Abstract
One of the key challenges in metagenomic forensics is to establish a microbial fingerprint that can help in the prediction of the geographical origin of the metagenomic samples. Understanding a combination of different aspects such as microbiome sample sources, sequencing technologies, bioinformatics processing, statistical and machine learning methods, play a vital role in a comprehensive analysis of metagenomic data. We demonstrate the analysis of metagenomic samples from 23 different cities around the world to construct classification models that can be utilized to predict the geographical location for unknown samples obtained from these regions. We also describe the bioinformatics pre-processing of the raw sequencing data and estimate the abundance profiles of microbes in the samples using multiple tools. For the prediction of the geographical origin of samples, we trained and evaluated a variety of supervised learning classifiers including an adaptive optimal ensemble classifier that performs well on different data generating procedures.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
1 Introduction
Samples for environmental microbiome analysis are collected from a variety of surfaces and environments such as plants, soil, ocean, public transit systems, public benches, stairwell handrails, elevators, and urban environments. Analysis that focuses on human microbiome relies on samples from different body sites such as skin, gut, tongue, buccal mucosa, stool, etc. Metagenomic experiments aim to describe microbial communities from these samples using high-throughput DNA sequencing, also known as next-generation sequencing (NGS) technologies. This has further helped scientists around the world to peek into a plethora of diversity of microbes in our environment. The data from these sequencing technologies pose various statistical and computational problems. Also, the sheer magnitude and special data characteristics make metagenomic data analysis a challenging task.
Metagenomic analysis has diverse applications and has led to foundational knowledge on various aspects of human lives. The composition of the human gut microbiome is associated with the physiological and psychological aspects of human health [28, 33, 61, 66, 67]. Metagenomic analysis has a wide-scale application in designing healthy urban environments [47] and discovering novel anti-resistant microbial strains [58]. Metagenomic analysis of microbial communities also provides a significant source of information in forensic science. One of the many questions in forensic studies that metagenomic analysis can answer is predicting the source origin of the metagenomic sample [10, 11, 15]. In this chapter, we discuss various classification methods that can be applied to achieve this goal.
Human Microbiome Project (HMP) [65] and Earth Microbiome Project (EMP) [26] are some of the large-scale initiatives that have offered a comprehensive database for microbiome research. The MetaSUB Consortium comprised of an international group of scientists is involved in the collection and sequencing of samples from numerous cities in different countries to understand the microbial signature across and within the public spaces of cities around the world. These large-scale data are published by the Critical Assessment of Massive Data Analysis (CAMDA) in the public domain to find innovative solutions to the pressing questions in modern life sciences. We use the data from CAMDA 2020 Geolocation Challenge and demonstrate a step-by-step approach for metagenomic data analysis. The analysis is divided into two parts, namely, upstream and downstream analysis. In the upstream analysis, we discuss the process of converting raw data of sequenced reads into an n × p data matrix ready for statistical analysis. This process involves quality control, taxonomic assignment, and estimation of taxonomic abundance of the sequenced reads from different samples. In the downstream analysis, we apply various classification methods and compare their performance for the prediction of the geographical location of microbial samples. Several supervised learning classifiers, such as Support Vector Machines (SVMs), Extreme Gradient Boosting (XGB), Random Forest (RF), and neural networks, can be applied to predict the geolocation of the metagenomic samples. Along with these classifiers, we describe the construction and implementation of an optimal ensemble classification algorithm proposed by Datta et al. [18], which combines several candidate classification algorithms and adaptively produces results that are better or as good as the best classifier included in the ensemble.
2 Bioinformatics Pipeline
2.1 Microbiome Data
Microbiome samples are sequenced using next-generation sequencing technologies. The two most widely used sequencing techniques are metataxonomics that use amplicon sequencing of the 16S rRNA marker genes and metagenomics that use random shotgun sequencing of DNA or RNA [8, 45]. Until recently, most studies sequenced the 16S ribosomal RNA gene that is present in bacterial species or focused on characterizing the microbial communities at higher taxonomic levels. Following the drop in the cost of sequencing, metagenomics studies have increasingly used shotgun sequencing that surveys the whole genome of all the organisms including viruses, bacteria, and fungi present in the sample [57].
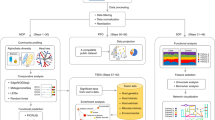
Metagenomic samples in our case study were sequenced using Illumina HiSeq next-generation shotgun sequencing technology, and the raw data for each sample was obtained in the form of paired-end .fastq files with forward and reverse reads. Fastq files contain both nucleotide sequences and their corresponding quality scores, known as Phred scores. These scores are used in the quality assessment of these sequencing reads. For the upstream analysis, we started with assessing the quality of the paired-end WGS (whole-genome sequencing) reads followed by their taxonomic classification. Please note the taxonomic classification should not be confused with the city-specific classification that we perform in the downstream analysis. Taxonomic classification refers to mapping the raw sequenced reads of a sample to an existing database of known genomic sequences to produce taxonomic abundance profiles for each sample. Figure 1 shows the schematic representation of the bioinformatics pipeline constructed for the analysis of the metagenomic data. The components of this pipeline are described in detail in the following sections. Table 1 provides information on the data set being analyzed in this chapter. The data set comprised 1065 samples collected from 23 cities around the world.
2.2 Quality Control
Raw NGS reads contain different types of contamination such as low-quality reads, adapter sequences, and host reads. It has been noticed that low-quality sequences can result in misleading inference from the downstream analysis [14, 71]. Hence, it is important to assess the quality of raw sequencing reads before moving ahead with the downstream analysis. If the metagenomic samples are contaminated due to the presence of host (human) sequences, it is necessary to identify and filter out the host reads.
There are a variety of computational tools that can be used for quality control for removing the contaminants and low-quality reads, such as FastQC [2], Cutadapt [46], Trimmomatic [4], and BBTools. The quality of reads from a sample can be assessed by using the diagnostics report generated by FastQC [2], and these quality assessment reports can be further aggregated into a single report using MultiQC [21] for multiple samples. Figure 2 shows the quality score plots from MultiQC for three arbitrarily selected cities from three continents in our study. The x-axis shows the positions of the bases, and the y-axis represents the Phred score. The Phred score (= −10log10 P) is an integer value representing the estimated probability P of error for identifying the bases generated by DNA sequencing technology. A Phred score of 40 of a base implies that the chance of this base being called incorrectly is 1 in 10,000 [22]. We employed KneadData (version 0.7.4) [49] for quality control analysis. KneadData invokes Trimmomatic [4] for quality trimming, filtering, and removal of adapter sequences. It further calls Bowtie2 [38], which maps the sample reads to a reference human genome database. We discard reads that map to the human genome database. The code snippets below demonstrate how we assessed quality using FASTQC and performed quality control using KneadData. In the pre-QC step, we analyze whether it is necessary to improve the quality of reads. Notice that some of the reads in the second and third columns of Fig. 2 have poor-quality scores (below 30). Hence, we choose to trim or drop poor-quality reads. Based on the pre-QC assessment, one can define various rules to improve the quality of the reads to be used for subsequent analysis. For example, in the quality control code, the parameter ILLUMINACLIP:NexteraPE-PE.fa:2:30:10:8:keepBothReads SLIDINGWINDOW:4:30 MINLEN:60 prompts Trimmomatic to remove adapters, defines a sliding window that cuts a read once the mean quality in a window of size 4 falls below a Phred score of 30, and retains sequences with a minimum length of 60. This procedure results in sequencing reads with reasonably good quality. We assessed the quality of the reads after quality control using MultiQC and noticed an obvious improvement in the quality of the reads when compared to the raw reads. The upper panel of Fig. 2 shows the reports from pre-QC analysis, and the lower panel of Fig. 2 shows the plots from the post-QC analysis. The code below can be used as a basic guideline for performing the bioinformatic pre-processing of raw sequenced reads. We encourage readers to make appropriate modifications to the parameters of the bioinformatics tools to suit the goal of their analysis. These tools are also constantly undergoing development. Consequently, it is recommended that the researcher works with the most recent versions of software and databases used for sequence mapping.



2.3 Taxonomic Profiling
After quality control of the sequencing reads, the next step is to estimate the taxonomic abundance of each sample. A taxonomic abundance table is an n × p matrix of absolute or relative abundance of p identified taxa in n samples. Taxonomic profiling of sequenced reads typically comprises two steps. First, the classification or the alignment of sequence reads to a database of microbial genomes. The second step involves the estimation of the abundance of each taxon (species, genus, etc.) in the metagenomic sample, i.e., estimating the number or percentage of reads belonging to each taxon. Various algorithms and tools have been developed to efficiently classify sequencing reads to known taxa with improved speed [70]. A variety of metagenomic profiling tools match sequences to known databases. These databases created at different times may have different contents as they go through regular updates with the addition of new sequences.
