Abstract
Machine learning (ML) and artificial intelligence (AI) applications in the field of neuroimaging have been on the rise in recent years, and their clinical adoption is increasing worldwide. Deep learning (DL) is a field of ML that can be defined as a set of algorithms enabling a computer to be fed with raw data and progressively discover—through multiple layers of representation—more complex and abstract patterns in large data sets. The combination of ML and radiomics, namely the extraction of features from medical images, has proven valuable, too: Radiomic information can be used for enhanced image characterization and prognosis or outcome prediction. This chapter summarizes the basic concepts underlying ML application for neuroimaging and discusses technical aspects of the most promising algorithms, with a specific focus on Convolutional Neural Networks (CNNs) and Generative Adversarial Networks (GANs), in order to provide the readership with the fundamental theoretical tools to better understand ML in neuroimaging. Applications are highlighted from a practical standpoint in the last section of the chapter, including: image reconstruction and restoration, image synthesis and super-resolution, registration, segmentation, classification, and outcome prediction.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
- Machine learning
- Deep learning
- Convolutional neural network
- Generative adversarial network
- Segmentation
- Classification
1 Introduction
Machine learning (ML) and artificial intelligence (AI) applications in the field of neuroimaging have been rising in recent years, and their adoption is increasing worldwide [1]. Due to the availability of extensive amounts of data, their inherent complexity, and the potentially unlimited applications, neuroimaging is particularly attractive for ML, since virtually every step in clinical imaging spanning from image acquisition and processing to disease detection, diagnosis, and outcome prediction can be the target of ML algorithms [2,3,4,5,6,7,8,9].
Deep learning (DL) is a field of ML that can be defined as a set of algorithms enabling a computer to be fed with raw data and to then progressively discover—through multiple layers of representation—more complex and abstract patterns in large data sets [10,11,12]. The reports of DL algorithms in imaging tasks have been increasing, with applications in the context of several diseases of neurosurgical relevance including but not limited to brain tumors [7, 9, 13,14,15], aneurysms [16,17,18] and spinal diseases [19, 20]. In addition to anatomical imaging, ML-augmented histological diagnosis has been investigated [21]. Another field of ML in neuroimaging is radiomics. The workflow underlying DL applications for radiomics is often complex and may appear confusing for those unfamiliar with the field. Even so, reports combining both radiomic feature extraction and ML are increasing [22,23,24].
In the present chapter, we provide clinical practitioners, researchers, and medical students with the necessary foundations in a rapidly developing area of clinical neuroscience. We highlight the basic concepts underlying ML applications in neuroimaging, and discuss technical aspects of the most promising algorithms adopted into this field—with a specific focus on Convolutional Neural Networks (CNNs) and Generative Adversarial Networks (GANs) [25,26,27]. While in the recent past, segmentation and classification tasks have attracted the most interest, many other tasks exist [8, 28,29,30,31]. These tasks can be considered to some extent overlapping, even if the underlying algorithms may be different. While the vast potential of ML and AI can still be considered early in its development, a clearer categorization of tasks and reporting standardization would be valuable in favoring reproducibility and performance comparison of different studies. At present, this technology is still mainly confined to academic research centers and industry. Still, it is reasonable to expect that the near future will witness a variable integration of ML-based computer-aided tasks in patient management [32]. For this reason, reported applications from a practical standpoint are introduced in the last section of the chapter including image reconstruction and restoration, image synthesis and super-resolution, registration, segmentation, classification, and outcome prediction.
2 The Radiomic Workflow
Radiomics can be defined as the extraction of a significant number of features from medical images applying algorithms for data characterization. “Radiomic features” have the potential to highlight characteristics that are not identifiable by conventional image analysis. The underlying hypothesis is that these distinctive imaging characteristics invisible to the naked eye may provide additional relevant information to be exploited for enhanced image characterization, which can then in turn be applied for enhanced prognosis or prediction. Importantly, recent advances have moved the field from the use handcrafted characteristics such as shape-based (shape, size, surface information), first-order (mean, median ROI value—no spatial relations) and second-order features (inter-voxel relationships) towards data-driven and ML-based approaches, which can automatically perform feature extraction and classification [22, 33, 34].
In general, the radiomic pipeline [35] consists of a series of consecutive steps that may be summarized as following (Fig. 17.1):
-
1.
Image Acquisition.
-
2.
Processing.
-
3.
Feature Selection/Dimensionality Reduction.
-
4.
Downstream Analysis.
Image acquisition protocols depend on chosen imaging technique (ultrasound, X-ray, computed tomography (CT), magnetic resonance imaging (MRI), positron emission tomography (PET)). An important limitation with this respect is represented by intra- and inter-institutional differences in hardware, acquisition and imaging processing techniques, which—by definition—affect image quality, noise, and texture. For practical reasons, it has proven difficult to reach standardization of such heterogeneous equipment and acquisition pipeline, although increasingly pursued by means of international consortia and consensus statements [36]. Corrections during pre-processing may be necessary, with methods specific to the imaging modality of choice. For example, CT uses Hounsfield units which are absolute and anchored to the radiodensity of water, while MRI—due to differing voxel intensities—requires normalization relative to another structure.
Then, a region of interest (ROI) that has to be radiomically analyzed has to be defined through either manual or (semi-) automatic segmentation. Segmentation can be achieved in two-dimensional (2D) space or volume of interest (VOI) segmentation can be achieved in three-dimensional (3D) space. This process is required to identify the area where the radiomic features are to be calculated. This process can be either manual (the traditional gold-standard, even if affected by inter and intra-rater variability), semi-automatic or fully automatic (by means of ML, also affected by a series of pitfalls such as artifact and noise disturbances) [22, 36]. Once segmented images are obtained, additional processing steps may be necessary before feature extraction and analysis such as interpolation to isotropic voxel spacing, range re-segmentation and intensity outlier filtering (normalization), discretization. For further details on this processing step please refer to van Timmeren et al. [35] Radiomic features to be extracted can be categorized into statistical —including histogram-based and texture-based—model-based, transformation-based, and shape-based [24]. The already introduced heterogeneity of the imaging modality—and therefore of their extracted features—have led to the recent introduction of recommendations, guidelines, definitions, and reference values for image features [37]. Interpretations of medical data remains to date largely in the hands of trained practitioners, with limitations due to inter-observer variability, complexity of the image, time constraints, and fatigue [5]. Conventional algorithms like Random Forest (RF), Support Vector Machine (SVM), Neural Networks (NN), k-Nearest Neighbor (KNN), and DL algorithms such as Convolutional Neural Networks (CNN), Recurrent Neural Networks (RNN), and Generative Adversarial Networks (GANs) have been investigated to overcome these drawbacks [5, 38]. Among DL-based approaches for imaging applications, which led to the most astonishing results, CNNs and GANs have attracted considerable attention and will be introduced in the next section.
