Abstract
Brain age inferred from neuroimaging data could reveal important information about the evolution of structural and functional cerebral features across the life span. This has important implications for understanding healthy aging and for identifying Imaging-Derived Phenotypes (IDPs) that characterise age-related neurodegenerative illnesses, such as Alzheimer’s and Parkinson’s disease. The so-called brain age delta refers to the difference between image-derived brain age and chronological age. Accelerated aging (positive delta) or resilience to aging (negative delta) have been found to be useful correlates of factors such as disease and cognitive decline. Multiple studies have proposed prediction models using brain IDPs as predictor variables, mostly relying on simple linear regression. However, methodological and population heterogeneity in these studies precludes definitive conclusions regarding the most informative modelling methodologies or predictor IDPs. To provide first hints in this respect, in this paper we propose to address three questions. First, four different state-of-the-art models are ranked based on well-known performance indices (e.g., mean absolute error) using the UK Biobank brain MRI data in different single/multi-modal settings. Second, for the best model, the association with individual IDPs are calculated to identify those that could play a prominent role in the aging process. Third, associations with non-brain variables are assessed as a first step towards a holistic approach. Our findings demonstrate a prominent role for dMRI IDPs in reducing the mean absolute error and rank high in the association study, dominating the first ten positions and being preceded only by three structural measures that are known to be related to the aging process. This provides evidence of the potential of dMRI IDPs as biomarkers of aging in health and disease.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Neuroimaging data have been extensively used to assess brain changes during aging, under both healthy and disease conditions. Moreover, they can be exploited to predict “brain age” which is the apparent biological age of an individual and depends on several endogenous (subject-specific) as well as exogenous (environmental) factors. Metrics derived from various brain magnetic resonance imaging (MRI) sequences have been adopted to estimate brain age, either using raw data or handcrafted features. Brain age delta (or relative brain age) is calculated by subtracting chronological age from the estimated one. While a younger-appearing brain might be the result of a healthy life style [1], having an older-appearing brain has been previously associated with poor future outcomes [2] and with an increased likelihood to develop neurodegenerative illnesses such as Alzheimer’s [3].
Statistical models for brain age estimation have been proven to be highly accurate, with prediction performance featuring high R\(^2\) values and low mean absolute error (MAE) in the range of 4–5 years [2]. Most of the studies have investigated this aspect with features derived from a single brain MRI technique, most commonly, conventional T1-weighted structural images (sMRI). Morphometric measures from sMRI, such as volume and thickness of grey matter (GM) structures, should not be overlooked as they provide information on the individual degree of brain atrophy that encodes aging-induced degeneration [4]. However, more recently, diffusion MRI (dMRI), susceptibility weighted imaging (SWI), and resting-state fMRI have been explored for potentially providing a richer set of IDPs bringing complementary information [2, 5]. Thus, consideration of IDPs derived from multiple brain MRI sequences would be the most desirable approach allowing deeper phenotyping and more complete capturing of the different factors shaping the aging process.
Regarding modeling approaches, the performance accuracy depends on the statistical method utilised, as demonstrated by several authors. In a recent paper, Jonsson and colleagues (2019) applied deep learning as well as eight different regression methods to sMRI-based features extracted from three well-known databases, showing notable differences in the performance parameters across the different models [3]. Niu et al. [1] report similar variation in model performance in their study of brain age estimation with four regression models using several neuroimaging variables (sMRI, dMRI, and resting-state fMRI) in healthy controls and patients with anxiety disorders. In addition, the authors showed the potential for superior prediction accuracy with a multi-modal versus single-modal approach.
The recent availability of large imaging databases has provided new opportunities to exploit the importance of a multi-modal approach for brain age prediction. In this context, the UK Biobank (UKB) represents an important resource thanks to its comprehensive repository with genetic and phenotypic data for 500000 subjects aged between 40 and 69 (at recruitment). The UKB imaging study includes detailed MRI, providing high quality multi-modal neuroimaging data including sMRI, dMRI, SWI and fMRI [6]. These data are linked to detailed clinical, biological and lifestyle information. The availability of such a rich research resource has motivated many researchers to focus on brain age estimation with promising results [2, 3, 5, 7].
Smith et al. [5] estimated brain age using simulated and real data by applying simple linear regression. With regard to real data, 2641 IDPs covering sMRI, fMRI and dMRI were used for 19000 participants. The results, among others, attained MAE = 3.6 years.