Taxonomic profiling tools use a variety of approaches such as alignment of marker genes (MetaPhlAn2 [59], mOTU [63], GOTTCHA [23]), k-mer mapping in WGS reads (Kraken [23], CLARK [53]), translating DNA into amino acid sequences, and mapping to protein databases (Kaiju [50], DIAMOND [9]).
This chapter does not pursue the goal of reviewing all of these taxonomic profiling tools. Several research papers provide discussion on the review and the comparison of these taxonomic profiling tools [3, 8, 43, 48]. Performance is usually compared on the basis of the proportion of mapped reads, run time, sensitivity, and other performance metrics. Since the evaluation of these tools is a complex task, no single metric is usually used to judge the performance; rather, multiple factors are examined. Considering that some tools utilize a limited set of marker genes while others use expansive databases, judging a profiling tool only by the proportion of reads mapped may not be adequate [43]. Since the application of any taxonomic profiling tool will potentially impact the results and conclusions of the metagenomic study, the selection of the appropriate tool should be based on performance metrics that suites the analyst’s scientific investigation. In this section, we describe and also discuss the implementation of three commonly used taxonomic profiling tools, namely MetaPhlAn2, Kraken2, and Kaiju.
2.3.1 MetaPhlAn2
We implement MetaPhlAn2 [59] for the quantitative taxonomic profiling of our quality-controlled sequenced reads. MetaPhlAn2 is computationally fast as it relies on the clade-specific marker genes approach for taxonomic profiling [59], and this approach is not expected to map all reads. Taxonomic assignment is attained by aligning the sequence reads to the marker set using Bowtie2 [38]. In the application, we used the default settings of MetaPhlAn2 to extract species-level relative abundances for each sample, and these values lie within [0, 1]. The relative abundances for each sample were then merged into a large relative abundance table using a custom MetaPhlAn2 script. After termination of MetaPhlAn2 procedure, we obtained a table of relative abundances of 1049 species for 1047 samples. In this setup, we have chosen to obtain species-level relative abundance. However, information for other taxonomic levels can be easily extracted from the output generated by MetaPhlAn2.

2.3.2 Kraken2
Kraken2 [69] is a rapid and highly accurate metagenomic classification tool that uses a k-mer approach. For assignment of sequence reads to taxonomic labels, it utilizes the k-mer information within each read, and each k-mer is mapped to the lowest common ancestor (LCA) of the genomes that contains the k-mer in a custom-built database. Lu et al. [44] point out that the LCA approach employed by the Kraken system means that the system is likely to underestimate the number of reads that are directly classified as species.
To overcome the issue of underestimation of taxonomic abundance by the Kraken system, Bracken [44] was developed. Bracken uses a Bayesian algorithm and the results from the Kraken2 classification for estimation of the relative abundance of a metagenomic sample at the user-specific taxonomic level. To illustrate the difference between these tools, the developers of Bracken report an instance [44] that we consider here. The genomes of Mycobacterium bovis and Mycobacterium tuberculosis are 99.95% identical. Since these species are very similar, Kraken classifies the vast majority of reads from either of them to their LCA, which in this case is the genus Mycobacterium. On the other hand, Bracken uses information on some reads from the species-specific portion of the genome along with the similarity information between close species to move reads from the genus level to the species level.
To estimate the abundance for each sample using the Kraken2–Bracken system, we employed a pre-computed standard database that consists of reference sequences from archaea, bacteria, and the human genome. Further, to generate the Bracken database file, the switch -t indicates the number of threads to use, and -l indicates the read length of the data. Since most of our data were 150 base pair (bp) reads, we set -l to be 150, and we use the default k-mer length of 35. Then, for each paired-end sample, we generate reports from the Kraken2 taxonomic assignment procedure, and these reports are then passed into the Bracken program for abundance estimation. Estimation of the abundance was carried out at the species level (-l S), with a default reads threshold of 10 (-t 10). Finally, we use a custom script to combine the Bracken output for all samples into a large single file. The column of interest in the Bracken output is the new_est_reads, which gives the newly estimated reads. After obtaining the abundance table, normalization was carried out using the cumulative sum scaling approach. This procedure was implemented with the metagenomeSeq [55] R package.

2.3.3 Kaiju
For the given DNA sequences, Kaiju [50] translates the reads into amino acid sequences and compares these reads against a reference database of protein sequences. It creates an efficient database structure by indexing the reference protein database using the Burrows–Wheeler transform (BWT) and saves each sequence in an FM-index (Full-text index in Minute space) table. It then searches for maximum exact matches between the reads and the reference database created. Kaiju’s developers [50] emphasize that protein-level classifiers such as Kaiju are more sensitive to novel or highly variable sequences because protein sequences are more conserved than the underlying DNA. Moreover, protein sequences are more tolerant to sequencing errors due to the lower mutation rate of amino acid sequences as compared with nucleotide sequences [1, 70].
To execute the taxonomic classification of sequencing reads using Kaiju, we used nr database as our reference database. Program kaiju-makedb downloads the source database of interest and constructs Kaiju’s index using BWT and FM-index. We observed that some tools used for quality control of the sequences may create disorder in the read names in both .fastq files. If the read names are not identical between the first and second files, program kaiju issues an error. We used Repair function from bbmap to fix this issue before moving ahead with the taxonomic classification of sequencing reads. For faster implementation, we used kaiju with multiple parallel threads using option -z 25 in MEM mode (-a mem). The output files obtained from program kaiju comprised 3 columns, classification status C/U for each read, read names, and NCBI taxon identifier of the assigned taxon. These output files were further summarized into a table using kaiju2table script, which gives read count (or percentage) for all samples and taxa in a long format. To process this data for the downstream analysis, we converted it into a wide format with taxa as rows and samples as columns using pivot_wider function from tidyverse package in R. Users may also choose to run Kaiju in greedy mode that yields a higher sensitivity as compared to the MEM mode, sometimes at the cost of increased run time.

As mentioned earlier, the choice of profiling tool may depend on multiple factors such as classification speed, proportion of mapped reads, output format, ease of use, and computational resources available. If the analyst has access to good computational resources with high amounts of available memory (>100Gb), then Kraken, Bracken, and Kaiju are useful options. If sufficient computational resources are not available, then MetaPhlAn is a viable alternative with fast classification speed. Kaiju, for instance, has a web server where one can upload the compressed .fastq files and select different options for taxonomic assignment for an easier implementation without running bash scripts via the command line. Simon et al. [70] provide an interesting and informative assessment of the performance of several metagenomic tools used for taxonomic profiling of real and simulated data sets.
2.4 Computing facilities
All bioinformatics procedures were performed using the University of Florida HiPerGator2 supercomputer. HiPerGator2 has 30,000 cores in Intel E5-2698v3 processors with 4 GB of RAM per core, and a total storage size of 2 petabytes (PB). Bash scripts and .fastq files were stored on the supercomputer’s parallel file system that offers high performance for data analysis. For the computing jobs submitted to the cluster, we typically requested an allocation of a single computing node, 20 cores per task, and 200 GB memory.
3 Methodology
In Sect. 2.3, we discussed several techniques for taxonomic profiling that comprised taxonomic classification/assignment and estimation of abundance. At the termination of each profiling technique presented, we obtained a species abundance table. Now, the rest of this chapter will focus on methods for classifying taxa abundances to known class labels. That is, we pursue the goal of modeling taxa abundances of metagenomic samples belonging to known class labels. Then, the model is used to predict class labels for new metagenomic samples based on their estimated abundances. For our analysis, the class labels are the source cities where samples originated. The classification of sequence reads to taxonomic labels should not be mistaken for the classification of abundance profiles to source cities. We stress that the term classification will refer to the latter described herein.
As we indicated in the previous paragraph, this section focuses on the supervised learning analysis of the pre-processed metagenomics data. We highlight methods for feature selection, present several classification algorithms that include the ensemble classifier, discuss techniques to overcome the problem of class imbalance, and finally discuss measures for evaluation of model performance.
3.1 Pre-Processing and Feature Selection
The species abundance matrix obtained after the taxonomic profiling contains a large set of features, i.e., taxa. For instance, 6152 taxa were obtained after taxonomic profiling with the Kraken2–Bracken system, while 1049 taxa and 32,146 taxa were obtained after profiling was, respectively, performed with MetaPhlAn2 and Kaiju.