3 Introduction to Deep Learning Algorithms for Imaging
3.1 Convolutional Neural Networks (CNNs)
3.1.1 Architecture
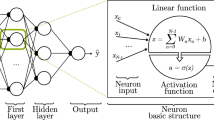
CNNs have been applied to several tasks in radiological image processing (segmentation, classification, detection, et cetera) [25, 28]. CNN architecture is derived from the neurobiology of the visual cortex and is composed of neurons, each having a learnable weight and bias. The structure itself is made up of an input layer, multiple hidden layers (convolutional layers, pooling layers, fully connected layers, and various normalization layers), and one output layer (Fig. 17.2).
The next sections will detail the foundational concepts of these layers in more detail. As a brief summary, the convolutional layer is meant to merge two sets of information. On the other hand, the pooling layer reduces dimensionality by associating the output of neuron clusters in one layer with the single neuron. Fully connected layers connect every neuron in one layer to every neuron in another layer. Its primary purpose is to classify the input images into several classes, based on the training datasets [25]. To simplify, it can be stated that each new CNN layer learns filters—or kernels—of increasing complexity. In a commonly reported and straightforward example, the first layers learn basic feature detection filters such as edges, corners and similar. The middle layers can detect higher-order features, for example, eyes or ears in facial recognition tasks. The higher the layer, the more complex features are recognized, such as differences between faces, et cetera.
3.1.2 Convolution and Kernels
The convolution operation allows the network to detect the same feature in different regions of the image and for this reason, the convolutional layer can be considered the crucial building block of a CNN [39, 40]. In mathematics, convolution between two functions results in a third function expressing how the shape of one function is modified by another. In practice, this operation allows feature extraction by applying a kernel (or filter) to the input (or tensor), both numeric in nature. The product of each element of the kernel and input tensor is derived at each location and added to generate feature maps. The process is repeated through the application of different kernels resulting in an arbitrary number of feature maps, each representing different features of the input tensors. For this reason, different kernels are regarded as different feature extractors [41].
A single CNN layer detects only local features of the image, while multilayer CNNs allow increasing the perception field and synthesizing the features extracted at previous layers. Moreover, CNNs reduce the number of weights by sharing them between the network’s neurons, which results in a considerable memory reduction.
3.1.3 Hyperparameter Optimization
CNNs aim to identify and “learn” the kernels that perform best for a chosen task based on a training dataset. Hyperparameter optimization of kernel size and number is crucial in defining the convolution operation. When visualizing the kernel as a matrix that moves over the input tensor, there are two other concepts that are relevant to be able to grasp how a CNN processes imaging data: padding and stride.
Given that the convolution operation does not center the kernel to overlap the boundaries of the input data, this would result in reduction of the dimension of the output feature map, leaving out the very border of the image. For this reason, to solve the so-called border effect problem, padding is applied. This consists of adding rows and columns of data to the frame of the input tensor, most commonly zero-padding, i.e. columns and rows of zeros, allowing the kernel center to fit on the outermost element of the input, i.e. more space for the kernel to cover the image, and maintain in-plane dimension when the convolution operation is performed [41, 42].
Stride can instead be defined as the distance between two successive kernel positions. For a thorough overview of stride and padding, readers are encouraged to refer to Doumolin and Visin [43].
Of note, kernel values are learned during the training process in the convolution layer (parameter). In contrast, kernel size and number, padding, and stride require being set before training, and are then adjusted during hyperparameter tuning.
Another hyperparameter to be selected is the batch size, namely the number of samples that will be propagated through the network before “updating” its kernels. To explain this concept, we hypothesize to have 500 training samples and to set the batch size as 50. The algorithm will train the network based on the first 50 samples (1–50). Then, it will train using samples 51–100, and so on. A different concept is instead represented by the epoch, which is defined as the number of passes through the training data. Of course, batch size can take values between 1 and the number of samples in the training dataset, while the number of epochs can take any integer value ≥1 [44].
3.1.4 Activation Function and Backpropagation
Outputs of the convolutions, which are a linear function, are passed through an activation function. Activation functions allow learning more complex functional mappings between the different layers. Examples of activation functions are the binary step function, a simple linear activation function, or nonlinear functions such as sigmoid, hyperbolic tangent, or rectified linear unit (ReLU), and leaky ReLU [45]. A binary step function, where activation is single-threshold-based, does not support multi-value output (i.e. multiple categories as output). A linear function on the contrary, after receiving the input (modified by the weight of each neuron) produces an output signal that is proportional to its input. Although smooth nonlinear functions have been extensively used given their similarity with physiological neuronal behavior, ReLU is now more commonly used. In simple words these functions are equations determining the activation (or firing) of a neuron. Specifically, a ReLU will output the input directly in a linear way if it is positive—otherwise, it will output zero. A leaky ReLU will allow a small positive gradient when the input is negative, i.e. changing the slope to a minimum in these cases.
Two major drawbacks of linear activations are the following: they cannot use backpropagation, because the derivative of the function is a constant and is thus not related to the input, preventing weight adjustment. Also, the neural network would be constituted by one collapsed layer as the last function would still be linear, making the NN a linear regression model [46]. On the contrary, nonlinear activation functions allow the model to identify complex relationships among inputs and outputs—an essential feature for complex (or multi-dimensional) data analysis. In this case, backpropagation and multilayer representation is possible (allowing hidden layers to achieve higher abstraction levels on complex data).
3.1.5 Backpropagation
We have just introduced the important concept of backpropagation. When fitting a feed-forward neural network, backpropagation allows descending the gradient with respect to all the weights simultaneously. By chaining the gradients found using the “chain rule,” backpropagation computes the gradient for any weight that is to be optimized—and consequently, can compute improvements with respect to the errors backwards towards the most upstream layer in the network [47, 48]. Due to its high efficiency, backpropagation is useful in many gradient descent methods for training multilayer networks, correcting weights to minimize loss. To better understand how this process works, we can describe that CNNs work in reverse. The gradient (updates to the weights) decreases closer to the input layer and increases towards the output layer as a result of backpropagation updating the weights from the final layer backwards towards the first. Minimization of error (loss) occurs at the final layer, where a higher level of abstraction is recognized and adjusted, tracing back through previous layers. Intuitively, starting from the input instead, a CNN can be described as progressively better at discriminating, e.g. an object that is to be identified, by stepping away from tiny details and looking instead at the “big picture” from a distance [40].
3.1.6 Optimization and Network Training
A loss (or cost) function computes the congruence between output predictions of the network through forward propagation and known ground truth labels. Loss functions are one of the hyperparameters to be determined according to the given task [41, 49]. The amount to which weights are updated during training is referred to as the step size or the “learning rate” [50]. This is an additional hyperparameter used in the training of neural networks, usually taking a small positive value.
A variety of algorithms can be applied for optimization of weights to reduce losses. These include gradient descent, stochastic gradient descent (SGD), mini-batch gradient descent, momentum, Nesterov-accelerated gradient, Adagrad, Adadelta, Adam, and RMSProp [51,52,53,54,55].
