Abstract
Preterm birth (PTB) (<37 weeks’ gestational age (GA)) is associated with increased risk of short- and long-term sequelae. Accurate predictive tools allow to improve the outcomes of those born preterm by offering early obstetric interventions to mothers at high-risk of PTB.
Methods: This study combines a wide range of structural and functional MRI parameters, from the fetal head, lung, placenta with clinically available Ultrasound and outcome data. A preprocessing pipeline adapted to the special requirements of the often incomplete and highly GA dependant data and a supervised machine learning model based on these derived markers derived is proposed. Data from 58 preterm and 217 term-born neonates were analysed.
Results: The best SVR model achieved an \(\text {R}^{2}\) value of 0.67 and correctly predicted 92% of true preterm cases using a combination of two maternal and four fetal features.
Conclusion: The significance of this study is uncovering the potential of markers derived from multi-modal imaging data in the prediction of PTB using large-scale fetal studies. This study paves the way for future studies focusing on at-risk women to further enhance the data set and thus predictive power.
Lisa Story and Jana Hutter are joint senior authors.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Preterm birth (PTB), affecting 8% of all deliveries in the UK, poses a significant challenge to healthcare services due to the complex and multifaceted nature of the condition. The burden is prevalent not only in the perinatal period but throughout life, with those born preterm having higher risk of neurodevelopmental delay and motor impairment compared to their term-born counterparts (Luu et al. 2017). Developing better diagnostic and predictive tools can help patients receive early, targeted support leading to improved outcomes (WHO 2020). However, current predictive capabilities are limited (Suff et al. 2019).
Most commonly, a history of previous PTB and cervical length (McIntosh et al. 2016) are used in a clinical setting. Recently, (Watson et al. 2019b) combined risk factors such as previous preterm births and multiple pregnancy (\(\ge \)2 fetuses), with clinical investigations such as fetal fibronectin values and cervical length measurements were employed to predict whether a woman is high risk for preterm birth (Watson et al. 2019b). For women with symptoms of threatened preterm labour, the model combining risk factors and fetal fibronectin predicted 77%–96% of the cases correctly depending on the GA.
However, most screening tools for preterm birth are limited to ultrasound (US) derived cervical length and biochemical markers and fail to match the complex etiology of PTB by not including placental or other fetal parameters. While US and Doppler US (DUS) are the mainstream screening techniques during pregnancy, they are operator-dependent methods that have limited utility in some clinical populations e.g. mother’s with increased body mass index (BMI). Fetal magnetic resonance imaging (MRI) is increasingly used both for research and clinical use especially in high risk populations (mother’s with increased BMI). It also provides both structural and functional information in an operator-dependent manner, covering the entire uterus even in late gestation. Studies using fetal MRI to investigate preterm birth have found decreased thymus volumes (Story et al. 2020b), smaller lungs (Story et al. 2020a) and a reduction in cortical and extra-axial cerebrospinal fluid volumes (Story et al. 2021) in fetuses who subsequently deliver preterm compared to those who deliver at term.
Previous in utero functional MRI studies have employed both diffusion MRI (Slator et al. 2021), which provides information about tissue microstructure and \(\text {T2}^{*}\) relaxometry, which provides an indirect measure of tissue oxygenation via the blood-oxygen-level-dependent (BOLD) effect (Sorensen et al. 2020). Decreased placental \(\text {T2}^{*}\) has been correlated with low birth weight (Sorensen et al. 2020), pre-eclampsia (Ho et al. 2020) and fetal growth restriction. However, there is a paucity of literature using in utero functional MRI to investigate preterm birth. To our knowledge no previous MRI studies have combined multiple functional and structural measures to predict preterm birth.
Data-driven methods are therefore ideally suited for the data set obtained. Identification of the features which hold the highest predictive power can provide valuable clinical insight and lead to improved targeting, monitoring and outcomes for high-risk women and their babies. This study aims to leverage the data available from large scale fetal MRI studies, together with available clinical background and US information, to build supervised machine learning models capable of predicting whether a fetus will be born preterm.
2 Methods
The steps in Fig. 1, from data collection to model evaluation will be detailed in the following.
2.1 Data
The data sets analysed here are combined from multiple ongoing large-scale fetal research studies with similar protocols. These studies are: the Cardiac and Placental Imaging Project (CARP), the Placental Imaging Project (PIP) and the infection study for patients with prolonged preterm rupture of membranes (PPROM). Data was collected from a combined total of 275 patients, and can be divided into the following five categories:
-
1.
Structural MRI data: automatic and manual segmentation of MRI scans to obtain imaging features e.g. volumes of different brain regions or bi-parietal diameter of the fetal head. (in red in Fig. 2B)
-
2.