Similar to the cases presented here, the most abundance data obtained from metagenomics samples are high-dimensional in nature, and it is usually desirable to extract only important features from the data. Common feature reduction techniques are based on the prevalence of the taxa in the abundance table. For instance, taxa with less than a specified number of reads, say 10, can be dropped. In addition, taxa that are present in less than, say, 1% of the samples may also be discarded. If these approaches are employed, then the resulting abundance table should be re-normalized.
Other advanced methods exist for feature selection, and in this section, we describe a couple of these techniques. In practice, feature selection aims at obtaining a reduced subset of relevant informative features that bolster the assignment of samples of known class labels based on their abundance information. However, from our experience and those of several research studies [54], feature selection may not provide a substantial improvement in the predictive ability of the fitted classification models due to the complex nature of microbiome data. Hence, even though fitting classification models on the data with a reduced feature space may be more computationally efficient, we recommend that analysts should also investigate the performance of such models when trained on the data with a complete feature space.
Among the other approaches to feature selection, first, features could be selected based on the importance scores returned after a supervised training of the Random Forest model on the data with a complete set of features. The features are ranked according to their importance scores, and the top k features are chosen as the set of informative features. The classification model of interest is then trained with the k selected importance features. In this setup, k is usually chosen from a set of a predetermined number of features via cross-validation, such that the number of features from the predetermined set that maximizes classification accuracy is chosen to be k. Pasolli et al. [54] utilized this method in their review study that assessed machine learning approaches for metagenomics-based prediction tasks.
In another heuristic approach, one may choose to use the Lasso [64] or ElasticNet [73] with a multinomial model for feature selection. However, the standard versions of penalized regression methods are not efficient for the analysis of relative abundance data because of the compositional nature of the data [26]. Owing to this fact, regression [31] and variable selection methods [41], which impose sum-to-zero constraints for the Lasso, have been developed for compositional data.
The hierarchical feature engineering (HFE) [52] technique is a recently developed tool for performing feature selection. To obtain a smaller set of informative microbial taxa, this tool uses information from the taxonomy table, the correlation between taxonomic labels, and the abundance data to exploit the underlying hierarchical structure of the feature space. At the termination of the algorithm after analyzing a species abundance table, it returns an OTU table that contains a combination of both species and other higher-level taxa. Fizzy [19] is another modern tool for feature selection. It is a collection of functions for performing widely implemented feature selection methods such as the Lasso, information-theoretic methods, and the Neyman–Pearson feature selection approach. Developers of the HFE used the predictive performance of several machine learning models to compare the HFE with other standard feature selection tools that do not account for the hierarchical structure of microbiome data. They reported that the HFE outperformed the other methods.
3.2 Exploration of Candidate Classifiers
In this section, we present brief descriptions of some supervised learning models commonly used for the classification of abundance values of metagenomics samples to known class labels. Our survey of algorithms will largely focus on supervised classifiers that are suitable for analyzing multiclass classification problems. These classifiers can be broadly partitioned into linear and non-linear classifiers.
Linear methods for classification such as linear discriminant analysis, quadratic discriminant analysis, regularized discriminant analysis, logistic regression, and SVM (without kernels) achieve classification of objects based on the value of a linear combination of features in the training data. These classifiers solve classification problems by partitioning the feature space into a set of regions that are defined by class membership of the objects in the training data. Also, the decision boundaries of the partitioned regions are linear [32]. Generally, these classifiers also take less time to train than non-linear classifiers. However, by using the so-called kernel trick, some linear classifiers can be converted into non-linear classifiers that operate on a different input scale.
In cases where the training data are not linearly separable (usually via a hyperplane), a linear classifier cannot perfectly distinguish classes of such data. For such cases, the non-linear classifiers will often provide better classification performance than the linear classifiers. Examples of non-linear classifiers commonly used for classification in metagenomics studies include the kernel SVM, Random Forest (RF), and neural networks (multilayer perceptron):
-
Recursive Partitioning (RPart)—A decision tree [7] is the fundamental element of the RPart model. A decision tree is grown by repeatedly splitting the training data set into subsets based on several dichotomous features. The recursive splitting from the root node to the terminal node is based on a set of rules determined by the features. The process is recursive in nature because each subset can be split an indefinite number of times until the splitting process terminates after a stopping criterion is reached. In the case where the target response is a unique set of labels, the tree model is called a classification tree. For the prediction of the class label of a new subject, the model runs the observation from the root node to the terminal node that assigns the class membership.
-
Random Forests (RF)—The idea of the RF classifier [6] is to grow a collection of trees by randomizing over subjects and features. That is, each tree in the forest is grown by using a bootstrap sample from the training data. Out-of-bag samples comprise samples that are not included in the bootstrap sample. These samples serve as a validation set. In contrast to bagging that uses all p predictors for splitting at each node, RF uses only m < p randomly selected features to obtain the best split. With the implementation of this step, the correlation between the trees is reduced. Also, it improves the classification performance obtained when a bagging procedure is implemented. Unlike decision trees, no pruning is performed for Random Forests, i.e., each tree is fully grown. For predicting the class of a new observation, each tree in the forest gives a class assignment, and majority voting is used to obtain the final prediction. Advantages of the RF include its robustness to correlated features, its applicability to high-dimensional data and the ability to handle missing data internally in an effective manner, and its use as a feature selection tool through its variable importance plot. Also, it offers competitive classification accuracy for most problems with little parameter tuning and user input.
-
Adaptive Boosting (AdaBoost)—In the boosting [24] procedure, many weak classifiers are sequentially combined to produce a strong learner. The procedure achieves this by repeatedly training many weak learners on modified versions of the data set, and then the strong learner is created by a weighted average of the weak classifiers. Note that a weak classifier is a learner whose performance is only slightly better than random guessing. Also, the weights used to fit each of the weak classifiers are functions of the prediction accuracy using some previous versions of the weak classifier. If we let G m(x), m = 1, …, M denote a sequence of weak classifiers trained with weighted versions of the training data, the final output of the AdaBoost classifier is a weighted sum of G m(x). In this case, weights w i, i = 1, …, N, that are updated iteratively are applied to the observations in the training set. At the first boosting iteration, m = 1, a base classifier, i.e., \(w_i=\frac {1}{n}\), is trained. Then, for m = 2, …, M, observations that were misclassified in the preceding iteration are given more influence than observations that were correctly classified. In this sense, the boosting procedure is adaptive because each subsequent classifier in the sequence is thereby forced to tweak its performance to favor observations that were misclassified by previous classifiers.
-
Extreme Gradient Boosting (XGBoost)—Gradient boosting [25], also referred to as gradient boosting machines (GBM), is another boosting algorithm that creates a strong learner from an ensemble of weak classifiers, typically decision trees. In the implementation, this machine combines a gradient descent optimization procedure with the boosting algorithm. The machine is constructed by fitting an additive model in a forward stage-wise manner. A weak learner is sequentially introduced at each stage to improve the performance of existing learners in classifying previously misclassified observations. These misclassified observations are determined by gradients, which in turn guide the improvement of the model. For XGBoost [13], trees are grown to have a varying number of terminal nodes. Contrasting to GBM that employs gradient descent, XGBoost employs Newton boosting that uses Newton–Raphson’s method to obtain the solution to the optimization problem. With set parameters, the XGB algorithm reduces the correlation among the trees grown, thus increasing classification performance. Further, the algorithm utilizes parallel and distributed computing that speeds up learning and enables quicker model exploration. Historically, this classifier has been popular among winning teams participating in machine learning competitions [13].
-
Support Vector Machines (SVM)—To understand the concept of the SVM [17], first, we consider a binary classification problem for which we intend to assign a new data point to either of two classes. The data point is treated as a p-dimensional vector, and the SVM algorithm aims at finding a (p − 1)-dimensional hyperplane that represents the largest separation between the two classes. Several hyperplanes may exist for partitioning the data. SVM selects the decision boundary that maximizes the distance to the nearest data point on each of its sides as the optimal hyperplane. SVMs are popular for solving classification problems because in the case where no linear decision boundary exists, they can allow for non-linear decision boundaries using the so-called “kernel trick.” Also, SVM solves a multiclass classification problem by decomposing the problem into multiple binary classification problems. In this sense, most SVM software constructs binary classifiers that distinguish between one of the class labels and the others (one-versus-all) or between every pair of classes (one-versus-one). In the latter approach, \(\frac {k(k-1)}{2}\) binary classifiers are constructed if the target variable is comprised of k classes. For the prediction of a new observation in the one-versus-all case, the binary classifier with the maximum output function decides the class label, while a majority voting strategy is used to assign the class label in the one-versus-one case.