Gradient descent is a first-order optimization algorithm dependent on the first-order derivative of a loss function. It aims to compute in which direction weights should be modified so that the function can reach a minimum (Fig. 17.3a). The loss is transferred from one layer to another by means of backpropagation, as discussed before, and the model’s parameters—or weights—are modified depending on the losses, so that loss itself can be minimized. Such optimization is performed after the gradient is calculated on the whole dataset. In addition to normal (batch) gradient descent, SGD and mini-batch descent are most commonly employed. SGD is particularly helpful to minimize the risk of reaching a local minimum (non-convex function) instead of the global minimum—one of the major drawbacks of normal gradient descent (Fig. 17.3b). In a commonly reported example, a normal gradient optimizes weights in a dataset with 1000 observations only after these are all analyzed (every epoch). In SGD, in contrast, the different data rows are analyzed individually, and thus model parameters are updated more often than in batch gradient descent. Of note, despite the higher fluctuations in updating weights, SGD requires significantly less time and less memory. In mini-batch gradient descent, model parameters are instead updated after every mini-batch (a certain subset of the training data). Normal batch gradient descent can be used for smoother curves. SGD can be used when the dataset is very large. In addition, batch gradient descent converges directly to minima, while SGD converges faster when datasets are very large.
Schematic representation of intuitions underlying: (a) gradient descent. Gradient descent is an optimization algorithm used to minimize a function by moving in the direction of steepest descent as defined by the negative of the gradient. In machine learning, it is used to update the parameters of the model; (b) stochastic gradient descent (SGD). While gradient descent risks to reach a local with respect to the global minimum, SGD fluctuations enable it to jump to new and potentially better local minima; (c) momentum. Momentum was introduced to limit the high fluctuations of SGD, allowing faster convergence in the right direction; (d) Nesterov-accelerated gradient (NAG). It can be used to modify a gradient descent-type method to improve its initial convergence
The advantages and disadvantages of other optimization techniques are briefly discussed. Given the high variance in SGD, momentum was introduced—with the need for an additional hyperparameter, namely γ—to accelerate descent in the right directions, and to limit fluctuation to the wrong one (Fig. 17.3c) [56]. A too high momentum may miss a minimum and start to ascend again. To address this problem, Nesterov-Accelerated Gradient (NAG)—or gradient descent with Nesterov momentum—was introduced (Fig. 17.3d). The intuition of NAG consists in anticipating when the slope is going to decrease. To achieve this, previously calculated gradients are considered for the calculation of the momentum, instead of current gradients. This process guarantees that minima are not missed, but makes the operation slower when minima are close.
Differently from the previously discussed optimizers, where the learning rate is constant, both with respect to parameters and cycle, Adagrad changes the learning rate, making smaller updates for parameters associated with frequently occurring features, and larger updates for ones occurring less often. One advantage of such an approach is that the learning rate does not require manual tuning. Unfortunately, squared gradients are accumulated in the denominator, causing the learning rate to continuously decrease reaching infinitesimally small values. For this reason, Adadelta was introduced, in which the sum of gradients is recursively defined as a decaying average of all past squared gradients. A similar rationale was the basis for the development of the RMSprop optimizer. Lastly, Adam (Adaptive Moment Estimation), in addition to storing an exponentially decaying average of past squared gradients like Adadelta and RMSprop, is also characterized by an exponentially decaying average of past gradients, similar to momentum. Intuitively, when visualizing momentum as a ball slope, Adam can be described as a slower ball with friction, which thus prefers flat minima in the error surface. Still other optimizers have been developed (AdaMax, Nadam, AMSGrad), but their discussion is out of the scope of this chapter [51, 54].
3.1.7 Pooling, Fully Connected Layers, and Last Activation Function
Convolutional layers are limited to the fact that a precise position of the feature map is recorded and small changes in the position of the feature in the input image will determine rather different feature maps. Pooling layers perform a downsampling operation which decreases in-plane dimensionality of feature maps obtained in the convolution. This layer lacks learnable parameters, while still maintaining other hyperparameters previously described. The aim of the operation is to reduce the spatial size of the input while maintaining volume depths. This results in a decrease of the number of learnable parameters. The final objective of this step, as described above, is to make the representation resilient to minor translations of the input. This resilience means that if we translate the input by a small amount, the values of most of the pooled outputs do not change [41, 42].
There are different pooling operations, such as maximum pooling and average pooling [42]. Average pooling calculates an average for each patch of the feature map according to pre-specified criteria. Maximum pooling instead calculates the maximum value in each specified patch. The results are downsampled to the pooled feature maps that highlight the most present feature in the patch, but not the average presence of the feature in the case of average pooling. This has been found to work better in practice than average pooling for computer vision tasks like image classification (Fig. 17.4).
Schematic representation of maximum and average pooling. Pooling reduced in-plane dimensionality of feature maps obtained in the convolution to make the representation become invariant to minor translations of the input (noise suppression). Average pooling calculates an average for each patch of the feature map according to pre-specified criteria. Maximum pooling, instead calculates the maximum value in each specified patch. Both approaches result in a downsampled feature map
At the fully connected layer level, feature maps of the last convolution/pooling layer are said to be “flattened,” i.e. converted into a one-dimensional vectors, in which every input is connected to every output by a learnable weight. The final fully connected layer typically has the same number of output nodes as the number of output classes. Their function is essentially to compile the data extracted by previous layers to arrive at the final output [41].
The activation function applied to the last fully connected layer is different from the previous ones and is selected depending on the task of interest (linear, sigmoid, softmax). Also, the loss function is selected according to the last activation function implemented (mean square error, cross-entropy). As an example, for multiclass classification, a softmax function is chosen which normalizes output values from the last fully connected layer to target class probabilities, where each value ranges between 0 and 1 and all values sum to 1 [41, 57].
3.1.8 Overfitting and Dropout
When training a ML model, one of the most important problems is overfitting (Fig. 17.5a). This phenomenon occurs when an algorithm “learns” training data too closely, subsequently failing to generate accurate predictions on new samples. Data are usually split into training and validation set, and performance is tested on this unseen validation set to determine generalizability.
Schematic representation of: (a) overfitting. In overfitting, algorithm training leads to a function that too closely fit a limited set of data, preventing generalizability on new unseen data; and selected regularization approaches, i.e. (b) dropout. Dropout allows to decrease complexity of the model by dropping a certain set of neurons chosen at random, forcing the network to rely on more robust feature for training; (c) early stopping. In early stopping, training stop as soon as the validation error reaches the minimum
Several strategies are available to help prevent overfitting, including increasing amounts of training data, data augmentation approaches, regularization (weight decay, dropout), batch normalization, early stopping, as well as reducing architectural complexity [41, 58]. Also, when a small training dataset is anticipated, novel approaches have focused on fine-tuning previously developed CNNs for adaptation to new applications in a process termed transfer learning, which is addressed in another paragraph below [14, 59, 60].
As stated, data augmentation may be required in the setting of limited sample availability. A variety of basic approaches have been used in the past, such as image flipping, rotation, scaling, cropping, translation, Gaussian noise, et cetera [61].