In [2], phenotypes from six different MRI modalities were chosen to estimate brain age for 17461 subjects, running a Least Absolute Shrinkage and Selection Operator (LASSO) regression for each modality (MAE range = 3.897–5.928 years, where minimum e maximum were found for dMRI and task fMRI, respectively). When all the IDPs were combined, age was more accurately predicted (MAE = 3.515 years). Thirty-four IDPs were deemed informative for the prediction of the brain age after bootstrapping, and were predominantly from sMRI and dMRI.
Ning et al. [7] aimed to assessing the correlation between brain age delta and alcohol intake, smoking and genetic variations. To this end, 403 morphometric measurements from sMRI were chosen along with LASSO regression (MAE = 3.8 years). A significant association between brain age delta and the consumption of alcohol and smoking could be demonstrated.
Finally in [3], sMRI data for 12395 subjects were used to estimate brain age using transfer learning and 3-D Convolutional Neural Network (CNN). In this study, two sequence variants were identified having a strong relation with the brain age delta. The MAE of the model was 3.63 years.
The great variability in the number of subjects, IDPs, MRI modalities and statistical models precludes a straightforward comparison of all the studies. However, existing work suggests that: (i) sMRI provides relevant IDPs for estimation of brain age; (ii) dMRI-based phenotypes are similarly informative and need to be further investigated; and (iii) a multi-modal approach can improve, in general, the estimation accuracy. To the best of our knowledge, a systematic comparison among different statistical methods has not been addressed in existing literature. Therefore, in this paper we compared four regression methods in combination with different IDPs for brain age prediction, aiming at providing a balanced comparison across different single-modal and multi-modal approaches. In particular, we focused on Simple Linear Regression (SLR), LASSO, Support Vector Regression (SVR) and Bayesian Ridge Regression (BRR), while the handcrafted features were derived from sMRI, dMRI and SWI with a clear numeric prevalence of dMRI. Model performance was assessed using several parameters including MAE, R\(^{2}\) and adjusted R\(^{2}\). Moreover, the associations between individual IDPs and brain age delta values were calculated for the best model. Finally, the association between brain age delta with selected biomedical and behavioral features was extracted to assess potential clinical/biological utility.
2 Data and Materials
Data were obtained from UKB. All the analyses here performed rely on the IDPs extracted centrally by researchers involved in the project [8]. Data were available from n = 16394 participants (age range = 40–70 years, n = 8652 females, n = 7742 males). This comprised a set of 714 IDPs for each subject, representing the summary metrics for sMRI, SWI and dMRI. From sMRI images, morphometric measures of brain volumes were reported as distinct IDPs, both normalised/not normalised for overall head size, in details: total brain volume (GM + white matter [WM]); volumes for WM, GM and cerebrospinal fluid (separately for each compartment); volume of peripheral cortical GM. Volume measures for subcortical structures were also calculated as further IDPs (e.g., thalamus, putamen, hippocampus), generally separated for left/right hemispheres and not normalised for head size. From SWI data, a T2* image was used and the median T2* value estimated as a separate IDP for each subcortical ROI identified from sMRI. Finally, several spatially-specific IDPs were extracted from dMRI data by following two different approaches. Indeed, nine dMRI-based indices derived from i) the diffusion tensor imaging (DTI), such as fractional anisotropy (FA) and mean diffusivity (MD), and ii) the neurite orientation dispersion and density imaging (NODDI) model, such as orientation dispersion (OD) and isotropic volume fraction (ISOVF), were calculated and averaged over specific areas/tracts. In the first approach, dMRI maps were aligned to a population-based WM tract skeleton and all the DTI/NODDI measures averaged over 48 regions defined using the Johns Hopkins University tract atlas [9]. In the second, probabilistic tractography was run for each subject and all the dMRI-based measures averaged within 27 distinct WM tracts. The final set of neuroimaging phenotypes included 25 IDPs from sMRI, 14 from SWI and 675 from dMRI. Full details on the acquisition protocols and image processing pipelines for the UKB brain data are available at https://biobank.ctsu.ox.ac.uk/crystal/crystal/docs/brain_mri.pdf.
The present analyses were conducted under data application number 2964. All participants provided formal consent, details on the UKB Ethics can be found at https://www.ukbiobank.ac.uk/the-ethics-and-governance-council.
3 Methods
3.1 Brain Age Estimation
Four different regression methods including LASSO [10], SLR [11], SVR [12] and BRR [13] were used to estimate the apparent brain age, all having chronological age as the dependent variable. All these models were implemented using Scikit-learn [14] library version 0.22.2 in Python 3.6.9.