-
Multilayer Perceptron (MLP)—Under the deep learning framework, MLP [32] is an interconnected network of neurons or nodes that have weights attached to the edges of the network. MLP utilizes an adaptive mathematical model that changes based on the information that is fed into the network. Using several layers of units, the network maps the input data to an output vector with length equal to the number of classes in the target variable. First, the input data are passed into an input layer. This layer emits a weighted output that is further passed into another hidden layer of units (there can be more than one hidden layer). In the final branch of the process, the output layer receives the weighted output from the hidden layer and assigns the network’s prediction.
Several classifiers provide the option to scale the features so that they have the same variance. This scaling procedure will destroy the compositional nature of the data, and hence, we suggest that scaling should not be done. From the documentation of most classification learning software, we can set the logical scale or standardize parameters that indicate whether scaling should be carried out. This parameter should be set to FALSE.
3.3 The Ensemble Classifier
In Sect. 3.2, we presented a variety of popular machine learning models that can be used to predict the source origin of metagenomics samples. These classifiers have been used to analyze data obtained from several experimental studies that aimed to explore associations between microbial imbalance and disease or environmental factors. The RF and SVM classifiers remain state-of-the-art for metagenomics-based classification purposes. In contrast, classifiers such as the AdaBoost and XGBoost that are based on boosting algorithms have not gained much traction in the metagenomics data classification.
Research papers such as Knights et al. [35], Moitinho-Silva et al. [51], and Zhou et al. [72] provide a review of a variety of supervised machine learning models commonly used for feature selection and classification in microbiota studies. The reviews on the classification of microbiota data often report microbiome–phenotype associations and host-microbiome and disease associations. Among other findings, several individual studies have utilized different pre-processing and analysis methods that yielded discrepant conclusions and difficulty of classification models to be generalized across research studies [20, 54, 72].
In the context of exploring the relationship between microbial samples and environmental factors, CAMDA had organized the Metagenomics Geolocation Challenge over the last three years. Participants who have worked on these challenges have used a combination of bioinformatics and machine learning techniques to build microbiome fingerprints for the prediction of the source origins of microbial samples. Neural networks, RF, and SVM are among commonly used machine learning techniques for the construction of such fingerprints. In particular, no single classifier has shown to give consistent optimal performance across these metagenomics studies. When addressing results from a classification competition based on proteomics data, Hand [29] points out this observation as well.
Several reasons may account for the inconsistencies and non-generalizability of machine learning models across microbiome studies. Potential factors that can elicit inconsistencies in microbiome studies include the nature of the data being studied, sample collection strategies, different sequencing techniques, and varying bioinformatics procedures. Furthermore, the performance of machine learning models is likely to depend on the techniques utilized during the pre-processing and taxonomic profiling of the microbial samples. In practice, it is generally impossible to know a priori which machine learning model will perform best for a given classification problem and data.
To create a more robust classifier, Datta et al. [18] proposed an ensemble classifier that combines a variety of classification algorithms in conjunction with dimension reduction techniques (if necessary) for classification-based problems. The ensemble classifier is constructed by bagging and weighted rank aggregation [56], and it flexibly combines the standard classifiers to yield classification performance that is at least as good as the best performing classifier in the set of candidate classifiers that define the ensemble. For any data set under investigation, the ensemble classifier excels in the sense that it adaptively adjusts its performance and attains that of the best performing individual performance without prior knowledge of such classifier(s). Hand [29] also states that the aggregation of results obtained from many fitted models serves to smooth the ensemble model away from a single model that is optimized on the training set, and therefore, the combination of models serves a role similar to regularization.
The ensemble classifier is itself a classification algorithm, and here, we describe the construction of this classifier. Consider the abundance matrix X = (x 1, …, x p) for n samples and p taxa, where each x j, j = 1, .., p, is normalized, and the target labels, y = (y 1, .., y n). The steps to build the ensemble classifier are as follows:
-
1.
Choose M candidate classifiers and K performance metrics. Then, for b = 1, …, B:
-
(i)
Draw a bootstrap sample \({\mathbf {Z}}_b^* = ({\mathbf {X}}_b^*, {\mathbf {y}}_b^*)\) of size n for the training data. Ensure samples from all classes are represented in \({\mathbf {Z}}_b^*\). OOB samples comprise all samples not included in \({\mathbf {Z}}_b^*\).
-
(ii)
Train each M classifier with the bootstrapped sample, \({\mathbf {Z}}_b^*\).
-
(iii)
Use each M classifier to predict the OOB samples.
-
(iv)
Based on the true values of the OOB set, and the predicted class labels, compute the K performance measures.
-
(v)
Perform weighted rank aggregation: The performance measures used in step (iv) rank the classifiers according to their performance under each measure, thereby producing K ordered lists, L 1, L 2, …, L K, each of size M. Using weighted rank aggregation, the ordered lists are aggregated to determine the best single performing classifier denoted as \(A^b_{(1)}\).
The ensemble is a set of \(\left \{A^1_{(1)},\ldots ,A^b_{(1)},\ldots ,A^B_{(1)} \right \}\) classifiers.
-
(i)
Notice that the algorithm evaluates the performance of each candidate classifier based on their prediction of the OOB samples. This protects the ensemble classifier from overfitting. Just like cross-validation, the classification performance based on the OOB samples is estimated using data that were not used when training the classifier. The OOB errors are not the same as the cross-validation errors, but in practical terms, they should be approximately close.
Given the abundance of a new sample, x 1xp, the ensemble classifier gives prediction for such sample using the following procedures:
-
1.
Each classifier, \(A^1_{(1)}, \ldots , A^B_{(1)}\), in the ensemble is used to predict the class label of x 1xp. Let \(\hat {y}_1, \ldots , \hat {y}_B\) denote the class predictions from the B models in the ensemble.
-
2.
The final prediction is obtained by majority voting, that is, the most frequent class label among the B predicted classes.
3.4 Class Imbalance
More often than not, metagenomics data are imbalanced. That is, at least one of the classes in the data is underrepresented. Data imbalance is likely to skew the performance of the classification models such that the models will be biased toward the majority classes. For instance, if there are disproportionately more samples from class A than there is from class B, the classification model is prone to assign a random label to class A than class B. Since classification algorithms aim to reduce the overall misclassification rate, rather than the error rate in majority classes, such models will not perform well for imbalanced data. Generally, classification algorithms are poised to perform better with nearly equal representation of classes in the training set.
The problem of class imbalance has received considerable attention in the machine learning literature, and a variety of methods exist to mitigate this problem. Some of these methods have also found application in the analysis of metagenomics data. In this section, we briefly describe the underpinnings of such procedures along with their pros and cons. The application of these methods does not improve the overall fit of the classification model discussed. When implemented, they aim to improve the prediction of samples in the minority classes. Roughly speaking, these methods are partitioned into down-sampling, over-sampling, hybrid, and weighting techniques:
-
(i)
Down-sampling techniques: This involves randomly removing samples from the majority classes until class frequencies are roughly balanced. One disadvantage of this technique is the loss of information in the majority classes since a large part of the majority classes will not be used to train the classifier.
-
(ii)
Over-sampling techniques: This involves the random replication of samples in the minority classes to attain approximately the same sample sizes in the majority classes. As noted by Chen et al. [12], more information is not added to the data by over-sampling; however by replication, the weight of the minority classes is increased. From our experience, down-sampling appears to be more computationally efficient since the classifier is trained on smaller data sets.
-
(iii)
Hybrid techniques: This class of techniques combines both over-sampling and down-sampling to artificially create a balance in the data set. For instance, SMOTE (and its variants), AdaSyn, DSRBF, and ProWSyn methods generate synthetic samples from the minority classes to balance class frequencies. Kovács [36] studied the performance of a variety of minority over-sampling techniques when applied to large imbalanced data sets. They report that no over-sampling technique gives consistent optimal performance. Hence, they suggest careful investigation when choosing the technique to use. The smote-variants [37] package provides Python implementation for a host of these hybrid techniques, while the UBL [5] package provides certain implementations in R. In the context of the analysis of microbiome data, a variety of user-specific hybrid over-sampling techniques have been employed. For instance, Knights et al. [35] used an artificial data augmentation approach to boost the representation of samples when analyzing microarray data. In their approach, they generate noisy replicates by adding a small amount of Gaussian noise to the OTU counts in each sample, with a threshold of zero to avoid negative values. The authors found that the difference in predicted error between their augmented and unaugmented model was at most 2% decrease in error. Also, Harris et al. [31] report an increment in classification accuracy from 83% to 91% after application of an optimized sub-sampling technique to address the problem of data imbalance in their analysis of metagenomics data aimed at predicting sample origins.