Functional MRI data: functional imaging features derived from the MRI data e.g. mean placental \(\text {T2}^{*}\) (in red, italic and bold in Fig. 2B).
-
3.
Ultrasonographic data: measurements such as the expected fetal weight (in blue in Fig. 2B)
-
4.
Medical history and demographic data: e.g. maternal age, previous preterm deliveries and smoking status from patient records.
-
5.
Pregnancy outcome data: gestational age at birth, birth weight, placental histopathology.
Structural and Functional MRI Data. After informed consent, all women where scanned in supine position on either a 3T Philips Achieva scanner or a 1.5 T Philips Ingenia scanner (Hughes et al. 2021) under constant monitoring of vital signs including blood pressure, oxygen saturation and heart rate, with frequent verbal interaction. After survey and calibration scans, T2-weighted Turbo Spin Echo images (\(1.25 \times 1.25 \times 2\) mm\(^3\) resolution) were acquired in 3–5 orientations, covering the uterus and fetal head in sagittal and coronal planes. A 30 s coronal Multi-Echo Gradient Echo scan (T2ME), covering the entire uterus (\(3 \times 3 \times 3\) mm\(^3\) resolution), was acquired. Furthermore, diffusion, perfusion, angiographic and other sequences were acquired, however, the present work here focuses on the T2 weighted and T2* scans.
The T2 weighted scans were employed to obtain 3D reconstructions of the brain and lung using slice-to-volume techniques (Uus et al. 2020). The T2ME data was fitted to the mono-exponential decay model, resulting in quantitative \(\text {T2}^{*}\) maps. These were either manually segmented (placenta, brain) or further processed by a 3D model (lungs) and then segmented.
Ultrasonographic Data and Medical History. A growth ultrasound was taken within one week of the MRI. In addition, the data from the screening and anomaly scan were available for this study. The following measurements were obtained: pulsatility indices of the uterine, umbilical and middle cerebral arteries and morphological measurements including abdominal circumference, femur length, expected fetal weight, head circumference and bi-parietal diameter.
Maternal age, body-mass index, parity, previous preterm birth, smoking status, medication status and diagnosis with gestational diabetes mellitus, fetal growth restriction or pre-eclampsia were recorded. At the time of delivery the birth weight, birth weight centile, head circumference and APGAR score at one and five minutes were included. Where available, histopathological information was recorded, most notably the placental weight, the presence of chorioamnionitis and maternal and fetal villi malperfusion.
2.2 Preprocessing
The main concerns for this specific dataset are the following: 1) a large proportion of missing values; 2) age-dependent features 3) imbalance in the dataset between preterm and term babies; 4) the relatively small size of the dataset. As the size of the dataset cannot be changed, it is important to preserve all present data points. The imbalance of the data will be dealt with during model training. The following describes the preprocessing performed for 1) and 2).
Z-scores were calculated (DeVore 2017) for all time-dependent variables using the control group as basis for the transformation. Z-score transformation was performed before imputation to ensure that only measured and no imputed values are included when finding the mean and standard deviation regression lines. This aims to limit any systematic error that could be introduced through Z-score transformation. k-nearest neighbour was then performed on all numerical features with missing values. Each missing value was thereby replaced with a weighted average value from the k-closest neighbours for that feature. Weighting by the Euclidean distance was required due to the imbalance in the dataset.
2.3 Model Optimisation and Implementation
Class imbalance (greater number of babies born at term compared to preterm) was addressed through weighted sampling, where weights were defined as the inverse of the class frequency. Two classification schemes were used: 1) term vs. preterm birth (binary categories), and 2) extremely preterm, very preterm, moderate-to-late preterm and term birth (four birth categories).
This study focuses on predicting GA as a continuous variable using a regression model. The results can then be categorised, allowing for comparison against a small number of existing studies with similar aims (Story et al. 2020a, Story et al. 2020b). Support vector regression (SVR) was chosen as it is captures non-linearity, is capable of dealing with many features and the flexibility to define error margins, which is essential when dealing with low signal-to-noise data such as fetal MRI. A split of 80/20 was used for stratified train/test. Feature selection and exploration was performed by computing the correlation between each feature and GA at birth and then converting into an F statistic. Features with the 19 highest F statistics, and parity (categorical), were selected. Next, Feature x feature interactions were explored between the top 20 features (19 continuous + 1 categorical). Features with >60% missing were excluded from further processing.