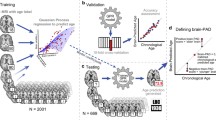
In order to examine the impact of different imaging modalities, each of the four methods was run with single-modal and multi-modal brain IDPs, leading to seven different combinations per method. All the imaging features (independent variables) were normalized to zero mean and unit variance to account for the different measurement scales, while the actual age was demeaned only [5]. Gender and education were considered as confound variables and regressed out of all IDPs as in [6, 15]. Data were randomly split into training (80%, n = 13115) and testing (20%, n = 3279) sets, respectively. The test set was used to predict brain ages on unseen data.
Hyper-parameters for BRR, LASSO and SVR were tuned on the training data (further split on 80% for training and 20% for validation) with GridSearchCV and the optimal model was retained. After the parameters were optimized from training data, the optimal model was applied to estimate brain age in the test set. The performance of each model was assessed using the Coefficient of Determination (R\(^{2}\)) and the MAE. Adjusted R\(^{2}\) was also calculated to account for the different number of predictors in each model.
Recent literature has demonstrated a proportional bias in brain age calculation, which might be caused by dilution bias of the prediction model [16, 17]. Moreover, this bias is also closely connected to the fact that brain age is overestimated in younger subjects and underestimated in older ones, while is more accurately predicted for participants whose actual ages are closer to the mean age of the training dataset [2, 5]. All these elements lead to a significant dependence of the brain age delta on chronological age, which resulted to be negatively correlated. Therefore, common practice is to apply a statistical age-bias correction procedure to overcome these limitations [5, 16] In this study, we adopted the procedure proposed by Beheshti et al. [16] that relies on a linear model given by the following equation:
where D is the brain age delta (estimated from training data), \(\varOmega \) is the chronological age of the training data, \(\alpha \) and \(\beta \) represent the slope and the intercept. These two measures are subsequently used to correct the brain age predictions in the test set as described in Eq. 2:
where CPBA stands for corrected predicted brain age.
After brain age was estimated and bias corrected in the test set, brain age delta was calculated for each subject. Pearson correlations for predicted brain age vs actual age (CPA) and brain age delta vs actual age (CBDA) were calculated twice for each model, before and after bias correction.
3.2 Associations with IDPs and Non-IDP Variables
For the best model results, Pearson correlations between brain age delta values and individual IDPs were calculated in order to identify the strongest associations, highlighting the features which contribute most to the modelling of the brain delta as suggested in [5]. The resulting p-values were Bonferroni-corrected for multiple comparison testing. Of note, the fully deconfounded versions of the IDPs were used in this step (including gender, education and age as confounds). As several studies demonstrated a significant association between brain and heart functionality, especially relying on brain volumetric measurements [18, 19], we also investigated whether a correlation between brain age delta and heart measures was present. In order to perform this analysis, five measures from Cardiac Magnetic Resonance (CMR) and eight Cardiovascular Risk Factors (CRFs) were considered. The correlation analysis was performed on a subgroup of the test set (n = 2730), as these measures were not available for all the test set subjects. CMR scans were performed on 1.5 T scanners using a standardised acquisition protocol [20]. The following indices derived for the left ventricle were retained: end-diastolic volume (LVEDV), end-systolic volume (LVESV), stroke volume (LVSV), mass (LVM), and ejection fraction (LVEF). Eight CRFs were also tested, covering biomedical and lifestyle measures: smoking status, material deprivation, body mass index, alcohol intake frequency, physical activity, diabetes diagnosis, presence of hypertension and high cholesterol. Smoking status and alcohol intake frequency were based on self-reports. Material deprivation was reported by UKB as the Townsend deprivation index. A continuous value for the amount of physical activity, measured in metabolic equivalent minutes/week, was calculated. Body mass index was derived from height and weight measures recorded at the baseline. Diabetes, hypertension, and hypercholesterolaemia were defined by cross-checking across self-report and blood biochemistry data. All the cardiac variables were initially normalized to zero mean and unit variance, and the main potential confounds (gender and age) regressed out from the data. Pearson correlation was finally computed between each of these measures and brain age delta values derived from the twenty-eight model combinations, and the results were Bonferroni-corrected to account for multiple comparison problems.
