Abstract
Glioma with a high incidence is one of the most common brain cancers. In the clinic, pathologist diagnoses the types of the glioma by observing the whole-slide images (WSIs) with different magnifications, which is time-consuming, laborious, and experience-dependent. The automatic grading of the glioma based on WSIs can provide aided diagnosis for clinicians. This paper proposes two fully convolutional networks, which are respectively used for WSIs and MRI images to achieve the automatic glioma grading (astrocytoma (lower-grade A), oligodendroglioma (middle-grade O), and glioblastoma (higher-grade G)). The final classification result is the probability average of the two networks. In the clinic and also in our multi-modalities image representation, grade A and O are difficult to distinguish. This work proposes a two-stage training strategy to exclude the distraction of the grade G and focuses on the classification of grade A and O. The experimental result shows that the proposed model achieves high glioma classification performance with the balanced accuracy of 0.889, Cohen’s Kappa of 0.903, and F1-score of 0.943 tested on the validation set.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Glioma is one common brain tumor, which occupies approximately 80\(\%\) of malignant brain tumors [8]. According to its severity degree evaluated on the pathology images, world health organization (WHO) categorizes glioma as three grades: astrocytoma (lower grade), oligodendroglioma (middle grade), and glioblastoma (higher grade). The lower grade has more optimistic and more survival years. However, the higher grades with worse prognosis are usually life-threatening. In the clinic, the glioma is diagnosed depending on the histopathology technique on the microscopic examination environment. The accurate diagnosis of lesions for pathologists is very time-consuming, laborious, and expertise-dependent. The computer-aided diagnosis is highly required to alleviate the difficulties of the pathologists.
The emergence of whole-slide images (WSIs) technology has realized the transformation from a microscopic perspective to a computer perspective and promoted the application of image processing technology to digital pathology. The digital pathology image analysis can help pathologists diagnose and provide quantitative information calculated from the WSIs, achieving the objectivity and reproducibility in the clinical diagnosis.
Magnetic resonance image (MRI) as a non-invasive imaging technique has been routinely used in the diagnosis of brain tumors. Multi-modalities with different brain tissue enhancement can be selected to make a clinical decision. Multi-modalities fusion technology can capture more abundant feature information to perform more precise tumor classification. MRI with the advantage of safety and non-invasion has been used to classify glioma [5, 6, 16, 18]. However, pathological information acquired by invasive methods is adopted as the gold standard in the current clinical environment. The integration of the two types of images could achieve higher glioma grading performance.
The CPM-RadPath 2020 MICCAI challenge releases a multi-modalities dataset which contains paired MRI scans and histopathology images of brain gliomas collected from the same patients. Their glioma classification annotations (astrocytoma, oligodendroglioma, and glioblastoma) have been provided for the training set. Based on this dataset, this work trains one 3D Densenet for MRI images classification and 2D fully constitutional networks (EfficientNet-B2, EfficientNet-B3, and SE-ResNext101) for pathology classification. The final glioma grading is determined by the average of the two types of models.
2 Related Work
In recent years, automatic glioma classification and grading have attracted widespread attention using machine learning and deep learning techniques.
A majority of these methods adopt the MRI image as their experiment data since MRI modality is a non-invasive and fast imaging technique and is routinely used for glioma diagnosis in the clinic. These MRI-based glioma grading methods can be mainly divided into two categories: hand-crafted feature engineering and deep learning-based feature representation. These hand-crafted features are usually extracted based on the ROI (region of interest) region that is delineated by experienced radiologists or some automatic image segmentation techniques. These extracted features comprise histogram-based features [7], shape features [19], texture features [13], contour feature[13], and wavelet features [15]. Based on these features, some machine learning techniques including SVM (support vector machines), RF (random forest), NN (neural network), and DT (decision tree) are used to achieve the automatic glioma classification or grading. However, these traditional feature engineering techniques fail to generate a robust and general feature parameter and are easily affected by the data variations (MRI scanner, data collection protocol, and image noises), which limits their promise for clinical application. Deep learning has the ability to learn high-level feature representation from the raw data through the network training procedure. Due to the 3D nature of MRI images, 2D and 3D models are alternative. These 3D models could contact the context information during each MRI scan, such as the 3-D multiscale CNN model [7] and the 3D residual network [5]. It is known that the 3D model extremely increases the number of parameters and requires a large amount of computation resources to support network training. These 2D models, such as the VGG model [1, 4] and Residual Network (ResNet) architecture [1, 9], ignore the connection between MRI slices and treat each MRI slice as an independent image to complete the classification task.