Regularization approaches to avoid overfitting include among others dropout and weight decay. The term “dropout” refers to dropping out units (hidden and visible) in a neural network. By dropping a unit out, we mean temporarily removing it from the network, along with all its incoming and outgoing connections. For this reason, this regularization technique can be described as a noise-adder to the hidden units. The choice of which units to drop at each iteration is random, and dropout probability is set as a hyperparameter [58, 62,63,64] (Fig. 17.5b).
Weight decay, reduces overfitting by penalizing the model’s weights so that the weights take only small values. This is obtained by adding an additional error, proportional to the sum of weights (L1 norm) or squared magnitude (L2 norm) of the weight vector, to the error at each node. L2 regularization is most commonly used as it strongly penalizes peaky weight vectors and prefers diffuse weight vectors. Due to multiplicative interactions between weights and inputs this system encourages the network to distribute little use of more inputs rather than high selective use of less of them. L1 regularization is a less common alternative. Simply stated, neurons with L1 regularization use only a sparse subset of their most important inputs and ignore noisy features. A combination of L1 with L2 regularizations is the elastic net regularization [58, 65, 66].
Batch normalization consists of a supplemental layer which adaptively normalizes (centering and scaling) the input values of the following layer, mitigating the risk of overfitting, as well as improving gradient flow through the network, allowing higher learning rates, and reducing the dependence on initialization. This allows the use of increased learning rates, and may eliminate the need for dropout and results in reduction of the number of training epochs needed to train the network. For a more structured overview on batch normalization, we advise consulting Ioffe and Szegedy [67], and of a simplified overview by Brownlee [50].
Lastly, early stopping can be considered a form of cross-validation strategy in which a part of the training set is used as a validation set. When the performance on this retained validation set starts to deteriorate, training of the model is interrupted (Fig. 17.5c).
3.1.9 2D vs. 3D CNN
Past image segmentation research has focused on 2D images. For MRI, for example, the approach has been individual segmentation for each slice followed by post-processing to connect 2D segmented slices in a 3D volume. Of course, this approach is prone to inhomogeneity in the reconstruction of the 3D images and loss of anatomical information [68]. Recent reductions in computational costs and the advent of graphics processing units (GPUs) in ML have allowed application of CNNs to 3D medical images using 3D deep learning. The mathematical formulation of 3D CNNs is very similar to 2D CNNs, with an extra dimension added. Here, a 3D convolution is different from the 2D one as the kernel slides in three dimensions as opposed to two dimensions (Fig. 17.6). The implications are particularly relevant for medical imaging when a model is constructed using 3D images voxels, granting increased precision and spatial resolution, higher data reliability at the expense of increased model complexity and slower computation [68,69,70]. For further readings on 3D CNN use for medical imaging, consult Singh et al. and Despotovic et al. [68, 70]
3.1.10 Transfer Learning
Recently the use of algorithms pre-trained for similar applications to be extended for other applications has proven valuable [60, 71]. This technique is named deep transfer learning (TL) and several reports in brain tumor research have been produced, for example, with CNNs [14, 59, 72, 73]. A pre-trained CNN has to be able to extract relevant features while maintaining irrelevant features and underlying noise. For a comprehensive overview of transfer learning, consult Zhuang et al. [71]
3.1.11 Available CNN Architectures
A variety of CNN architectures have been developed and are being extensively exploited in imaging applications: LeNet, AlexNet, GoogLeNet, ResNet, SENet, VGG16, VGG19 [74]. For a comprehensive overview of pre-trained CNN architectures we refer the readers to Khan et al. [75]
3.2 Generative Adversarial Networks
The basic function of GANs is to train a generator and discriminator in an adversarial way. Based on different requirements, either a stronger generator or a more sensitive discriminator is designed as the target goal [26, 76, 77]. These two models are typically implemented by neural networks such as CNNs. The generator tries to capture the distribution of true examples for new data example generation. The discriminator is usually a binary classifier, discriminating generated examples from the true examples as accurately as possible (Fig. 17.7). With improving generator performance, discriminator performance worsens. For this reason, GAN optimization is said to be a “minimax optimization problem.” The optimization terminates at a saddle point (convergence) that is a minimum in terms of error with respect to the generator and a maximum in terms of error with respect to the discriminator [26]. Past the transitory convergent state, model training may continue with the discriminator providing only random feedback (50/50 or coin tossing), implying for the generator to train on meaningless feedback. This of course would result in decreased performance of the generator.
The contribution of GANs to medical imaging is therefore twofold. The generative part can help in exploring hidden structures in the training data leading to new image synthesis with valuable implications for addressing issues such as lack of data and privacy concerns. The discriminative part can be instead considered as a “learned prior” for normal images, so that it can be used as a regularizer or detector when presented with abnormal images [27].
3.3 Data Availability and Privacy
We have already mentioned how, to some extent, the “firepower” granted by DL techniques is difficult to implement due to the poor availability of training data. Morever, the sensitive nature of patient medical information, data safety practices such as deidentification (anonymization and pseudo-anonymization) are crucial [78]. One solution to the lack of data availability has been proposed using ML approaches such as artificial image synthesis for data augmentation [79, 80]. Another option is federated learning, in which an algorithm is trained at various sites locally, without exchanging data—exchanging only the weights of the further trained model [81].
3.4 Deep Learning-Based Tasks in Imaging
The number of tasks that can be performed by DL in imaging is vast and intrinsically problem-oriented. A major distinction consists in supervised versus unsupervised machine learning approaches. In supervised learning, training data are given with known labels for which the correct outputs are already known, differently from unsupervised learning in which labels are not available, e.g. clustering [82]. Each of these methods carries its own advantages and disadvantages. Regardless of the approach, practical applications derive from widely appreciated clinical problems such as suboptimal image acquisition, time-consuming image analysis, and long learning curves for clinical experts or inter-observer variability in disease diagnosis and classification. In the next paragraphs, we aim to provide an overview of some clinical problems and the ML-based approaches that have been applied to tackle them. For descriptive purposes we identified the following tasks subgroups: image reconstruction and restoration, synthesis and super-resolution, registration, detection and classification, outcome prediction.
3.4.1 Image Reconstruction and Restoration
Image reconstruction refers to several scenarios where high-quality images are obtained from incomplete data or partial signal loss. The underlying issues are technique-dependent and can vary in different imaging modality for e.g. MRI, PET-CT, CT [33, 83]. Such problems are intimately connected to image restoration, whose aim is to improve the quality of suboptimal images acquired because of technical limitations or patient-related factors (e.g. respiration, discomfort, radiation doses). Other terminology to indicate issues of pertaining to image restoration are “denoising” and, more broadly, also artifact detection can be considered in this area. Few examples are here presented.