4 Results
4.1 Brain Age Estimation
Results are summarised in Tables 1 and 2 reporting the overall performance of the four regression methods combined with the different IDPs. Table 1 reports the estimation performance for the test subjects in terms of MAE values before bias correction, as this represents the actual model performance. Results demonstrated that using all the 714 IDPs from the three imaging modalities provided the best model performance in terms of MAE for all regression methods. In particular, BRR gave the best results (MAE = 3.482 years), closely followed by LASSO (MAE = 3.483 years), while SVR performed less accurately among the four tested methods. When considering the different feature types, the performance of the models using SWI only was worst (MAE \(\approx \) 6.0 years) compared to the other single-modal approaches that is sMRI (MAE \(\approx \) 4.5 years) and especially dMRI (MAE \(\approx \) 3.7 years). When considering the multi-modal models, adding dMRI phenotypes improved the accuracy of all methods.
These results were further confirmed by the R\(^{2}\) and adjusted R\(^{2}\) parameters (Table 2), for which the lowest value was reached using the SWI IDPs (R\(^{2}\) = 0.075–0.085). When the IDPs from sMRI and dMRI were used jointly in the model, the performance was improved and very close to the one reached by using all the IDPs, and this finding held for all the four regression methods. For the sake of completeness, CPA and CBDA were calculated before and after bias correction, leading to the results summarised in Table 3. When using all the 714 IDPs, the correlation between brain age delta and actual age decreases towards zero after applying the bias correction steps. Conversely, CPA increased after bias correction in all four methods.
4.2 Association with Brain IDPs
Considering that the BRR method combined with all the IDPs reached the lowest MAE and highest R\(^{2}\)/adjusted R\(^{2}\)r, here we report associations between individual IDPs and brain age delta values estimated from this model. In particular, Table 4 shows the first ten significant correlations (after correction for multiple comparisons), revealing a strong and significant association between these IDPs and the brain age delta. As further note, the association between brain age delta values and individuals IDPs were largely overlapped for the other regression methods, especially concerning the features in the top 10 positions. The order of the most significant features that are associated with the brain age delta is similar in the four methods, although the correlation values changed across them.
As it can be appreciated, the volumetric measurements from sMRI such as GM volume and volume of peripheral cortical GM (both normalised for head size) were negatively correlated with the brain age delta. Diffusion measures from DTI (such as MD, L1, L2 and L3) and from NODDI (such as ISOVF) in fornix were positively correlated with the brain age delta, while FA revealed an opposite pattern.
4.3 Association with Cardiac Variables
Table 5 reports Pearson correlations between brain age delta derived from the winning model (BRR with all IDPs) and CMR/CRFs measures (test set). For CMR, no significant associations were found after multiple comparison correction (p\(_{FDR}\) > 0.05), and only the correlation with the left ventricular mass (LVM) was significant before correction (p = 0.021). Regarding CRFs, all parameters except exercise and alcohol were significantly associated with the brain age delta (p\(_{FDR}\) < 0.05) .
5 Discussion
In this study, we investigated whether chronological age could be accurately predicted using brain MRI IDPs as predictor variables in various statistical models using data in the UKB. In particular, we focused on four well-known regression methods (SLR, SVR, LASSO and BRR) and considered measures from sMRI, SWI and dMRI as IDPs, either alone or in combination. Regarding the regression methods, overall, BRR achieved the highest accuracy as measured by MAE, R2 and adjusted R2 values. In particular, when dealing with a relatively small number of IDPs (< 50), for example in models with sMRI/SWI features only, better results were obtained using SVR and SLR. Conversely, in cases where a greater number of features was included, BRR reached the best performance, possibly because of its ability to handle multicollinearity between IDPs [21, 22].
Previous studies addressing modelling brain age using UKB data report MAE values between 3.5–3.8 years. Of note, Peng et al. [23] achieved the lowest MAE (2.14 years) although leveraging from deep CNN model, Simple Fully Convolutional Network, using sMRI from UKB for 14503 participants. In our study, the accuracy reached by BRR model in the different conditions was comparable (and even better in some cases) to such benchmarks, despite the generally lower number of subjects and MRI features.
Regarding the imaging predictors, models including all the 714 IDPs from the three brain MRI sequences had the best performance. However, when considering models with single-modal IDPs, dMRI reached the highest accuracy in terms of MAE values (MAE \(\approx \) 3.7 years) compared to sMRI (MAE \(\approx \) 4.5 years) and especially SWI (MAE \(\approx \) 6.02 years), and this was further confirmed by the R\(^2\)/adjusted R\(^2\) values. This might indicate that age-related alteration of brain can be better detected by dMRI, in agreement with literature findings [24]. A previous study also found similar results and further confirm that dMRI phenotypes are more informative than SWI IDPs in predicting brain age [2]. Phenotypes from sMRI and dMRI were generally the most informative for age prediction, as further supported by the correlation analysis between delta values and IDPs. Indeed, the strongest associations were found for features based on these modalities. In particular, our study revealed that brain age delta was negatively correlated with volumetric measures, while positively correlated with both ISOVF and diffusivities in the fornix.
