Abstract
People affected by diabetes are at a high risk of developing diabetic nephropathy, which, in turn, is the leading cause of end-stage chronic kidney disease worldwide. Predicting the onset of renal complications as early as possible, when kidney function is still intact, is of paramount importance for therapy selection due to existence of a class of antidiabetic agents (SGLT2 inhibitors) with known nephroprotective properties.
In the present work, we study the anthropometric and laboratory data of 28,955 diabetic patients followed for a median of 6.6 years (IQR 4.7–7.8) by 14 Italian diabetes outpatient clinics. We develop a deep learning model, based on the incorporation of variable-length longitudinal baseline data via recurrent layers, to predict the onset of impaired kidney function (KDOQI stage ≥ 3). We adopt a multi-label output-coding system to address the irregularity and sparsity in the sampling of endpoints induced by the real-life structure of the data.
Using the cumulative/dynamic AUROC with respect to a variable prediction horizon of 1 to 7 years, we compare the proposed model against the predictor of imminent deterioration of kidney function used in clinical practice, i.e., the estimated glomerular filtration rate (eGFR), and a set of year-specific logistic regressions trained on a single baseline visit.
The proposed deep learning model generally outperforms both benchmarks, especially in the medium-to-long term, with AUROC ranging from 0.841 to 0.895. Supplementary analyses confirm the effective encoding of sequence data within the network.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
People affected by diabetes, a chronic, incurable disease characterised by elevated blood glucose concentration levels, often experience a broad range of macro- and micro-vascular complications. Among the latter, diabetic nephropathy is the leading cause of end-stage chronic kidney disease (CKD) worldwide [1]. Indeed, it is estimated that the prevalence of CKD among people with diabetes may be as high as double that in the general population [2]. Key intervention targets include improvements in glycaemic control, blood pressure, and lipid profile, which, combined with frequent monitoring via routine check-ups, appropriate therapeutic choices, and positive lifestyle changes, have been shown to delay the onset and slow the progression of diabetic nephropathy [3, 4]. Recently, a novel class of antidiabetic agents known as sodium-glucose cotransporter 2 inhibitors (SGLT2is) have demonstrated marked nephroprotective properties in diabetic patients with pre-existing albuminuria or reduced estimated glomerular filtration rate (eGFR) [5,6,7,8]. However, as a much greater number of diabetic patients with preserved kidney function would need to be treated with SGLT2is to prevent even a single case of nephropathy [5, 9], suboptimal resource allocation remains a concern, and there is no clear indication for specific CKD-preventing therapies in subjects at non-immediate risk.
In light of these considerations, it is apparent that early prediction of impaired renal function is a crucial target with notable ramifications not only on individual quality of life, but also on resource allocation with respect to the early identification of potential candidates for innovative anti-CKD therapy. Recent research in this direction has highlighted that machine learning models based on routinely acquired real-world data have a great potential as tools to aid in the prediction of future CKD [10, 11]. Oftentimes, however, data collection objectives for clinical practice and model development do not align. This is the case, e.g., of routine check-up visits, where different batteries of laboratory tests are usually performed at a physician’s discretion, resulting, on the one hand, in the potentially advantageous acquisition of additional longitudinal information, but, on the other, in incomplete or sparsely sampled data points, which might render baseline definition and outcome adjudication more difficult.
Taking into account this inherent divergence of purposes, in the present work, we develop a deep learning model based on recurrent neural networks to predict the onset of impaired renal function using the routine check-up data of 28,955 patients, acquired in 14 Italian diabetes outpatient clinics. In doing so, we address two main challenges related to model development with this type of data: 1) the incorporation of longitudinal baseline data in the form of the sequence of anthropometric and laboratory information collected during a series of past visits; and 2) the highly irregular sampling of endpoints that is ill-suited to traditional methods.
2 Prediction Target and Study Population
2.1 Prediction Target: Impaired Kidney Function on the KDOQI Scale
The prediction target was the onset of impaired kidney function, i.e., stage ≥ 3 on the Kidney Disease Outcomes Quality Initiative (KDOQI) scale [12]. As only stages ≥ 3 meet the criteria for CKD, we will refer to “CKD onset” and “impaired kidney function onset” interchangeably. Operatively, in terms of outcomes, we distinguished between subjects with preserved renal function, i.e., eGFR ≥ 60 (KDOQI stages 1 and 2) and those with eGFR < 60 (KDOQI stages 3a, 3b, 4, and 5).