3 Results
3.1 Preprocessing
Results from before (Fig. 3) and after (Fig. 4) Z-score transformation, demonstrated exemplarily for placental mean \(\text {T2}^{*}\), illustrate the change from a negative linear relationship to close to constant evolution over GA.
Feature selection was performed in three steps, with the mean placental \(\text {T2}^{*}\) performing best. The predicted GA at birth for all cases in the test dataset using the best model was further divided into four birth categories (\({<}28^{+0}, 28^{+0}-33^{+6}, 34^{+0}-36^{+6}\) and \({\ge } 37^{+0}\) weeks’ GA) and binary birth categories (preterm vs. term) to show the confusion matrices in Fig. 5 and Fig. 6 respectively. Figure 5 indicates that the only fetus in the test set born extremely preterm was correctly predicted by the model. For the very preterm group, one fetus was correctly predicted by the model while two instances were predicted to be born late preterm rather than very preterm. For the late preterm group, seven out of eight children were correctly predicted by the model. 65.5% of the instances were correctly predicted by the model to be term babies while 12.7% who were also term-born were incorrectly predicted to be late preterm. Similarly, when the prediction results were divided into term and preterm, the number of correctly diagnosed term instances was 36 or 65.5% while there were 11 or 20% of correctly diagnosed preterm instances. The number of false positives or instances which were predicted to be preterm but were actually born at term was seven or 12.7%. Only one instance or 1.8% was predicted to be term while they were actually born preterm.
Figure 7 illustrates the \(\text {R}^{2}\) values using the best model for all possible combinations of two features among the best 19 continuous features and the categorical parity feature. The mean placental T2* score (\(\text {R}^{2}\in \) [0.34,0.6]) followed by the pulsatility index of the uterine artery and the body volume with \(\text {R}^{2}\) values \(\in \) [0.1–0.5]. A number of features, which did not individually result in high \(\text {R}^{2}\) scores display high \(\text {R}^{2}\) scores when combined with other features. An example of this would be the mean brain T2* value, which alone yields a \(\text {R}^{2}\) value of 0.018 but paired with the placental mean T2* score the \(\text {R}^{2}\) increases to 0.4. The most extreme case is the parity which raises its \(\text {R}^{2}\) value from −0.031 to 0.54 if combined with the mean placental T2* score.
Results of the SVR model with the highest \(\text {R}^{2}\) score (C = 100, \(\epsilon \) = 0.1, \(\gamma \) = 0.1, kernel = sigmoid). Predicted GA from the best SVR model is plotted vs true GA at birth for the test dataset. The features used were the placenta T2* mean, the head and abdominal circumference, the femur length and the pulsatility index of the right uterine artery from the growth ultrasound and the number of previous preterm births.
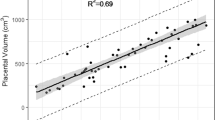
From the top 19 continuous features with the highest \(\text {R}^{2}\) scores and the parity feature, any feature with a missing value percentage of more than 60% was removed. The top 10 features were then the following: Mean placental T2* score, CPR score, head circumference, abdominal circumference, femur length, pulsatility index uterine artery, bi-parietal diameter from the growth ultrasound and the parity. For the top 10 features, models were trained and tested with all combinations of features, with the number of features ranging from 1 to 10. The best five models all used the same following parameters: C = 100, degree = 2, \(\gamma \) = 0.1 and kernel = sigmoid. The best model uses six features and results in a \(\text {R}^{2}\) of 0.665 and a mean absolute error of 1.6 weeks. Figure 8 shows the predicted GA at birth for the test set using the best model compared to the true GA. The \(\text {R}^{2}\) value for the best model was 0.665, the mean absolute error was 1.6 weeks and the root mean squared error was 2.0 weeks.
4 Discussion and Conclusion
The present study exploits a comprehensive dataset containing clinical, US and multimodal fetal MRI data to predict the GA, and thus ultimately preterm birth. The results reflect that, in order to accurately predict preterm birth, acquiring datasets that capture the multifactorial nature of preterm birth are essential. As preterm birth is still poorly understood, acquiring detailed datasets provides an opportunity to better investigate the aetiology and pathophysiology of preterm birth. This study is however merely a first attempt to combine such large and diverse derived parameters.
There are a number of important limitations. These include the number of available datasets. While the collection is big for obstetric comprehensive datasets, it is small for ML standards. It is not well balanced between PTB and term-born cases and includes data from a range of different pregnancy complications, all with their own disease aetiology and progression. The required and here developed pre-processing pipeline reflects these challenges and works towards overcoming them. Future studies should include a higher number of women with threatened PTB to allow to stratify these cases further. Another limitation of this study is the choice of simple imputation method. Next steps can include recently proposed methods such as graph-based imputation techniques (You et al. 2020). The dataset contains both cases of spontaneous and iatrogenic PTB, both with distinct aetiology. The GA at birth prediction results thus also include this information and larger studies are required to treat these as different entities. A further significant limitation of this study is the fact that cervical length was not included. Tools are currently been developed to add this into a future study. Further second order derived quantities can also be included in a next step.