GM volume was the most informative phenotype, in line with previous studies [2, 5]. This might be related to the fact that brain volume changes considerably over time and decreases during the aging process, causing atrophy [25] and macroscopic variations. Our analysis highlighted a prominent role of dMRI IDPs. Differences in diffusion properties across the life span have been demonstrated along specific WM tracts [24]. Diffusivity and FA values across the fornix spanned the first 10 ranking position, preceded only by atrophy measures in GM and WM. Noteworthy, the fornix is among those tracts that mature very early [26]. The IDPs that are present in the top 10 association ranking are, besides FA and MD, ISOVF, that is the isotropic volume fraction as estimated by the NODDI model, and the three tensor eigenvalues L1, L2 and L3 that represent the axial (L1) and transversal (L2, L3) diffusivities. A reduction in the FA and an increase in diffusivity, as indicated by a positive correlation of ISOVF, MD, L1, L2 and L3 with delta age, could indicate impaired WM integrity. Moreover, myelin breakdown might be measured by radial diffusivity (L2 and L3) alterations, while increasing in apparent diffusivity value might be a sign of axonal disruptions [27]. Furthermore, AD (L1) and RD (L2 and L3) have been observed to increase in elderly people which may be a signal of deterioration of the WM fibers [28]. Noteworthy, ISOVF has been observed to increase in older people in most of the major tracts, pointing to a disrupted integrity [29]. Our results are in agreement with such findings reporting a negative association of FA and a positive association of L1 , L2, L3 and MD in fornix with brain delta age. Fornix tracts have a vital role in memory tasks, specially episodic memory. Alteration in diffusion measures during aging process might be good biomarkers for neurological diseases that are related to memory impairments [26]. This could indicate that such IDPs are more prone to alteration over the life span of an individual at least over specific WM tracts, making them potential biomarkers for the aging process in health and disease.
Regarding the associations with CMR measures, our study revealed a significant association with LVM, however there was loss of statistical significance after multiple comparison correction. A previous study reported association of increase in LVM with alterations in WM microstructure in elderly people [30]. In our study, the limited age range in the UKB did not permit consideration of relationship in very old individuals. Among the CRFs, all measures except exercise and alcohol were significantly correlated with brain age delta (p\(_{FDR}\) < 0.05), inline with what described by Cole et al [7, 31], despite using a different number of IDPs and subjects for estimating brain delta. In conclusion, results suggest that dMRI IDPs play a prominent role in reducing the MAE and rank high in the association study, providing evidence of the potential of dMRI IDPs as biomarkers of aging in health and disease. Future work will investigate the integration of other IDPs such as functional MRI, graph-based measures from brain connectomes as well as the genetic information to pursue the holistic path.