2.2 Study Population and Dataset Split
The primary source for the present study was a multi-centre database comprising the data of 28,955 subjects treated at 14 diabetes outpatient clinics in the Veneto region between 1st January 2010 and 14th May 2019 (median observation time: 6.6 years; IQR 4.7–7.8). For each subject, a number of routine check-up visits, recorded with an irregular (on average, yearly) sampling rate, were available. At each visit, demographic, anthropometric, and laboratory data were collected as part of the subjects’ regularly scheduled monitoring sessions. The complete list of variables comprised sex, age, diabetes duration, body-mass index (BMI), systolic and diastolic blood pressures, fasting glucose, glycated haemoglobin (HbA1c), total and HDL cholesterol levels, triglycerides, aspartate transaminase (AST), alanine transaminase (ALT), creatinine, and eGFR for a total of 15 variables (14 dynamic, 1 static). All subjects met the following inclusion criteria.
-
1.
At least three visits with known eGFR (at least two to serve as a sequential input, and at least one more to determine the output).
-
2.
At least two consecutive visits with eGFR ≥ 60.
-
3.
No evidence of CKD at database entry.
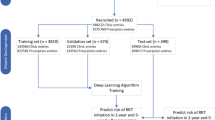
We split the total cohort of 28,955 patients into a training, validation, and test sets, comprising, respectively, 80% (23,164), 10% (2,895), and 10% (2,896) of the subjects. We define the most recent visit in the baseline sequence, i.e., the latest one before the start of the follow-up period, as the end-of-baseline (EOB) visit. The EOB visit is the one that would be used for prediction in the absence of sequence data. Table 1 summarises the characteristics of the study population at the EOB visit (see Sect. 3.1). The average subject had a 60% chance of being male, was 66 years old, had had diabetes for 9 years, a BMI of 29.5, a blood pressure of 140/80 mmHg, a fasting glucose of 145 mg/dL, and an HbA1c of 7.2%. The average eGFR was 86.1 mL/min/1.73 m2.
Missing data were present (except for sex, age, and diabetes duration), but their proportion was small, i.e., <3.5% at the EOB visit.
3 Methods
3.1 Input Data Preparation
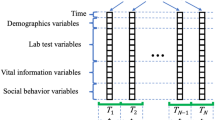
The input data preparation process was guided by our stated objective of incorporating longitudinal baseline data into the model development pipeline. In summary, we identified each patient via a multidimensional sequence of data corresponding to a variable-length sequence of baseline routine check-ups, and a single static feature, i.e., sex. The minimum number of baseline visits was 2, as per the inclusion criteria in Sect. 2.2, thus avoiding the degenerate case of 1-visit sequences. The actual number was subject-specific, i.e., between 2 and the minimum between: a) the number of available visits minus one (at least one was needed for the outcome, as per inclusion criterion 1); b) the number of consecutive outcome-free visits; and c) an arbitrary threshold of 6.
We formatted each subject’s baseline data according to model requirements (see Sect. 3.3), thus obtaining a 14-variable × 6-visit padded matrix and a scalar value (technically, a 1-dimensional vector) encoding the static sex variable. Missing data in the matrix were set to “0” if they were, in fact, missing in the original dataset, whereas “-1” was the masking indicator to distinguish between informative and padded portions of the variable-length sequence. Additionally, to aid in data description and benchmarking, we created a static version of the dataset comprising only each subject’s (unmodified) EOB visit and the “sex” variable.
3.2 Output Coding
The irregular and relatively sparce sampling rate induced by the real-life configuration of the data source prevented us from encoding outcome occurrence via the typical (event indicator, censoring time) tuples used in survival analysis. Indeed, survival analysis requires that exact information on outcome occurrence be known and that there be no gaps in the observation of follow-up. On the contrary, here, outcome information was only available via inspection of the eGFR values collected during each follow-up visit, meaning that status changes between two visits were inherently unknowable, and so was the exact time or reason for right censoring.
To overcome this limitation, we cast the problem of predicting impaired kidney function at different prediction horizons as a multi-label classification problem with a 7-dimensional output. Each of the 7 elements of the outcome vector, say j, encoded the answer to the question “Was there evidence of CKD onset by the end of the j-th year?” Hence, if an eGFR < 60 was recorded between the start of follow-up and the end of the j-th yeah, the j-th element of the outcome vector was equal to 1; if there was evidence of eGFR ≥ 60 after the end of the j-th year (but no evidence to the contrary before then) the j-th element of the outcome vector was equal 0; in all other cases, patient status was unknown and the j-th element was encoded as “NA” (note that this may happen both in the “natural” case of right censoring and due to gaps in eGFR sampling).