-
(iv)
Weighting: A cost-sensitive approach to fitting classification models is to train them using class weights. In this approach, the algorithms place heavier weights on the minority classes and will penalize the classifier for misclassifying the minority classes. The weighted Random Forest [12] is an example of a classification model that implements class weighting.
To avoid overfitting the data, these techniques for addressing class imbalance are generally applied only to the training set. Further, if a resampling technique (bootstrap or cross-validation) is used for model evaluation during analysis, the over-sampling procedure should be performed inside the resampling technique. This approach is followed because if an over-sampling is done before, for instance, cross-validation is performed, the model is likely to have glanced at some samples in the hold-out set during model fitting; therefore, the hold-out set is not truly unknown to the model. This implementation will result in overly optimistic estimates of model performance.
3.5 Performance Measures
In this section, we focus on measures used for evaluating the performance of classification algorithms on imbalanced data. In such scenarios, the overall classification accuracy is often not an appropriate measure of performance since rare classes have little impact on accuracy than majority classes [34]. Other performance metrics such as recall (or sensitivity), precision (or positive predictive value, PPV), F-measure, and G-mean are commonly used single-class metrics in binary classification problems. These metrics can also be used to assess the prediction of individual class labels in multiclass problems. These metrics are defined as the following:
For evaluating the overall performance of the classifiers for imbalanced learning, multiclass extensions of the G-mean [62] and AUC [30], as well as Cohen’s Kappa [16], are commonly used metrics.
where K is the number of classes, Recalli is the recall for class i, P 0 is the relative observed agreement among classifiers (i.e., the overall accuracy of the model), and P E is the probability that agreement is due to chance. G-mean is the geometric mean of recall values for all classes, while MAUC is the average AUC for all pairs of classes. Apparently, the G-mean will be equal to 0 if the recall for any class is 0. These three performance measures were used in constructing the ensemble classifier that will be implemented in our analysis.
3.6 Data Analysis
In this section, we lay out some analytical techniques for the pre-processed species abundance table. These techniques focus on training supervised machine learning models for the classification of the OTU abundance to known class labels. Here, our analysis will be based on the species abundance tables obtained after bioinformatics pre-processing and taxonomic profiling of the WGS data gotten from the 2020 CAMDA Forensic Challenge, see Sect. 2.1. In Sect. 2.3, we used three different taxonomic profiling tools to obtain the species abundance table, and the supervised algorithms for classification will be applied to each data set. The primary objective of the analysis lies in predicting the source origins of given metagenomics samples from 23 cities across the globe.
First, we fitted ten candidate classifiers. The candidate classifiers consist of all classifiers discussed in Sect. 3.2 together with certain modifications of these classifiers. For instance, we considered the RF classifier with principal component terms (denoted as PCA+RF) and partial least squares terms (PLS+RF). And we also trained the AdaBoost, XGBoost, RPart classifiers each with PLS terms (PLS+ADA, PLS+XGB, PLS+RPart). Furthermore, we trained the ensemble classifier for which the ensemble constitutes the mentioned candidate classifiers. Candidate classifiers with different parameter combinations can also be included in the ensemble; however, we constructed the ensemble classifier such that no candidate classifier is represented more than once in the candidate set. Also, hyperparameters of the candidate classifiers can be tuned, but we have chosen to use mostly default parameters of the candidate parameters. In the case where the default value of a parameter is not used, the value was chosen based on our experience in the analysis of metagenomic data. Nonetheless, since the default hyperparameters in some machine learning libraries may not be optimized for the classification problem at hand, we encourage analysts to consider tuning such parameters during analysis.
Furthermore, to evaluate the performance of the techniques discussed in Sects. 3.1 and 3.4 for feature selection and to overcome class imbalance, respectively, we will apply these methods to the species abundance table obtained from the Kraken2–Bracken system. The construction of the ensemble classifier can easily be modified to accommodate the implementation of these techniques.
4 Results
Here, we present results for the analysis described in Sect. 3.6. First, we describe the results obtained from the analysis of the species abundance tables obtained after taxonomic profiling was performed with MetaPhlAn2 (MP), Kraken2–Bracken (KB), and Kaiju (KJ), respectively. For each abundance table, further downstream pre-processing as discussed in Sect. 3.1 was carried out, and we obtained 1029, 4770, and 25,750 taxa for MP, KB, and KJ data, respectively. We performed a 10-fold split of the abundance data into 80% training and 20% test sets. For each split, we ensured each class was represented by at least three samples in both the training and test sets. The classification analysis was conducted by training the classifiers mentioned in Sect. 3.6 on the training set, while the test set was used to evaluate the performance of the models.
We used a consistent framework for the analysis of the respective abundance tables, that is, a pre-specified set of candidate classifiers and classifier parameters, performance measures, and resampling techniques were consistently employed across the analysis for each abundance table. However, we excluded the RPart classifier from the set of candidate classifiers when analyzing the KJ data; the classifier could not handle the vast number of features in this particular training set. Also, for the construction of the ensemble classifier, the number of bootstrap samples to be drawn, B, was set to be 50, while Kappa, multiclass G-mean, and MAUC were the performance measures used for performing weighted rank aggregation.
For the analysis of the abundance tables obtained from the respective taxonomic profiling tools, Table 2 shows the mean performance measures for each classification algorithm. Based on the results from all performance measures, and across the analysis for each profiling tool, the ensemble classifier yields classification results that are as good as the best candidate classifier. Furthermore, the candidate classifiers perform differently for each abundance data. For instance, based on the Kappa statistics, the MLP, PLS+RF, RF, and XGB were the best performing candidate classifiers for the analysis of the KB and KJ data, while the RF and XGB gave the most promising results for the analysis of the MP data. These classifiers proved to be the most competitive in the set of candidate classifiers; hence, the ensemble of classifiers across the analysis for each data set was mostly dominated by the MLP, PLS+RF, RF, and XGB classifiers. For each sub-table in Table 2, the last column shows the number of times each candidate classifier was the best performing local classifier in 500 instances (10 replications with 50 bootstrap iterations each).
Furthermore, the SVM with a radial basis kernel and the RPart classifiers yield moderate classification performance. Classifiers trained with integrated PLS terms performed better than classifiers with PCA terms; we observed that the PCA+RF classifier yields the poorest classification results among all candidate classifiers. Also, the PLS+RF classifier performed better than its RF counterpart for the analysis of the KB data, and the two classifiers have closely related results for the analysis of the KJ data, while the RF outperforms the PLS+RF classifier for the analysis of the MP data. In general, the trained classifiers yielded better performance results for the KB and KJ data than for the MP data.
For the second phase of our analysis, we sought to investigate the impact of both dimension reduction and techniques for handling class imbalance on the classification performance of the classifiers. In this regard, we have applied these methods solely for the analysis of the KB data. For each application, we follow a similar design of the analysis presented in the first paragraph of this section. For the weighted classifiers, class weights were computed as w c = 1∕n c, where n c is the number of samples in class c. While for over-sampling, the Gauss Noise (introduces Gaussian noise for the generation of synthetic samples) [39] over-sampling procedure was implemented. The HFE described in Sect. 3.1 was employed for dimension reduction. Table 3 shows the mean performance measures for the set of candidate classifiers and the ensemble classifier, and the classification results for the data with a reduced feature space are shown in parentheses. First, by contrasting the classification performance for the HFE and non-HFE data across the three different techniques shown in the sub-tables of Table 3, notice that there is little or no improvement in classification results for the feature-reduced data. For most of the results reported, the classifiers performed slightly better on the non-HFE data.
Also, for comparison of classification results across the methods used to address the problem of class imbalance and the standard classifiers, we find that there is no substantial improvement in classification performance. Figure 3 shows the mean multiclass AUC scores for the standard classifiers as well as the classifiers trained with class weights and oversampled data. The classifiers are trained on both the non-HFE and HFE data. For each classifier, the multiclass AUCs reported for all three approaches are very similar. This finding is consistent with the description that the class weighting and over-sampling techniques do not improve the overall fit of the models.