Future work will expand the achieved results into multiple directions. Further models will be explored, direct prediction on the imaging data will be explored to include whether further characteristics such as the heterogeneity of the placenta further increases the ability to predict PTB and further cohorts will be recruited, such as these with previous cervical surgeries or overt signs of inflammation.
References
DeVore, G.: Computing the Z score and centiles for cross-sectional analysis: a practical approach. J. Ultrasound Med. 36, 459–473 (2017)
McIntosh, J., Feltovich, H., Berghella, V., Manuck, T., Society for Maternal-Fetal Medicine (SMFM): The role of routine cervical length screening in selected high- and low-risk women for preterm birth prevention. Am. J. Obstet. Gynecol. 215, B2–B7 (2016)
Hughes, E.J., Price, A.N., McCabe, L., et al.: The effect of maternal position on venous return for pregnant women during MRI. NMR Biomed. 34, e4475 (2021)
Story, L., et al.: Brain volumetry in fetuses that deliver very preterm. NeuroImage Clin. 30, 102650 (2021)
Story, L., et al.: Foetal lung volumes in pregnant women who deliver very pretermy. Pediatr. Res. 87, 1066–1071 (2020a)
Uus, A., et al.: Deformable slice-to-volume registration for reconstruction of quantitative T2* placental and fetal MRI. In: Hu, Y., et al. (eds.) ASMUS/PIPPI -2020. LNCS, vol. 12437, pp. 222–232. Springer, Cham (2020). https://doi.org/10.1007/978-3-030-60334-2_22
Story, L., Zhang, T., Uus, A., et al.: Antenatal thymus volumes in fetuses that delivered \(<\)32 weeks’ gestation: an MRI pilot study. Acta Obstet. Gynecol. Scand. 100, 1040–1050 (2020b). https://doi.org/10.1111/aogs.13983
Suff, N., Story, L., Shennan, A.: The prediction of preterm delivery: what is new? Semin. Fetal Neonatal Med. 24(1), 27–32 (2019)
Luu, T.M., Rehman Mian, M.O., Nuyt, A.M.: Long-term impact of preterm birth: neurodevelopmental and physical health outcomes. Clin. Perinatol. 44(2), 305–314 (2017). Delivery in the Periviable Period
You, J., Ma, X., Yi, D., Ding, Y., Kochenderfer, M., Leskovec, J.: Handling missing data with graph representation learning. In: NeurpIPS 2020 Proceedings (2020)
Slator, P.J., et al.: Data-driven multi-contrast spectral microstructure imaging with InSpect. Med. Image Anal. 71, 102045 (2021)
Sørensen, A., Hutter, J., Seed, M., Grant, P.E., Gowland, P.: T2*-weighted placental MRI: basic research tool or emerging clinical test for placental dysfunction? Ultrasound Obstet. Gynecol. 55, 293–302 (2020)
Ho, A.E.P., et al.: T2* placental magnetic resonance imaging in preterm preeclampsia an observational cohort study. Hypertension 75, 1523–1531 (2020)
Watson, H., et al.: Development and validation of predictive models for QUiPP App vol 2: tool for predicting preterm birth in asymptomatic high?risk women. USOG 55, 348–356 (2019b)
WHO: Preterm birth (2018). https://www.who.int/en/news-room/fact-sheets/detail/preterm-birth. Accessed 24 Jan 2021
WHO: The top 10 causes of death (2020). https://www.who.int/news-room/fact-sheets/detail/the-top-10-causes-of-death. Accessed 28 Mar 2021
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 Springer Nature Switzerland AG
About this paper
Cite this paper
Heinsalu, R. et al. (2021). Predicting Preterm Birth Using Multimodal Fetal Imaging. In: Sudre, C.H., et al. Uncertainty for Safe Utilization of Machine Learning in Medical Imaging, and Perinatal Imaging, Placental and Preterm Image Analysis. UNSURE PIPPI 2021 2021. Lecture Notes in Computer Science(), vol 12959. Springer, Cham. https://doi.org/10.1007/978-3-030-87735-4_27
Download citation
DOI: https://doi.org/10.1007/978-3-030-87735-4_27
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-87734-7
Online ISBN: 978-3-030-87735-4
eBook Packages: Computer ScienceComputer Science (R0)