A study by Schramm et al. investigated anatomically-guided PET reconstruction aiming to improve bias-noise characteristics in brain PET imaging using a CNN. By applying a dedicated data augmentation during the training phase they showed encouraging results which could be generated in virtually real-time [84]. Yan et al. [85] trained a GAN algorithm to generate BOLD signals that were lost for technical reasons during fMRI. Intriguingly, reconstructed signals closely resembled the uncompromised signals and were coherent with each individual’s functional brain organization. Kidoh et al. [86] have reported in five patients artificial noise addition to brain MRI, and training of a CNN to perform image reconstruction. The authors reported that their algorithm significantly reduced image noise while preserving image quality for brain MR images. CNNs have been most commonly reported for this task. Despite the preliminary encouraging results, recent reports point at instabilities in deep learning based methods raising concern on artifacts formation, failure to recover structural changes (from complete removal of details to more subtle distortions and blurring of the features) and others [87]. Additional applications are related to 3D reconstruction of anatomical regions. In spine surgery, DL can substitute manual segmentation and 3D reconstruction to aid surgical planning [88].
3.4.2 Image Synthesis and Super-Resolution
The applications of image synthesis are different and can be categorized in unconditional synthesis and cross-modality synthesis (image conversion), with the former meaning image generation from random noise without conditional information and the latter being instead referred to, for example, obtaining CT-like images from MRI or more in general to derive new parametric images or new tissue contrast [27, 89, 90].
This latter application has also been referred to as image super-resolution whose aim is to reconstruct a higher resolution image or image sequence from the observation of low-resolution images [91].
Especially for ML modeling, this allows training data to be augmented without recurring to traditional methods such as scaling, rotation, flipping, translation, and elastic deformation which do not account for variations resulting from different imaging protocols or sequences, not to mention variations in the size, shape, location, and appearance of specific pathology [27, 80]. Some examples of past studies are here discussed. Liu et al. [91] reported super-resolution reconstruction of experiments real datasets of MR brain images and demonstrated that multiscale fusion convolution network was able to recover detailed information from MR images outperforming traditional methods. A recent small preliminary study reported training of GANs to generate MRI T2w images from CT spine slices, obtaining far from optimal results [29]. The potential advantages of unconditional synthesis are related to overcoming privacy issues related to medical imaging use and the insufficient cases of patients positive for a given pathology [27, 79]. Generative Adversarial Networks (GANs) and Convolutional Neural Networks (CNNs) have been studied for this application.
3.4.3 Image Registration
Registration establishes anatomical correspondences between two images by mapping source and reference volume to the same coordinates [31, 92]. This task is required for intraoperative navigation, 3D reconstruction, multimodality image mappings, atlas construction, and arithmetic operations such as image averaging, subtraction, and correlation [31]. Implications are clear: Intraoperative neuronavigation requires mapping of a preoperative image onto an intraoperative image by registration. Another clinically relevant application in neuro-oncology is found in the context of rapid brain tumor growth, which requires longitudinal evaluation for disease evolution and for treatment results monitoring—both of which may greatly benefit from accurate registration to improve intra-individual imaging comparison [92]. Traditional methods can be summarized in deformable or elastic registration and linear registration or graph-based approaches [92].
Investigators have used a variety of approaches, with different degrees of manual interaction, to perform image registration. These approaches use either information obtained about the shape and topology of objects in the image or the presumed consistency in the intensity information from one slice to its immediate neighbor or from one brain or image set to another [31].
Despite the several strategies proposed, this task remains challenging due to the computational power needed, high-dimensional optimization, and task-dependent parameter tuning [93]. Recently, Fan et al. [93] reported the use of dual-supervised fully convolutional networks for image registration by predicting deformation from image appearance and showed promising registration accuracy and efficiency compared with the state-of-the-art methods. Estienne et al. [92] recently reported the introduction of DL-based framework to address segmentation and registration simultaneously.
3.4.4 Image Segmentation, Classification, and Outcome Prediction
Segmentation can be described as the process of partitioning an image into multiple non-overlapping regions that share similar attributes, enabling localization and quantification. Both supervised and unsupervised learning can play a role in segmentation tasks [12]. Segmentation from MR images is useful for diagnosis, growth prediction, and treatment planning. Its results are labels identifying each homogeneous region or a set of contours describing the region limits [68]. Of course, the higher the lesion complexity, the more problematic the segmentation. Well-defined lesions are easier to segment, while infiltrative, diffuse lesions are more daunting. Other obstacles to successful segmentation are represented by lesion variable shape, size, and location in addition to unstandardized voxel values in different modalities [28]. Segmentation applications have been reported for acute ischemic lesion segmentation [94], brain tumor (gliomas, meningiomas, metastases) [9, 15, 28, 79, 95,96,97], spine [19, 98], and aneurysms [4, 99] have been reported. Segmentation and classification are always intimately connected as segmentation implies a classification, while an imaging classifier implicitly segments an image. The segmentation results can be further used in several applications such as for analysis of anatomical structures, for the study of pathological regions, for surgical planning, et cetera [68]
The research area of disease detection, classification, and grading through machine learning based methods has also been referred as computer-aided diagnosis (CAD) [14]. A few examples are here discussed together with clinical implications. Deepak et al. [14] reported an automatic classification system designed for three brain tumor types (glioma, meningioma, and pituitary tumor) using a deep transfer learned CNN model for feature extraction from brain MRI images and classified using a SVM algorithm with high accuracy and AUC. CAD of a brain tumor can have a significant impact on clinical practice. For example, in the context of metastatic disease, early and accurate identification of brain metastases is crucial for optimal patient management. Given their small size, similarity to blood vessels and low technical contrast to background ratio, computer-assisted detection by means of DL algorithms can provide a valuable tool for early lesion identification [9]. Also, glioma recurrence can be difficult to identify at MRI due to post-treatment changes such as pseudo-progression and radiation necrosis and DL-based classification of these two lesions would be highly clinically relevant [13]. In the field of vascular neurosurgery, CNNs have proven useful in improving aneurysms detection at neuroimaging [100, 101]. Stemming from segmentation and classification tasks, outcome prediction – such as survival - has also been assessed preliminary by some studies [15, 102, 103].
4 Conclusions
The present chapter introduces ML applications in neuroimaging in a step-wise manner. The concept of radiomics has significantly increased expectations deriving from image analysis with respect to enhanced lesion diagnosis, characterization, segmentation, classification, outcome prediction, and prognosis evaluation. The computational power granted by ML—and DL in particular—has convincingly demonstrated preliminary potential to significantly impact patient management. CNNs and GANs, among other algorithms, constitute flexible tools to tackle multiple different ML tasks. Successful application in a variety of tasks spanning from image reconstruction and restoration, image synthesis and super-resolution, segmentation, classification, and outcome prediction have been introduced. Technical and ethical challenges posed by this technology are yet to be solved, with future research expected to improve upon the current limitations—Especially regarding explainable learning. Foundational knowledge of this field of ML by clinicians is required to safely guide the next medical revolution, truly introducing ML into neuroimaging.
References
Staartjes VE, Stumpo V, Kernbach JM, et al. Machine learning in neurosurgery: a global survey. Acta Neurochir. 2020;162(12):3081–91. https://doi.org/10.1007/s00701-020-04532-1.