Mean multiclass AUC measures for ten standard classifiers, and an ensemble classifier comprising of the standard classifiers. These classifiers were trained with the species abundance table obtained after taxonomic profiling was done with the Kraken2–Bracken system. The training data with a set of complete features comprise 4770 taxa that were obtained after downstream pre-processing, while the data with a reduced feature space comprise 796 taxa, on average
We further investigated the performance of the classifiers when predicting the known class labels in the primary data. The classifiers had a varied performance for prediction of the sample origins. Figure 4 shows a boxplot of the positive predictive values (PPV) based on the classification results from the standard ensemble classifier (i.e., class weighting and over-sampling procedure were not applied) trained on a full feature space. The PPV results described here were obtained for the analysis of KB data discussed in the first paragraph of this section. The classifier yields near perfect prediction for samples obtained from Barcelona, Berlin, Denver, Doha, Kuala Lumpur, Offa, Santiago, Sendai, San Francisco, and Tokyo. The average PPV for prediction of these sample origins was at least 95%. In contrast, the ensemble classifier does not yield good classification performance for the prediction of samples that originated from Kiev, Lisbon, Offa, and Singapore. The average PPV for these cities ranges from 60% to 74%. The poor performance of the classifier in predicting certain cities will negatively impact the overall classification performance of the classifier. Thus, it is worthwhile to investigate the reasons for the poor predictive ability of the classifier for these cities. For instance, we observed that the classifier had trouble discriminating between Kiev and Zurich. Certain factors could influence the sub-par ability of the classifier in discriminating between cities. The proximity of source cities is an obvious factor. Naturally, we can expect the classifiers to misclassify cities in close proximity to one another. For instance, Offa and Ilorin are geographically close, and the classifier, in several cases, misclassified Offa as the Ilorin.
Boxplot showing the positive predictive value for all cities represented in the training data. The results are based on predictions from a standard ensemble classifier that was trained on the full feature space of the species abundance data obtained after taxonomic profiling was performed with Kraken2–Bracken
The boxplots in Fig. 5 show some of the top microbial species that were found to be differentially abundant across various cities. The left panel of Fig. 5 shows the feature importance plot of the top 20 species from RF classifier in the ensemble. Variable importance plot consists of many species belonging to genus Bradyrhizobium that is a soil bacteria and is also found in the roots and stems of plants [27]. Pseudomonas.sp..CC6.YY.74 species belongs to genus Pseudomonas that is a common genus of bacteria that resides on moist surfaces, soil, and water [42].
5 Discussion
We have presented a practical workflow for the analysis of microbiome data that are based on samples that are usually collected from the different body and environmental sites. This workflow was partitioned into two sections—pre-processing of raw WGS data and downstream analysis. For the raw WGS data pre-processing of the microbiome data, we constructed a standard pipeline using a variety of bioinformatics tools for quality control and taxonomic profiling. The taxonomic profiling involves classifying sequence reads to taxonomic labels and estimation of species abundance, and this was performed with three widely used profiling tools, namely, MetaPhlAn2, Kraken2–Bracken, and Kaiju. At the termination of the bioinformatics pipeline, we obtain species abundance tables from each of the respective profiling tools, and these abundance tables were passed into the downstream analysis.
The downstream analysis of the data comprised fitting supervised learning models for the classification of the species abundance of the samples to known class labels. We have evaluated several machine learning approaches to the metagenomics-based classification of sample origins. For this purpose, we adopted a robust ensemble classifier that uses species-level abundance as features, a user-specific set of supervised learning models as candidate classifiers, and user-defined performance metrics for model evaluation. The ensemble classifier is an adaptive classification algorithm that can perform well on different data structures. This classifier utilizes performance on OOB samples to guard against overfitting. The ensemble classifier gives classification performance better or as good as the best performing candidate classifier in its ensemble.
Across many metagenomics studies, we noticed a great deal of variation in classification results presented by different researchers working in this area. One natural explanation for this variation in results stems from the bioinformatics and data generation procedures employed in these studies. Since standard classification models will perform differently when trained on different data structures, restricting the classification problem to a single classifier may not be a practical approach. For a given classification problem, the analyst is expected to try out a variety of classifiers, judging each one according to a set of user-defined performance metrics. In this sense, the analyst will likely begin their exploration with simple models before trying out more complex models. With the application of the ensemble classifier described here, the analyst can automate the process and achieve a near optimal performance.
In this chapter, we have trained the ensemble classifier with only the classifiers discussed in Sect. 3.2. However, the ensemble need not be restricted to these models but could include any reasonable user-specified classifier. For instance, we notice that the XGBoost classifier that is popular among competing teams solving data science problems has been rarely used in the analysis of metagenomics data. Results from classification performance presented in this chapter showed that the XGBoost performs almost as well as the RF classifier. Therefore, in a future analysis of these types of data, we may choose to include XGBoost in our ensemble.
The best classification results for the prediction of source cities were obtained when the classifiers were trained on the full data set rather than on the feature-reduced version. This explains the complex nature of metagenomics data where a plethora of taxa are needed to characterize the variation among sample origins; hence, building a model with only a subset of these taxa may not sufficiently explain such variations.
In addition to fitting an ensemble of classifiers, we also highlight other techniques that may improve the classification of metagenomics data. Since most machine learning models tend to lean toward predicting the majority classes over the minority classes, balancing the class frequencies of samples in the training data is an ideal method to incorporate in the analytical pipeline. The application of an optimal minority over-sampling scheme and class weighting in the training of the classifiers only marginally impacted the performance of the classifiers presented in this chapter. These techniques can easily be incorporated while constructing the ensemble classifier. We notice that training the classifier with class weights is computationally more efficient than utilizing an over-sampling scheme.
An obvious drawback of the ensemble classifier is that it is computationally intensive. It would take more time to train an ensemble classifier than it would for a stand-alone classifier. The computing times of the ensemble classifier are mainly impacted by the number of bootstrap samples that the individual classifiers are trained on, the number and complexity of user-specified candidate classifiers, and the performance measures that are used to compute the weighted rank aggregation. However, the computing times can be appreciably reduced if the ensemble classifier is trained using parallel computational approaches on a computing cluster. When we selected 10 candidate classifiers (i.e., the candidate classifiers presented in Sect. 4), three performance measures (namely, Cohen’s Kappa coefficient, multiclass G-mean, and AUC) for computing weighted rank aggregation, and 50 bootstrap samples for the construction of the ensemble classifier, the construction procedure took an average time of 9.47 h (wall-clock time). This procedure was done on a University computing cluster for which 12 CPU cores and 40GB of memory were allocated to the job.
The downstream classification analysis presented here can be extended in two different directions. Each of these extensions requires the knowledge of additional information besides the microbiome data—such information are often present in the form of geographic location of the training cities or the weather information in both training and test cities and so on. In the former case, we can build a potentially improved classifier that effectively utilizes a larger collection of features. In the later situation, one may be able to predict the city of origin in a bigger list than what was provided in the training data. These extensions may be pursued elsewhere.
6 Data Acknowledgement
All analyses presented in this chapter are based on the raw WGS metagenomics data provided as part of the 2020 CAMDA Metagenomic Geolocation Challenge. The primary data along with other supplementary data is publicly available on the challenge’s website. We participated in this challenge and presented our classification results at the 2020 Intelligent Systems for Molecular Biology (ISMB) conference. An extensive report on the results from our analysis will be published in the conference proceedings.
7 Code Availability
Bash scripts for each procedure performed in the bioinformatics pipeline and R scripts for building an ensemble of standard classifiers are available at https://github.com/samuelanyaso/metagenomic_data_analysis. The sample code below shows a standard interface to analyze an abundance matrix. The code calls the ensemble.R script for training an ensemble classifier, predicts test cases, and evaluates the performance of the ensemble classifier along with other candidate classifiers in the ensemble.
