 255:E41–E45
Masri S, Sassone-Corsi P (2018) The emerging link between cancer, metabolism, and circadian rhythms. Nat Med 24:1795–1803
McCarthy JJ, Andrews JL, McDearmon EL et al (2007) Identification of the circadian transcriptome in adult mouse skeletal muscle. Physiol Genomics 31:86–95
Miller BH, McDearmon EL, Panda S et al (2007) Circadian and CLOCK-controlled regulation of the mouse transcriptome and cell proliferation. Proc Natl Acad Sci U S A 104:3342–3347
Mirizio GG, Nunes RSM, Vargas DA et al (2020) Time-of-day effects on short-duration maximal exercise performance. Sci Rep 10:9485
Mohawk JA, Green CB, Takahashi JS (2012) Central and peripheral circadian clocks in mammals. Annu Rev Neurosci 35:445–462
Moore-Ede MC (1986) Physiology of the circadian timing system: predictive versus reactive homeostasis. Am J Physiol Integr Comp Physiol 250:R737–R752
Mure LS, Le HD, Benegiamo G et al (2018) Diurnal transcriptome atlas of a primate across major neural and peripheral tissues. Science (80-) 359:eaao0318
Peek CB, Levine DC, Cedernaes J et al (2017) Circadian clock interaction with HIF1α mediates oxygenic metabolism and anaerobic glycolysis in skeletal muscle. Cell Metab 25:86–92
Perrin L, Loizides-Mangold U, Chanon S et al (2018) Transcriptomic analyses reveal rhythmic and CLOCK-driven pathways in human skeletal muscle. elife 7:e34114
Persson PB, Bondke Persson A (2019) Circadian rhythms. Acta Physiol 225:e13220
Pittendrigh CS, Daan S (1976) A functional analysis of circadian pacemakers in nocturnal rodents. J comp Physiol ? A 106:333–355
Pizarro A, Hayer K, Lahens NF, Hogenesch JB (2012) CircaDB: a database of mammalian circadian gene expression profiles. Nucleic Acids Res 41:D1009–D1013
Ramanathan C, Kathale ND, Liu D et al (2018) mTOR signaling regulates central and peripheral circadian clock function. PLoS Genet 14:e1007369
Reilly T, Waterhouse J (2009) Sports performance: is there evidence that the body clock plays a role? Eur J Appl Physiol 106:321–332
Richter EA, Ruderman NB (2009) AMPK and the biochemistry of exercise: implications for human health and disease. Biochem J 418:261–275
Saggerson D (2008) Malonyl-CoA, a key signaling molecule in mammalian cells. Annu Rev Nutr 28:253–272
Sato S, Basse AL, Schönke M et al (2019) Time of exercise specifies the impact on muscle metabolic pathways and systemic energy homeostasis. Cell Metab 30:92–110.e4
Schmidt I, Herpin P (1998) Carnitine Palmitoyltransferase I (CPT I) activity and its regulation by Malonyl-CoA are modulated by age and cold exposure in skeletal muscle mitochondria from newborn pigs. J Nutr 128:886–893
Schmutz I, Ripperger JA, Baeriswyl-Aebischer S, Albrecht U (2010) The mammalian clock component PERIOD2 coordinates circadian output by interaction with nuclear receptors. Genes Dev 24:345–357
Schroeder AM, Truong D, Loh DH et al (2012) Voluntary scheduled exercise alters diurnal rhythms of behaviour, physiology and gene expression in wild-type and vasoactive intestinal peptide-deficient mice. J Physiol 590:6213–6226
Sedliak M, Finni T, Cheng S et al (2009) Effect of time-of-day-specific strength training on muscular hypertrophy in men. J Strength Cond Res 23:2451–2457
Sedliak M, Zeman M, Buzgó G et al (2018) Morphological, molecular and hormonal adaptations to early morning versus afternoon resistance training. Chronobiol Int 35:450–464
Small L, Altıntaş A, Laker RC et al (2020) Contraction influences Per2 gene expression in skeletal muscle through a calcium-dependent pathway. J Physiol 598:5739–5752
Souissi N, Gauthier A, Sesboüé B et al (2004) Circadian rhythms in two types of anaerobic cycle leg exercise: force-velocity and 30-s Wingate tests. Int J Sports Med 25:14–19
Syamsunarno MRAA, Iso T, Hanaoka H et al (2013) A critical role of fatty acid binding protein 4 and 5 (FABP4/5) in the systemic response to fasting. PLoS One 8:e79386
Takahashi JS (2017) Transcriptional architecture of the mammalian circadian clock. Nat Rev Genet 18:164–179
Thosar SS, Herzig MX, Roberts SA et al (2018) Lowest perceived exertion in the late morning due to effects of the endogenous circadian system. Br J Sports Med 52:1011–1012
Tipton CM (2003) Exercise physiology. Springer, New York, New York, NY
Um J-H, Pendergast JS, Springer DA et al (2011) AMPK regulates circadian rhythms in a tissue- and isoform-specific manner. PLoS One 6:e18450
van Moorsel D, Hansen J, Havekes B et al (2016) Demonstration of a day-night rhythm in human skeletal muscle oxidative capacity. Mol Metab 5:635–645
Viijlar-Palasí C, Guinovart JJ (1997) The role of glucose 6-phosphate in the control of glycogen synthase. FASEB J 11:544–558
Voigt ED, Engel P, Klein H (1968) On the daily operation of physical work capacity. Int Z Angew Physiol 25:1–12
Wahlberg I, Astrand I (1973) Physical work capacity during the day and at night. Work Environ Health 10:65–68
Wolff G, Esser KA (2012) Scheduled exercise phase shifts the circadian clock in skeletal muscle. Med Sci Sports Exerc 44:1663–1670
Wu Y, Tang D, Liu N et al (2017) Reciprocal regulation between the circadian clock and hypoxia signaling at the genome level in mammals. Cell Metab 25:73–85
Wu R, Dang F, Li P et al (2019) The circadian protein Period2 suppresses mTORC1 activity via recruiting Tsc1 to mTORC1 complex. Cell Metab 29:653–667.e6
Youngstedt SD, Elliott JA, Kripke DF (2019) Human circadian phase–response curves for exercise. J Physiol 597:2253–2268
Zhang R, Lahens NF, Ballance HI et al (2014) A circadian gene expression atlas in mammals: implications for biology and medicine. Proc Natl Acad Sci 111:16219–16224
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this chapter
Cite this chapter
Wolff, C.A., Hesketh, S.J., Esser, K.A. (2022). Circadian Rhythms and Exercise Metabolism. In: McConell, G. (eds) Exercise Metabolism. Physiology in Health and Disease. Springer, Cham. https://doi.org/10.1007/978-3-030-94305-9_16
Download citation
DOI: https://doi.org/10.1007/978-3-030-94305-9_16
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-94304-2
Online ISBN: 978-3-030-94305-9
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)