Akeret K, Stumpo V, Staartjes VE, et al. Topographic brain tumor anatomy drives seizure risk and enables machine learning based prediction. NeuroImage Clin. 2020;28:102506.
Lubicz B, Levivier M, Francois O, Thoma P, Sadeghi N, Collignon L, Baleriaux D. Sixty-four-row multisection CT angiography for detection and evaluation of ruptured intracranial aneurysms: interobserver and intertechnique reproducibility. Am J Neuroradiol. 2007;28(10):1949–55.
Park A, Chute C, Rajpurkar P, et al. Deep learning–assisted diagnosis of cerebral aneurysms using the HeadXNet model. JAMA Netw Open. 2019;2(6):e195600.
Razzak MI, Naz S, Zaib A. Deep learning for medical image processing: overview, challenges and future. In: Dey N, Ashour A, Borra S, editors. Classification in BioApps. Lecture notes in computational vision and biomechanics, vol. 30. Cham: Springer; 2017.
Song J, Yin Y, Wang H, Chang Z, Liu Z, Cui L. A review of original articles published in the emerging field of radiomics. Eur J Radiol. 2020;127:108991.
Swinburne NC, Schefflein J, Sakai Y, Oermann EK, Titano JJ, Chen I, Tadayon S, Aggarwal A, Doshi A, Nael K. Machine learning for semiautomated classification of glioblastoma, brain metastasis and central nervous system lymphoma using magnetic resonance advanced imaging. Ann Transl Med. 2019;7(11):232.
Zacharaki EI, Wang S, Chawla S, Soo Yoo D, Wolf R, Melhem ER, Davatzikos C. Classification of brain tumor type and grade using MRI texture and shape in a machine learning scheme. Magn Reson Med. 2009;62(6):1609–18.
Zhang M, Young GS, Chen H, Li J, Qin L, McFaline-Figueroa JR, Reardon DA, Cao X, Wu X, Xu X. Deep-learning detection of cancer metastases to the brain on MRI. J Magn Reson Imaging. 2020;52(4):1227–36.
Djuric U, Zadeh G, Aldape K, Diamandis P. Precision histology: how deep learning is poised to revitalize histomorphology for personalized cancer care. NPJ Precis Oncol. 2017;1(1):22.
LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521(7553):436–44.
Lundervold AS, Lundervold A. An overview of deep learning in medical imaging focusing on MRI. Z Med Phys. 2019;29(2):102–27.
Bacchi S, Zerner T, Dongas J, Asahina AT, Abou-Hamden A, Otto S, Oakden-Rayner L, Patel S. Deep learning in the detection of high-grade glioma recurrence using multiple MRI sequences: a pilot study. J Clin Neurosci. 2019;70:11–3.
Deepak S, Ameer PM. Brain tumor classification using deep CNN features via transfer learning. Comput Biol Med. 2019;111:103345.
Sun L, Zhang S, Chen H, Luo L. Brain tumor segmentation and survival prediction using multimodal MRI scans with deep learning. Front Neurosci. 2019;13:810.
Hainc N, Mannil M, Anagnostakou V, Alkadhi H, Blüthgen C, Wacht L, Bink A, Husain S, Kulcsár Z, Winklhofer S. Deep learning based detection of intracranial aneurysms on digital subtraction angiography: a feasibility study. Neuroradiol J. 2020;33(4):311–7.
Shi Z, Hu B, Schoepf UJ, Savage RH, Dargis DM, Pan CW, Li XL, Ni QQ, Lu GM, Zhang LJ. Artificial intelligence in the management of intracranial aneurysms: current status and future perspectives. Am J Neuroradiol. 2020;41(3):373–9.
Sichtermann T, Faron A, Sijben R, Teichert N, Freiherr J, Wiesmann M. Deep learning–based detection of intracranial aneurysms in 3D TOF-MRA. Am J Neuroradiol. 2019;40(1):25–32.
Huang J, Shen H, Wu J, Hu X, Zhu Z, Lv X, Liu Y, Wang Y. Spine explorer: a deep learning based fully automated program for efficient and reliable quantifications of the vertebrae and discs on sagittal lumbar spine MR images. Spine J. 2020;20(4):590–9.
Jamaludin A, Kadir T, Zisserman A. SpineNet: automated classification and evidence visualization in spinal MRIs. Med Image Anal. 2017;41:63–73.
Hollon TC, et al. Near real-time intraoperative brain tumor diagnosis using stimulated Raman histology and deep neural networks. Nat Med. 2020;26(1):52–8.
Avanzo M, Wei L, Stancanello J, Vallières M, Rao A, Morin O, Mattonen SA, El Naqa I. Machine and deep learning methods for radiomics. Med Phys. 2020;47(5):e185–202. https://doi.org/10.1002/mp.13678.
Lambin P, et al. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol. 2017;14(12):749–62.
Mayerhoefer ME, Materka A, Langs G, Häggström I, Szczypiński P, Gibbs P, Cook G. Introduction to Radiomics. J Nucl Med. 2020;61(4):488–95.
Dhillon A, Verma GK. Convolutional neural network: a review of models, methodologies and applications to object detection. Prog Artif Intell. 2020;9(2):85–112.
Gui J, Sun Z, Wen Y, Tao D, Ye J. A review on generative adversarial networks: algorithms, theory, and applications. arXiv:2001.06937 [cs, stat]. 2020.
Yi X, Walia E, Babyn P. Generative adversarial network in medical imaging: a review. Med Image Anal. 2019;58:101552.
Havaei M, Davy A, Warde-Farley D, Biard A, Courville A, Bengio Y, Pal C, Jodoin P-M, Larochelle H. Brain tumor segmentation with deep neural networks. Med Image Anal. 2017;35:18–31.
Lee JH, Han IH, Kim DH, Yu S, Lee IS, Song YS, Joo S, Jin C-B, Kim H. Spine computed tomography to magnetic resonance image synthesis using generative adversarial networks: a preliminary study. J Korean Neurosurg Soc. 2020;63(3):386–96.
Li Y, Sixou B, Peyrin F. A review of the deep learning methods for medical images super resolution problems. IRBM. 2020;42(2):120–33.
Toga AW, Thompson PM. The role of image registration in brain mapping. Image Vis Comput. 2001;19(1–2):3–24.
Topol EJ. High-performance medicine: the convergence of human and artificial intelligence. Nat Med. 2019;25(1):44–56.
Hammernik K, Knoll F. Machine learning for image reconstruction. Handbook of medical image computing and computer assisted intervention. Amsterdam: Elsevier; 2020. p. 25–64.
Rizzo S, Botta F, Raimondi S, Origgi D, Fanciullo C, Morganti AG, Bellomi M. Radiomics: the facts and the challenges of image analysis. Eur Radiol Exp. 2018;2(1):36.
van Timmeren JE, Cester D, Tanadini-Lang S, Alkadhi H, Baessler B. Radiomics in medical imaging—“how-to” guide and critical reflection. Insights Imaging. 2020;11(1):91.