Table 2 shows the absolute frequencies of patient status across the 7 time points. We observe an expected, progressive inversion of the ratio between 1s and 0s as the prediction horizon moves forward into the future: as time goes on, follow-up visits that confirm undeteriorated renal function become rarer and rarer, whereas the number of CKD onsets accumulates. Predictably, “NA” values start appearing immediately after the start of follow-up, demonstrating the presence of subjects for whom endpoint information is temporarily unclear in addition to truly right-censored subjects.
3.3 Model Architecture and Development
Using a typical train/validate/test scheme, we developed a deep learning model based on the incorporation of longitudinal baseline data via a recurrent layer. Operatively, we optimised the network’s weights on the training set, selected the best combination of hyperparameters via the validation set, and evaluated performance on the previously unseen test set. We carried out weight estimation via the ADAM optimiser with a fixed learning rate of 0.0005 for a maximum of 100 epochs. The cost function was a modified version of the binary cross-entropy where “NA” labels did not contribute to weight update via back-propagation (this is done, e.g., by artificially setting the missing prediction to the currently predicted value, resulting in a null contribution to the gradient).
As shown in Fig. 1, the proposed neural network initially handles sequence data via a recurrent layer, namely a gated recurrent unit (GRU) [13]. The objective, here, is encoding the variable-length multi-dimensional sequence as a fixed-length vector that can be concatenated with the static “sex” variable. In this portion of the network, the hyperparameters were the number of GRU units (16, 32, or 48), and the dropout fractions related to the inputs and recurrent connections (possible values for both: 0, 0.05, 0.1, 0.2, 0.3, 0.5). The result of this dynamic-to-static encoding step is then concatenated with the static “sex” variable and sent to a cascade of fully connected layers. The hyperparameters at this stage were the number of post-concatenation, pre-output layers (2 or 3) and their dimensions (valid combinations: {64, 32}, {48, 24}, {32, 16}, {16, 8}, {64, 32, 16}, {48, 24, 12}, {32, 16, 8}, {16, 8, 4}). The fully connected cascade ends on the 7-dimensional output layer. Finally, to obtain a more robust scalar score for each prediction horizon, we implemented a cumulative summation step such that each prediction at j years was the sum of the first j output neurons.
We carried out the hyperparameter selection phase in two steps. First, for each hyperparameter combination, we selected the set of weights that minimised the binary cross-entropy on the validation set, thus obtaining a set of 864 candidate models. Second, we computed the cumulative/dynamic areas under the receiver-operating characteristic curve (AUROC) [14] at 1 to 7 years on the validation set, and ranked all 864 candidates according to their predictive ability at each prediction horizon. The final model was the one with the minimum year-wise median rank.
3.4 Performance Evaluation and Secondary Analyses
In our primary performance analysis, we evaluated the discrimination power of the proposed model on the unseen test set via our target metrics, i.e., the seven AUROCs corresponding to the 1- to 7-year prediction horizons.
Our first secondary analysis was meant to challenge the hypothesis that the deep learning model was effectively encoding the sequence of visits comprising the longitudinal baseline. Hence, we measured the model’s prediction ability on a modified version of the test set where we artificially inverted the order of the visits comprising each subject’s longitudinal baseline.
In another secondary analysis, we compared the proposed model to a trivial model returning the eGFR collected at the time of the EOB visit, the known predictor of imminent renal function deterioration used in clinical practice [15].
In a third set of secondary analyses, we compared the proposed model with a battery of year-specific logistic regressions trained with the full EOB visit as the input and with each year’s status (whenever available) as the output. The minority of missing values was imputed via mean imputation.
In all analyses, we estimated 95% confidence intervals (CIs) via the DeLong estimator [16], and assessed statistical significance at the 0.05 level.
4 Results
The hyperparameter selection process resulted in the identification of the optimal architecture as the one having 32 GRU units with standard and recurrent drop-out fractions of 0.05 and 0.1, and three fully connected layers of sizes 16, 8, and 4.