References
Niu, X., et al.: Improved prediction of brain age using multimodal neuroimaging data. Hum. Brain Mapp. 41(6), 1626–1643 (2020)
Cole, J.H.: Multi-modality neuroimaging brain-age in UK Biobank: relationship to biomedical, lifestyle and cognitive factors. Neurobiol. Aging (2020)
Atli Jónsson, B. et al.: Brain age prediction using deep learning uncovers associated sequence variants. Nat. Commun. 10(1), 1–10 (2019)
Franke, K., Gaser, C.: Ten years of BrainAGE as a neuroimaging biomarker of brain aging: What insights have we gained? Frontiers Neurology 10, 789 (2019)
Smith, S.M. et al.: Estimation of brain age delta from brain imaging. Neuroimage 200, 528–539 (2019)
Elliott, L.T. et al.: Genome-wide association studies of brain imaging phenotypes in UK Biobank. Nature 562(7726), 210–216 (2018)
Ning, K., et al.: Association of relative brain age with tobacco smoking, alcohol consumption, and genetic variants. Sci. Rep. 10(1), 1–10 (2020)
UK Biobank.: About UK Biobank (2014). Available at https://www.uk-biobank.ac.uk/about-biobank-uk
Hua, K., et al.: Tract probability maps in stereotaxic spaces: analyses of white matter anatomy and tract-specific quantification. Neuroimage 39(1), 336–347 (2008)
Tibshirani, Robert: Regression shrinkage and selection via the lasso. J. R. Stat. Soc. Ser. B (Methodological) 58(1), 267–288 (1996)
Kenney, J.F., Keeping, E.S.: Linear regression and correlation. Math. Stat. 1, 252–285 (1962)
Drucker, H. et al.: Support vector regression machines. Adv. Neural Inf. Process. Syst. 155–161 (1997)
MacKay, D.J.C.: Bayesian interpolation. Neural Comput. 4(3), 415–447 (1992)
Pedregosa, F., et al.: Scikit-learn: machine learning in python. J. Mach. Learn. Res. 12, 2825–2830 (2011)
Miller, K.L. et al.: Multimodal population brain imaging in the UK Biobank prospective epidemiological study. Nat. Neurosci. 19(11), 15231536 (2016)
Beheshti, I. et al.: Bias-adjustment in neuroimaging-based brain age frameworks: a robust scheme. NeuroImage: Clin. 24, 102063 (2019)
Le, T.T et al.: A nonlinear simulation framework supports adjusting for age when analyzing BrainAGE. Frontiers Aging Neuroscience 10, 317 (2018)
van der Velpen, I.F. et al.: Impaired cardiac function and cognitive brain aging. Canadian J. Cardiol. 33(12), 1587–1596 (2017)
Jefferson, A.L. et al.: Cardiac index is associated with brain aging: the framingham heart study. Circulation 122(7), 690 (2010)
Petersen, S.E. et al.: UK Biobank’s cardiovascular magnetic resonance protocol. J. Cardiovascular Mag. Reson. 18(1), 8 (2015)
Assaf, A.G., Tsionas, M., Tasiopoulos, A.: Diagnosing and correcting the effects of multicollinearity: Bayesian implications of ridge regression. Tourism Manag 71, 1–8 (2019)
Efendi, A., Effrihan.: A simulation study on Bayesian Ridge regression models for several collinearity levels. In: AIP Conference Proceedings, vol. 1913. 1, p. 020031. AIP Publishing LLC (2017)
Peng, H. et al.: Accurate brain age prediction with lightweight deep neural networks. BioRxiv (2019)
Yao, X. et al.: Evaluation of human brain aging via diffusion tensor imaging tract characteristics. In: 2017 13th International Conference on Natural Computation, Fuzzy Systems and Knowledge Discovery (ICNC-FSKD), pp. 2651–2655. IEEE (2017)
Driscoll, I., et al.: Longitudinal pattern of regional brain volume change differentiates normal aging from MCI. Neurology 72(22), 1906–1913 (2009)
Douet, V., Chang, L.: Fornix as an imaging marker for episodic memory deffcits in healthy aging and in various neurological disorders. Frontiers Aging Neurosci. 6, 343 (2015)
Liu, H., et al.: Aging of cerebral white matter. Ageing Res. Rev. 34, 64–76 (2017)
Chen, D.Q. et al.: Age-related changes in diffusion tensor imaging metrics of fornix subregions in healthy humans. Stereotactic Funct. Neurosurg. 93(3), 151–159 (2015)
Cox, S.R. et al.: Ageing and brain white matter structure in 3,513 UK Biobank participants. Nat. Commun. 7(1), 1–13 (2016)
Moore, E.E., et al.: Increased left ventricular mass index is associated with compromised white matter microstructure among older adults. J. Am. Heart Assoc. 7(13), e009041 (2018)
Cole, J.H. et al.: Body mass index, but not FTO genotype or major depressive disorder, inuences brain structure. Neuroscience 252, 109–117 (2013)
Acknowledgements
This work was funded by INVITE program co-financed by the European Union within the Horizon 2020 Programme and by the Regione del Veneto.
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Salih, A. et al. (2021). Multi-modal Brain Age Estimation: A Comparative Study Confirms the Importance of Microstructure. In: Gyori, N., Hutter, J., Nath, V., Palombo, M., Pizzolato, M., Zhang, F. (eds) Computational Diffusion MRI. Mathematics and Visualization. Springer, Cham. https://doi.org/10.1007/978-3-030-73018-5_19
Download citation
DOI: https://doi.org/10.1007/978-3-030-73018-5_19
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-73017-8
Online ISBN: 978-3-030-73018-5
eBook Packages: Mathematics and StatisticsMathematics and Statistics (R0)