WD <- "/path/to/data/and/source/scripts" setwd(WD) df <- read.delim("abundanceTable.txt", header = TRUE, sep = "\t", dec = ".") df$class <- factor(df$class) # class labels ## Begin training Models num.class <- length(levels(df$class)) idx <- 1:nrow(df) # row indices ## loads the ensemble function source("ensemble.R") Result1 <- list() Result2 <- list() bestAlg <- list() confMat <- list() reps <- 10 # number of replications set.seed(2021) for(r in 1:reps){ repeat{ ## repeat partitioning of the data into train and test set until ## all classes are present in both test and train set inTraining <- createDataPartition(df$class,p = 0.9,list = FALSE) shuf <- sample(inTraining[,1],replace = FALSE) # train set shufT <- sample(idx[which(!idx %in% inTraining[,1])], replace = FALSE) # test set # partitions the dataset dat.train <- df[shuf,] dat.test <- df[shufT,] if(all(table(dat.train$class) >= 1) & all(table(dat.test$class) >= 1)){ break } } ## Train set y <- dat.train$class y <- as.factor(as.numeric(y)-1) # Factor levels should begin from 0 x <- data.matrix(dat.train[,!(names(dat.train) %in% c("class"))]) ## Test set yTest <- dat.test$class yTest <- as.factor(as.numeric(yTest)-1) # Factor levels should begin from 0 xTest <- data.matrix(dat.test[,!(names(dat.test) %in% c("class"))]) cat("Started Replication: ",r," of ",reps,"\n ") ens <- ensembleClassifier(x, y, M=50, ncomp=30, train = dat.train, test = dat.test, algorithms=c("svm","rang","pls_rf", "pca_rf","rpart", "pls_rpart", "xgb","pls_xgb","mlp"), levsChar =as.character(levels(dat.train$class))) # the names of the best local classifiers bestAlg[[r]] <- ens$bestAlg ## predict using the test data pred <- predictEns(ens, xTest, yTest, test = dat.test, dlEnsPath = "dl_ens_time.h5", dlIndPath = "dl_ind_time.h5") # Saves the results Result1[[r]] <- pred$ensemblePerf Result2[[r]] <- pred$indPerf ## predicted class yPred <- pred$yhat ## confusion matrix confMat[[r]] <- caret::confusionMatrix(yPred,yTest) # displays the truth and predictions for each of the "best" algorithms dfPred <- data.frame(truth=yTest, ensemble=yPred, pred$pred) dfPred <- as.list(dfPred) # convert numeric factors to character factors dfPred <- lapply(dfPred,function(x) as.character(num2charFac(x,char.levs = as.character(levels(dat.train$class))))) names(dfPred) <- c("truth","ensemble",ens$bestAlg) dfPred <- as.data.frame(dfPred) cat("Predictions for the best individual models for iteration: ",r," of ",reps,"\n ") print(dfPred) cat("Completed Replication: ",r," of ",reps,"\n ") } # save performance results saveRDS(Result1,"ensClassifPerf.RDS") saveRDS(Result2,"indClassifPerf.RDS") saveRDS(bestAlg,"bestAlg.RDS") saveRDS(confMat,"confMat.RDS") warnings()
References
Altschul, S.F., Gish, W., Miller, W., Myers, E.W., Lipman, D.J.: Basic local alignment search tool. J. Mol. Biol. 215(3), 403–410 (1990)
Andrews, S.: FastQC. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/. Accessed: 2020-08-28
Bharti, R., Grimm, D.G.: Current challenges and best-practice protocols for microbiome analysis. Brief. Bioinform. 22(1), 178–193 (2019). https://doi.org/10.1093/bib/bbz155
Bolger, A.M., Lohse, M., Usadel, B.: Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics 30(15), 2114–2120 (2014)
Branco, P., Ribeiro, R.P., Torgo, L.: UBL: an R package for utility-based learning. Preprint (2016). arXiv:1604.08079
Breiman, L.: Random forests. Mach. Learn. 45(1), 5–32 (2001)
Breiman, L., Friedman, J., Stone, C.J., Olshen, R.A.: Classification and regression trees. CRC Press, Boca Raton, FL (1984)
Breitwieser, F.P., Lu, J., Salzberg, S.L.: A review of methods and databases for metagenomic classification and assembly. Brief. Bioinform. 20(4), 1125–1136 (2019)
Buchfink, B., Xie, C., Huson, D.H.: Fast and sensitive protein alignment using DIAMOND. Nature Methods 12(1), 59–60 (2015)
Casimiro-Soriguer, C.S., Loucera, C., Perez Florido, J., López-López, D., Dopazo, J.: Antibiotic resistance and metabolic profiles as functional biomarkers that accurately predict the geographic origin of city metagenomics sample. Biology Direct 14, 15 (2019)
Chase, J., Fouquier, J., Zare, M., Sonderegger, D.L., Knight, R., Kelley, S.T., Siegel, J., Caporaso, J.G.: Geography and location are the primary drivers of office microbiome composition. mSystems 1(2), e00022-16 (2016)
Chen, C., Liaw, A., Breiman, L.: Using random forest to learn imbalanced data. Statistics Department of University of California at Berkeley, Berkeley. Technical Report 666 (2004)
Chen, T., Guestrin, C.: XGBoost: A Scalable Tree Boosting System. In: Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining (New York, NY, USA, 2016), KDD ’16, pp. 785–794. Association for Computing Machinery (2016)
Claesson, M.J., Clooney, A.G., O’Toole, P.W.: A clinician’s guide to microbiome analysis. Nat. Rev. Gastroenterol. Hepatol. 14(10), 585–595 (2017)
Clarke, T.H., Gomez, A., Singh, H., Nelson, K.E., Brinkac, L.M.: Integrating the microbiome as a resource in the forensics toolkit. Forensic Sci. Int. Genet. 30, 141–147 (2017)
Cohen, J.: A coefficient of agreement for nominal scales. Educ. Psychol. Meas. 20(1), 37–46 (1960)
Cortes, C., Vapnik, V.: Support-vector networks. Machine Learning 20(3), 273–297 (1995)
Datta, S., Pihur, V., Datta, S.: An adaptive optimal ensemble classifier via bagging and rank aggregation with applications to high dimensional data. BMC Bioinformatics 11, 427 (2010)
Ditzler, G., Morrison, J.C., Lan, Y., Rosen, G.L.: Fizzy: feature subset selection for metagenomics. BMC Bioinformatics 16(1), 358 (2015)
Duvallet, C., Gibbons, S.M., Gurry, T., Irizarry, R.A., Alm, E.J.: Meta-analysis of gut microbiome studies identifies disease-specific and shared responses. Nature Communications 8(1), 1–10 (2017)
Ewels, P., Magnusson, M., Lundin, S., Käller, M.: MultiQC: Summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32(19), 3047–3048 (2016)
Ewing, B., Green, P.: Base-calling of automated sequencer traces using Phred. II. Error probabilities. Genome Res. 8(3), 186–194 (1998)
Freitas, T.A.K., Li, P.-E., Scholz, M.B., Chain, P.S.G.: Accurate read-based metagenome characterization using a hierarchical suite of unique signatures. Nucleic Acids Res. 43(10), e69 (2015)
Freund, Y., Schapire, R.E.: A decision-theoretic generalization of on-line learning and an application to boosting. J. Comput. Syst. Sci. 55(1), 119–139 (1997)
Friedman, J.H.: Greedy function approximation: a gradient boosting machine. Ann. Stat., 1189–1232 (2001)
Gilbert, J.A., Jansson, J.K., Knight, R.: The earth microbiome project: successes and aspirations. BMC Biology 12(1), 69 (2014)
Giraud, E., Xu, L., Chaintreuil, C., Gargani, D., Gully, D., Sadowsky, M.J.: Photosynthetic Bradyrhizobium sp. strain ORS285 is capable of forming nitrogen-fixing root nodules on soybeans (glycine max). Appl. Environ. Microbiol. 79(7), 2459–2462 (2013)
Grice, E.A., Segre, J.A.: The human microbiome: Our second genome. Annu. Rev. Genomics Hum. Genet. 13(1), 151–170 (2012). PMID: 22703178
Hand, D.J.: Breast cancer diagnosis from proteomic mass spectrometry data: A comparative evaluation. Stat. Appl. Genet. Mol. Biol. 7(2), Article 15 (2008)
Hand, D.J., Till, R.J.: A simple generalisation of the area under the ROC curve for multiple class classification problems. Machine Learning 45(2), 171–186 (2001)
Harris, Z.N., Dhungel, Eliza Mosior, M., Ahn, T.-H.: Massive metagenomic data analysis using abundance-based machine learning. Biology Direct 14(12), Article 12 (2019)
Hastie, T., Tibshirani, R., Friedman, J.: The Elements of Statistical Learning. Springer Series in Statistics. Springer New York, New York, NY, USA (2001)