As shown in the second column of Table 3, model performance was satisfactory across the board (AUROC always > 0.84), and particularly promising in the medium term, where it ranged from year 5’s 0.853 (CI: 0.828–0.878) to year 7’s 0.895 (CI: 0.852–0.937). The performance comparison with the artificially inverted version of the test set (first secondary analysis) strongly suggests that the model’s good behaviour was at least in part attributable to a fruitful encoding of temporal relationships between the longitudinal baseline’s visits. This is apparent from the substantially (and significantly, except at the 7-year mark) diminished performance of the model when confronted with improperly ordered sequences (third column of Table 3). Had order been irrelevant, we would have expected to see a negligible difference.
As expected, the proposed model always outperformed EOB eGFR in terms of discrimination power. Interestingly, however, the AUROC difference at the 1-year mark (0.15) was only nominally greater than 0, suggesting that eGFR alone might be a sufficiently effective predictor of imminent deterioration in renal function, while additional information should be collected for longer-term prediction.
The comparison with the battery of year-specific logistic regressions (third secondary analysis, fourth column of Table 3) also yielded encouraging results. Except at the 1-year prediction horizon, the proposed model always outperformed logistic regression, and exhibited the most notable and statistically significant performance gains at 4, 5, and 6 years (respectively, AUROC 0.844 vs. 0.829, 0.853 vs. 0.830, and 0.874 vs. 0.839). Regrettably, despite the proposed model’s excellent AUROC of 0.895 (CI: 0.852–0.937), the 7-year comparison was underpowered and failed to detect statistically significant differences. Overall, it appears that the inclusion of longitudinal baseline data, possibly combined with increased model capacity and with the simultaneous learning from different prediction horizons (via the proposed multi-label coding scheme), was beneficial to long-term prediction. While, under the current experimental framework, it is difficult to disentangle the contributions of these factors, it is notable that our deep learning model, i.e., a single, one-size-fits-all model, was able to compete with and generally outperform individual models specifically trained on the expected outcome distributions observed at each prediction horizon (recall the inversion of the 1:0 ratio shown in Table 2).
5 Discussion and Conclusions
An early prediction of CKD onset in people affected by diabetes but whose renal function is still satisfactory could be extremely useful in reconciliating therapeutic intervention with patient needs and resource allocation constraints. Motivated by previously reported, promising results obtained using machine learning and real-world data [10], in the present work we demonstrated the feasibility and potential benefit of developing a predictive model of impaired kidney function (KDOQI stage 3) using deep learning to integrate longitudinal information on routine check-ups. Thus, we obtained a well-performing model that yielded AUROC values between 0.841 (1-year prediction horizon) and 0.895 (7-year prediction horizon), generally and often significantly outperforming the tested benchmarks.
From a methodological perspective, our study showcases a fruitful approach to utilise routine data whose natural format is suboptimal for traditional survival analysis or classification approaches. Indeed, at variance with most similar models [17, 18], which attempt to recreate the clinical trial setting by predicting a well-behaved outcome via one-shot baseline data, here, we embraced the longitudinal vocation of routine diabetes check-ups by incorporating a sequence of past visits via a recurrent layer, and offset the inconsistent sampling scheme of the CKD endpoint using a multi-label framework and an opportunely modified cost function.
The main limitation of our study was the impossibility of disentangling the contributions to performance improvement of 1) adding baseline sequence data (although we showed that sequence order was effectively encoded by the model), 2) increasing model capacity with respect to traditional techniques such as logistic regression, and 3) casting the problem as a multi-label task. Future research will revolve around the systematic testing of the proposed architecture (or variants thereof, e.g., using different recurrent units, such as LSTMs [19]) against a stronger set of literature and custom-made benchmarks to determine the key factors in achieving high discrimination ability.