Kocak B, Durmaz ES, Ates E, Kilickesmez O. Radiomics with artificial intelligence: a practical guide for beginners. Diagn Interv Radiol. 2019;25(6):485–95.
Zwanenburg A, Leger S, Vallières M, Löck S. Image biomarker standardisation initiative. Radiology. 2020;295(2):328–38.
Lambin P, Rios-Velazquez E, Leijenaar R, et al. Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer. 2012;48(4):441–6.
Brownlee J. How do convolutional layers work in deep learning neural networks? Machine Learning Mastery; 2019.
Convolutional neural networks—basics · machine learning notebook. https://mlnotebook.github.io/post/CNN1/. Accessed 27 Jan 2021.
Yamashita R, Nishio M, Do RKG, Togashi K. Convolutional neural networks: an overview and application in radiology. Insights Imaging. 2018;9(4):611–29.
Brownlee J. A gentle introduction to padding and stride for convolutional neural networks. Machine Learning Mastery; 2019.
Dumoulin V, Visin F. A guide to convolution arithmetic for deep learning. arXiv:1603.07285 [cs, stat]. 2018.
Brownlee J. Difference between a batch and an epoch in a neural network. Machine Learning Mastery; 2018.
Dutta-Roy T. Medical image analysis with deep learning—II. In: Medium. 2018. https://medium.com/@taposhdr/medical-image-analysis-with-deep-learning-ii-166532e964e6. Accessed 27 Jan 2021.
7 types of activation functions in neural networks: how to choose? In: MissingLink.ai. https://missinglink.ai/guides/neural-network-concepts/7-types-neural-network-activation-functions-right/. Accessed 28 Jan 2021.
Agarwal M. Back propagation in convolutional neural networks—intuition and code. In: Medium. 2020. https://becominghuman.ai/back-propagation-in-convolutional-neural-networks-intuition-and-code-714ef1c38199. Accessed 27 Jan 2021.
Backpropagation - Wikipedia. Accessed 26 Sep 2021. https://en.wikipedia.org/wiki/Backpropagation.
Brownlee J. Loss and loss functions for training deep learning neural networks. Machine Learning Mastery; 2019.
Brownlee J. How to configure the learning rate when training deep learning neural networks. Machine Learning Mastery; 2019.
Doshi S. Various optimization algorithms for training neural network. In: Medium. 2020. https://towardsdatascience.com/optimizers-for-training-neural-network-59450d71caf6. Accessed 27 Jan 2021
Peixeiro M. The 3 best optimization methods in neural networks. In: Medium. 2020. https://towardsdatascience.com/the-3-best-optimization-methods-in-neural-networks-40879c887873. Accessed 27 Jan 2021.
Smolyakov V. Neural network optimization algorithms. In: Medium. 2018. https://towardsdatascience.com/neural-network-optimization-algorithms-1a44c282f61d. Accessed 27 Jan 2021.
Soydaner D. A comparison of optimization algorithms for deep learning. Int J Patt Recogn Artif Intell. 2020;34(13):2052013.
MLTut. What Is Stochastic Gradient Descent- A Super Easy Complete Guide! 2020. https://www.mltut.com/stochastic-gradient-descent-a-super-easy-complete-guide/.
Bushaev V. Stochastic gradient descent with momentum. In: Medium. 2017. https://towardsdatascience.com/stochastic-gradient-descent-with-momentum-a84097641a5d. Accessed 28 Jan 2021.
Deep learning: which loss and activation functions should I use? | By Stacey Ronaghan | towards data science. https://towardsdatascience.com/deep-learning-which-loss-and-activation-functions-should-i-use-ac02f1c56aa8. Accessed 28 Jan 2021.
Analytics Vidhya. Regularization Techniques | Regularization In Deep Learning. 2018. https://www.analyticsvidhya.com/blog/2018/04/fundamentals-deep-learning-regularization-techniques/.
Ahmed KB, Hall LO, Goldgof DB, Liu R, Gatenby RA. Fine-tuning convolutional deep features for MRI based brain tumor classification. In: Armato SG, Petrick NA, editors. Orlando, Florida, United States. 2017. p 101342E.
Tajbakhsh N, Shin JY, Gurudu SR, Hurst RT, Kendall CB, Gotway MB, Liang J. Convolutional neural networks for medical image analysis: full training or fine tuning? IEEE Trans Med Imaging. 2016;35(5):1299–312.
Data Augmentation | How to Use Deep Learning When You Have Limited Data. Accessed 26 Sep 2021. https://nanonets.com/blog/data-augmentation-how-to-use-deep-learning-when-you-have-limited-data-part-2/.
Hinton GE, Srivastava N, Krizhevsky A, Sutskever I, Salakhutdinov RR. Improving neural networks by preventing co-adaptation of feature detectors. arXiv:1207.0580 [cs]. 2012.
Srivastava N, Hinton G, Krizhevsky A, Sutskever I, Salakhutdinov R. Dropout: a simple way to prevent neural networks from overfitting. J Mach Learn Res. 2014;15(1):1929–58.
Wu H, Gu X. Towards dropout training for convolutional neural networks. Neural Netw. 2015;71:1–10.
Murugan P, Durairaj S. Regularization and optimization strategies in deep convolutional neural network. arXiv:1712.04711 [cs]. 2017.
Nagpal A. L1 and L2 regularization methods. In: Medium. 2017. https://towardsdatascience.com/l1-and-l2-regularization-methods-ce25e7fc831c. Accessed 28 Jan 2021.
Ioffe S, Szegedy C. Batch normalization: accelerating deep network training by reducing internal covariate shift. arXiv:1502.03167 [cs]. 2015.
Despotović I, Goossens B, Philips W. MRI segmentation of the human brain: challenges, methods, and applications. Comput Math Methods Med. 2015;2015:1–23.
Ji S, Xu W, Yang M, Yu K. 3D convolutional neural networks for human action recognition. IEEE Trans Pattern Anal Mach Intell. 2013;35(1):221–31.
Singh SP, Wang L, Gupta S, Goli H, Padmanabhan P, Gulyás B. 3D deep learning on medical images: a review. arXiv:2004.00218 [cs, eess, q-bio]. 2020.
Zhuang F, Qi Z, Duan K, Xi D, Zhu Y, Zhu H, Xiong H, He Q. A comprehensive survey on transfer Learning. arXiv:1911.02685 [cs, stat]. 2020.
Mehrotra R, Ansari MA, Agrawal R, Anand RS. A transfer learning approach for AI-based classification of brain tumors. Mach Learn Appl. 2020;2:100003.
Liu R, Hall LO, Goldgof DB, Zhou M, Gatenby RA, Ahmed KB. Exploring deep features from brain tumor magnetic resonance images via transfer learning. In: 2016 international joint conference on neural networks (IJCNN). IEEE, Vancouver, BC, Canada, 2016. p. 235–42.
Chelghoum R. Transfer learning using convolutional neural network architectures for brain tumor classification from MRI images.
Khan A, Sohail A, Zahoora U, Qureshi AS. A survey of the recent architectures of deep convolutional neural networks. Artif Intell Rev. 2020;53(8):5455–516.