Huttenhower, C., et al.: Structure, function and diversity of the healthy human microbiome. Nature 486(7402), 207–214 (2012)
Joshi, M.V., Kumar, V., Agarwal, R.C.: Evaluating boosting algorithms to classify rare classes: Comparison and improvements. In: Proceedings 2001 IEEE International Conference on Data Mining, San Jose, CA, USA pp. 257–264. IEEE (2001)
Knights, D., Costello, E.K., Knight, R.: Supervised classification of human microbiota. FEMS Microbiol. Rev. 35(2), 343–359 (2011)
Kovács, G.: An empirical comparison and evaluation of minority oversampling techniques on a large number of imbalanced datasets. Appl. Soft Comput. 83, 105662 (2019a)
Kovács, G.: Smote-variants: a python implementation of 85 minority oversampling techniques. Neurocomputing 366, 352–354 (2019b)
Langmead, B., Salzberg, S.L.: Fast gapped-read alignment with Bowtie 2. Nature Methods 9(4), 357–359 (2012)
Lee, S.S.: Regularization in skewed binary classification. Computational Statistics 14, 277–292 (1999)
Li, H.: Microbiome, metagenomics, and high-dimensional compositional data analysis. Annu. Rev. Stat. Appl. 2, 73–94 (2015)
Lin, W., Shi, P., Feng, R., Li, H.: Variable selection in regression with compositional covariates. Biometrika 101(4), 785–797 (2014)
Lin, X., Zhang, Z., Zhang, L., Li, X.: Complete genome sequence of a denitrifying bacterium, Pseudomonas sp. CC6-YY-74, isolated from Arctic Ocean sediment. Marine Genomics 35, 47–49 (2017)
Lindgreen, S., Adair, K.L., Gardner, P.P.: An evaluation of the accuracy and speed of metagenome analysis tools. Scientific Reports 6, 19233 (2016)
Lu, J., Breitwieser, F.P., Thielen, P., Salzberg, S.L.: Bracken: estimating species abundance in metagenomics data. PeerJ Comput. Sci. 3, e104 (2017)
Marchesi, J.R., Ravel, J.: The vocabulary of microbiome research: a proposal. Microbiome 3(1), 31 (2015)
Martin, M.: Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17(1), 10–12 (2011)
Mason, C., Hirschberg, D., Consortium, T.M.I.: The metagenomics and metadesign of the subways and urban biomes (MetaSub) International Consortium inaugural meeting report. Microbiome 4(1), 24 (2016)
McIntyre, A.B., Ounit, R., Afshinnekoo, E., Prill, R.J., Hénaff, E., Alexander, N., Minot, S.S., Danko, D., Foox, J., Ahsanuddin, S., et al.: Comprehensive benchmarking and ensemble approaches for metagenomic classifiers. Genome Biology 18(1), 182 (2017)
McIver, L.J., Abu-Ali, G., Franzosa, E.A., Schwager, R., Morgan, X.C., Waldron, L., Segata, N., Huttenhower, C.: bioBakery: a metaá’omic analysis environment. Bioinformatics 34(7), 1235–1237 (2017)
Menzel, P., Ng, K.L., Krogh, A.: Fast and sensitive taxonomic classification for metagenomics with Kaiju. Nature Communications 7, 11257–11257 (2016)
Moitinho-Silva, L., Steinert, G., Nielsen, S., Hardoim, C.C., Wu, Y.-C., McCormack, G.P., López-Legentil, S., Marchant, R., Webster, N., Thomas, T., et al.: Predicting the HMA-LMA status in marine sponges by machine learning. Front. Microbiol. 8, 752 (2017)
Oudah, M., Henschel, A.: Taxonomy-aware feature engineering for microbiome classification. BMC Bioinformatics 19, 227 (2018)
Ounit, R., Wanamaker, S., Close, T.J., Lonardi, S.: Clark: fast and accurate classification of metagenomic and genomic sequences using discriminative k-mers. BMC Genomics 16(1), 236 (2015)
Pasolli, E., Truong, D.T., Malik, F., Waldron, L., Segata, N.: Machine learning meta-analysis of large metagenomic datasets: tools and biological insights. PLoS Comput. Biol. 12(7), e1004977 (2016)
Paulson, J.N., Pop, M., Bravo, H. C.: metagenomeSeq: Statistical analysis for sparse high-throughput sequencing Bioconductor package 1(0), 191 (2013)
Pihur, V., Datta, S., Datta, S.: Weighted rank aggregation of cluster validation measures: a Monte Carlo cross-entropy approach. Bioinformatics 23(13), 1607–1615 (2007)
Ranjan, R., Rani, A., Metwally, A., McGee, H.S., Perkins, D.L.: Analysis of the microbiome: Advantages of whole genome shotgun versus 16s amplicon sequencing. Biochem. Biophys. Res. Commun. 469(4), 967–977 (2016)
Schmieder, R., Edwards, R.: Insights into antibiotic resistance through metagenomic approaches. Future Microbiology 7, 73–89 (2012)
Segata, N., Waldron, L., Ballarini, A., Narasimhan, V., Jousson, O., Huttenhower, C.: Metagenomic microbial community profiling using unique clade-specific marker genes. Nature Methods 9, 811–814 (2012)
Shi, P., Zhang, A., Li, H., et al.: Regression analysis for microbiome compositional data. Ann. Appl. Stat. 10(2), 1019–1040 (2016)
Singh, R.K., Chang, H.-W., Yan, D., Lee, K.M., Ucmak, D., Wong, K., Abrouk, M., Farahnik, B., Nakamura, M., Zhu, T.H., Bhutani, T., Liao, W.: Influence of diet on the gut microbiome and implications for human health. J. Transl. Med. 15(1), 73 (2017)
Sun, Y., Kamel, M.S., Wang, Y.: Boosting for learning multiple classes with imbalanced class distribution. In: Sixth International Conference on Data Mining (ICDM’06), Hong Kong, China. pp. 592–602. IEEE (2006)
Sunagawa, S., Mende, D.R., Zeller, G., Izquierdo-Carrasco, F., Berger, S.A., Kultima, J.R., Coelho, L.P., Arumugam, M., Tap, J., Nielsen, H.B., Rasmussen, S., Brunak, S., Pedersen, O., Guarner, F., de Vos, W.M., Wang, J., Li, J., Doré, J., Ehrlich, S.D., Stamatakis, A., Bork, P.: Metagenomic species profiling using universal phylogenetic marker genes. Nature Methods 10(12), 1196–1199 (2013)
Tibshirani, R.: Regression shrinkage and selection via the Lasso. J. Roy. Stat. Soc. B (Methodol.) 58(1), 267–288 (1996)
Turnbaugh, P.J., Ley, R.E., Hamady, M., Fraser-Liggett, C.M., Knight, R., Gordon, J.I.: The human microbiome project. Nature 449(7164), 804–810 (2007)
Wade, W.: The oral microbiome in health and disease. Pharmacological Research 69(1), 137–143 (2013). Copyright 2012 Elsevier Ltd. All rights reserved
Wang, W.-L., Xu, S.-Y., Ren, Z.-G., Tao, L., Jiang, J.-W., Zheng, S.-S.: Application of metagenomics in the human gut microbiome. World J. Gastroenterol. 21(3), 803–814 (2015)
Wood, D.E., Salzberg, S.L.: Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biology 15, Article R46 (2014)
Wood, D.E., Lu, J., Langmead, B.: Improved metagenomic analysis with Kraken 2. Genome Biology 20(1), 257 (2019)
Ye, S.H., Siddle, K.J., Park, D., Sabeti, P.C.: Benchmarking metagenomics tools for taxonomic classification. Cell 178, 779–794 (2019)
Zhou, Q., Su, X., Ning, K.: Assessment of quality control approaches for metagenomic data analysis. Scientific Reports 4(1), 6957 (2014)
Zhou, Y.-H., Gallins, P.: A review and tutorial of machine learning methods for microbiome host trait prediction. Front. Genet. 10, 579 (2019)
Zou, H., Hastie, T.: Regularization and variable selection via the ElasticNet. J. Roy. Stat. Soc. B (Stat. Methodol.) 67(2), 301–320 (2005)
Acknowledgements
This work was partially supported by the National Science Foundation under Award DMS-1461948 to SG.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 Springer Nature Switzerland AG
About this chapter
Cite this chapter
Anyaso-Samuel, S., Sachdeva, A., Guha, S., Datta, S. (2021). Bioinformatics Pre-Processing of Microbiome Data with An Application to Metagenomic Forensics. In: Datta, S., Guha, S. (eds) Statistical Analysis of Microbiome Data. Frontiers in Probability and the Statistical Sciences. Springer, Cham. https://doi.org/10.1007/978-3-030-73351-3_3
Download citation
DOI: https://doi.org/10.1007/978-3-030-73351-3_3
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-73350-6
Online ISBN: 978-3-030-73351-3
eBook Packages: Mathematics and StatisticsMathematics and Statistics (R0)