References
Koye, D.N., Magliano, D.J., Nelson, R.G., Pavkov, M.E.: The global epidemiology of diabetes and kidney disease. Adv. Chron. Kidney Dis. 25, 121–132 (2018). https://doi.org/10.1053/j.ackd.2017.10.011
Ene-Iordache, B., et al.: Chronic kidney disease and cardiovascular risk in six regions of the world (ISN-KDDC): a cross-sectional study. Lancet Global Health 4, e307–e319 (2016). https://doi.org/10.1016/S2214-109X(16)00071-1
Lin, Y.-C., Chang, Y.-H., Yang, S.-Y., Wu, K.-D., Chu, T.-S.: Update of pathophysiology and management of diabetic kidney disease. J. Formosan Med. Assoc. 117, 662–675 (2018). https://doi.org/10.1016/j.jfma.2018.02.007
Andrésdóttir, G., et al.: Improved survival and renal prognosis of patients with type 2 diabetes and nephropathy with improved control of risk factors. Diab. Care 37, 1660–1667 (2014). https://doi.org/10.2337/dc13-2036
Tuttle, K.R., et al.: SGLT2 inhibition for CKD and cardiovascular disease in type 2 diabetes: report of a scientific workshop sponsored by the national kidney foundation. Diabetes 70, 1–16 (2021). https://doi.org/10.2337/dbi20-0040
Perkovic, V., et al.: Canagliflozin and renal outcomes in type 2 diabetes and nephropathy. N. Engl. J. Med. (2019). https://doi.org/10.1056/NEJMoa1811744
Wanner, C., et al.: Empagliflozin and progression of kidney disease in type 2 diabetes. N. Engl. J. Med. 375, 323–334 (2016). https://doi.org/10.1056/NEJMoa1515920
Neal, B., et al.: Canagliflozin and cardiovascular and renal events in type 2 diabetes. N. Engl. J. Med. 377, 644–657 (2017). https://doi.org/10.1056/NEJMoa1611925
Yin, W.L., Bain, S.C., Min, T.: The effect of glucagon-like peptide-1 receptor agonists on renal outcomes in type 2 diabetes. Diab. Ther. 11, 835–844 (2020). https://doi.org/10.1007/s13300-020-00798-x
Ravizza, S., et al.: Predicting the early risk of chronic kidney disease in patients with diabetes using real-world data. Nat. Med. 25, 57–59 (2019). https://doi.org/10.1038/s41591-018-0239-8
Yang, C., Kong, G., Wang, L., Zhang, L., Zhao, M.-H.: Big data in nephrology: Are we ready for the change? Nephrology 24, 1097–1102 (2019). https://doi.org/10.1111/nep.13636
National Kidney Foundation: K/DOQI clinical practice guidelines for chronic kidney disease: evaluation, classification, and stratification. Am. J. Kidney Dis. 39, S1–266 (2002)
Gal, Y., Ghahramani, Z.: A theoretically grounded application of dropout in recurrent neural networks. arXiv:1512.05287 [stat]. (2016)
Bansal, A., Heagerty, P.J.: A tutorial on evaluating the time-varying discrimination accuracy of survival models used in dynamic decision making. Med. Decis. Making 38, 904–916 (2018). https://doi.org/10.1177/0272989X18801312
Mayer, G., et al.: Systems biology-derived biomarkers to predict progression of renal function decline in type 2 diabetes. Diab. Care 40, 391–397 (2017). https://doi.org/10.2337/dc16-2202
DeLong, E.R., DeLong, D.M., Clarke-Pearson, D.L.: Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics 44, 837–845 (1988). https://doi.org/10.2307/2531595
Dagliati, A., et al.: Machine learning methods to predict diabetes complications. J. Diab. Sci Technol. 12, 295–302 (2018). https://doi.org/10.1177/1932296817706375
Retnakaran, R., Cull, C.A., Thorne, K.I., Adler, A.I., Holman, R.R.: Risk factors for renal dysfunction in type 2 diabetes: U.K. Prospective diabetes study 74. Diabetes 55, 1832–1839 (2006). https://doi.org/10.2337/db05-1620
Gers, F.A., Schmidhuber, J., Cummins, F.: Learning to forget: continual prediction with LSTM. Neural Comput. 12, 2451–2471 (1999)
Acknowledgments
This work was partly supported by MIUR (Italian Ministry for Education) under the initiative “Departments of Excellence” (Law 232/2016).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 Springer Nature Switzerland AG
About this paper
Cite this paper
Longato, E., Fadini, G.P., Sparacino, G., Avogaro, A., Di Camillo, B. (2021). Recurrent Neural Network to Predict Renal Function Impairment in Diabetic Patients via Longitudinal Routine Check-up Data. In: Tucker, A., Henriques Abreu, P., Cardoso, J., Pereira Rodrigues, P., Riaño, D. (eds) Artificial Intelligence in Medicine. AIME 2021. Lecture Notes in Computer Science(), vol 12721. Springer, Cham. https://doi.org/10.1007/978-3-030-77211-6_37
Download citation
DOI: https://doi.org/10.1007/978-3-030-77211-6_37
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-77210-9
Online ISBN: 978-3-030-77211-6
eBook Packages: Computer ScienceComputer Science (R0)