Goodfellow IJ, Pouget-Abadie J, Mirza M, Xu B, Warde-Farley D, Ozair S, Courville A, Bengio Y Generative adversarial networks. arXiv:1406.2661 [cs, stat]. 2014.
Lan L, You L, Zhang Z, Fan Z, Zhao W, Zeng N, Chen Y, Zhou X. Generative adversarial networks and its applications in biomedical informatics. Front Public Health. 2020;8:164.
Montagnon E, Cerny M, Cadrin-Chênevert A, Hamilton V, Derennes T, Ilinca A, Vandenbroucke-Menu F, Turcotte S, Kadoury S, Tang A. Deep learning workflow in radiology: a primer. Insights Imaging. 2020;11(1):22.
Mok TCW, Chung ACS. Learning data augmentation for brain tumor segmentation with coarse-to-fine generative adversarial networks. arXiv:180511291 [cs]. 2019;11383:70–80.
Nalepa J, Marcinkiewicz M, Kawulok M. Data augmentation for brain-tumor segmentation: a review. Front Comput Neurosci. 2019;13:83.
Yang Q, Liu Y, Chen T, Tong Y. Federated machine Learning: concept and applications. arXiv:1902.04885 [cs]. 2019.
Kotsiantis S, Zaharakis I, Pintelas P, et al. Supervised machine learning: a review of classification techniques. Emerging Artificial Intelligence Applications in Computer Engineering. 2007;160(1):3–24.
Zhang H, Dong B. A review on deep learning in medical image reconstruction. arXiv:1906.10643 [physics]. 2019.
Schramm G, Rigie D, Vahle T, Rezaei A, Van Laere K, Shepherd T, Nuyts J, Boada F. Approximating anatomically-guided PET reconstruction in image space using a convolutional neural network. NeuroImage. 2021;224:117399.
Yan Y, Dahmani L, Ren J, et al. Reconstructing lost BOLD signal in individual participants using deep machine learning. Nat Commun. 2020;11(1):5046.
Kidoh M, Shinoda K, Kitajima M, Isogawa K, Nambu M, Uetani H, Morita K, Nakaura T, Yamashita Y, Yamashita Y. Deep learning based noise reduction for brain MR imaging: tests on phantoms and healthy volunteers. Magn Reson Med Sci. 2020;19(3):195–206.
Antun V, Renna F, Poon C, Adcock B, Hansen AC. On instabilities of deep learning in image reconstruction and the potential costs of AI. Proc Natl Acad Sci U S A. 2020;117(48):30088–95.
Fan G, Liu H, Wu Z, Li Y, Feng C, Wang D, Luo J, Wells WM, He S. Deep learning–based automatic segmentation of lumbosacral nerves on CT for spinal intervention: a translational study. Am J Neuroradiol. 2019;40(6):1074–81.
Nie D, Cao X, Gao Y, Wang L, Shen D. Estimating CT image from MRI data using 3D fully convolutional networks. In: Deep learning and data labeling for medical applications—1st international workshop, LABELS 2016, and 2nd international workshop, DLMIA 2016 held in conjunction with MICCAI 2016, proceedings. 2016. https://doi.org/10.1007/978-3-319-46976-8_18.
Staartjes VE, Seevinck PR, Vandertop WP, van Stralen M, Schröder ML. Magnetic resonance imaging–based synthetic computed tomography of the lumbar spine for surgical planning: a clinical proof-of-concept. Neurosurg Focus. 2021;50(1):E13.
Liu C, Wu X, Yu X, Tang Y, Zhang J, Zhou J. Fusing multi-scale information in convolution network for MR image super-resolution reconstruction. Biomed Eng Online. 2018;17(1):114.
Estienne T, Lerousseau M, Vakalopoulou M, et al. Deep learning-based concurrent brain registration and tumor segmentation. Front Comput Neurosci. 2020;14:15.
Fan J, Cao X, Yap P-T, Shen D. BIRNet: brain image registration using dual-supervised fully convolutional networks. Med Image Anal. 2019;54:193–206.
Chen L, Bentley P, Rueckert D. Fully automatic acute ischemic lesion segmentation in DWI using convolutional neural networks. NeuroImage Clin. 2017;15:633–43.
Bennai MT, Guessoum Z, Mazouzi S, Cormier S, Mezghiche M. A stochastic multi-agent approach for medical-image segmentation: application to tumor segmentation in brain MR images. Artif Intell Med. 2020;110:101980.
Laukamp KR, Thiele F, Shakirin G, Zopfs D, Faymonville A, Timmer M, Maintz D, Perkuhn M, Borggrefe J. Fully automated detection and segmentation of meningiomas using deep learning on routine multiparametric MRI. Eur Radiol. 2019;29(1):124–32.
Zhou T, Canu S, Ruan S. Fusion based on attention mechanism and context constraint for multi-modal brain tumor segmentation. Comput Med Imaging Graph. 2020;86:101811.
Fan G, Liu H, Wang D, et al. Deep learning-based lumbosacral reconstruction for difficulty prediction of percutaneous endoscopic transforaminal discectomy at L5/S1 level: a retrospective cohort study. Int J Surg. 2020;82:162–9.
Shahzad R, Pennig L, Goertz L, Thiele F, Kabbasch C, Schlamann M, Krischek B, Maintz D, Perkuhn M, Borggrefe J. Fully automated detection and segmentation of intracranial aneurysms in subarachnoid hemorrhage on CTA using deep learning. Sci Rep. 2020;10(1):21799.
Duan H, Huang Y, Liu L, Dai H, Chen L, Zhou L. Automatic detection on intracranial aneurysm from digital subtraction angiography with cascade convolutional neural networks. Biomed Eng Online. 2019;18(1):110.
Nakao T, Hanaoka S, Nomura Y, Sato I, Nemoto M, Miki S, Maeda E, Yoshikawa T, Hayashi N, Abe O. Deep neural network-based computer-assisted detection of cerebral aneurysms in MR angiography. J Magn Reson Imaging. 2018;47(4):948–53.
Bhandari A, Koppen J, Agzarian M. Convolutional neural networks for brain tumour segmentation. Insights Imaging. 2020;11(1):77.
Lao J, Chen Y, Li Z-C, Li Q, Zhang J, Liu J, Zhai G. A deep Learning-based radiomics model for prediction of survival in glioblastoma multiforme. Sci Rep. 2017;7(1):10353.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Stumpo, V. et al. (2022). Machine Learning Algorithms in Neuroimaging: An Overview. In: Staartjes, V.E., Regli, L., Serra, C. (eds) Machine Learning in Clinical Neuroscience. Acta Neurochirurgica Supplement, vol 134. Springer, Cham. https://doi.org/10.1007/978-3-030-85292-4_17
Download citation
DOI: https://doi.org/10.1007/978-3-030-85292-4_17
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-85291-7
Online ISBN: 978-3-030-85292-4
eBook Packages: MedicineMedicine (R0)