The previously mentioned MRI-based studies have achieved limited performance since the gold standard for the glioma diagnosis is from the pathology images. Thus, a combination of the MRI and pathology images may provide complementary information and achieve finer classification results. The previous CPM challenge has reported four solutions for the glioma grading by combining MRI and pathology images [10, 12]. Pei et al. [12] used an Unet-like model to segment ROI and a 3D CNN to classify the glioma types focusing on the extracted course tumor region. Their experimental results reported that using MRI sequences alone can realize better performance than using the pathology alone or the combination of MRI and pathology images. Ma et al. [10] applied a 2D ResNet-based model to classify glioma based on pathology and a 3D DenseNet-based model to classify glioma based on MRI images. Then, a simple regression model was used to achieve the ensemble of the two models. Chan et al. [2] extracted features by using two CNN networks (VGG16 and ResNet 50) and then classified three types of brain tumors based on several clustering methods (K-means and random forest). Xue et al. [17] trained a 2D ResNet18 and a 3D ResNet18 to classify three types of gliomas based on the pathology and MRI images, respectively. The features in fully connected layers of the two models were concatenated together as the input of the following softmax layer to achieve the classification of brain tumors.
Our work is different from the above-mentioned methods. We apply a two-stage classification algorithm to first detect the glioblastoma and then pay more attention to the distinction between the astrocytoma and oligodendroglioma. Sine the glioblastoma is defined as the Grade IV and astrocytoma and oligodendroglioma are both defined as the Grade II or III by WHO. Thus, astrocytoma and oligodendroglioma can be more difficult to separate. Our two-stage training has the ability to improve classification performance for astrocytoma and oligodendroglioma cases.
3 Method
This paper applies two fully convolutional networks to achieve a feature-independent end-to-end glioma grading. In the following, we introduce the image preprocessing and network framework in detail.
3.1 Data Preprocessing
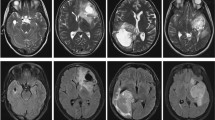
The data used in this paper includes paired pathology and multi-sequence MRI (T2-FLAIR, T1, T1ce, and T2) images. Figure 1 and Fig. 2 illustrate the pathology and corresponding four modalities MRI images in terms of three glioma types (astrocytoma, glioblastoma, and oligodendroglioma).
For the preprocessing procedure of the pathology images, the OTSU method is firstly adopted to remove the non-tissue region [10]. The original WSIs with large size (e.g., 50000 \(\times \) 50000) can not be directly fed into the network as the input, we apply a sliding window with no overlap to segment the WSI into image patch with the size of 1024\(\times \)1024 (Fig. 3). Not every path contains tumor tissue, so we set some rules for each path to exclude these useless patches. First, the mean value of the patch is limited from 100 to 200. Second, the standard deviation of each patch is limited to greater than 20. Third, in the color space HSV (hue, saturation, value), the mean of channel 0 is set as greater than 50. In the preprocessing procedure of MRI images, the four MRI modalities images are all cropped to 128\(\times \)192\(\times \)192.
3.2 Model Details
Figure 4 illustrates the overall architecture of our proposed glioma grading system which is composed of two types of networks: 2D CNN for pathology image classification (Fig. 5) and 3D CNN for MRI image classification (Fig. 6). Each type of network performs the two-stage classification. Since the astrocytoma and oligodendroglioma appear as more similar, the first stage achieves the detection of glioblastoma, after that, the second stage distinguishes the astrocytoma and oligodendroglioma.
For the 2D pathology image classification network, the input image is a small WSI patch that is segmented from a large WSI in the preprocessing procedure. The backbone networks include EfficientNet-B2, EfficientNet-B3, and SE-ResNext101. Before the fully connected layer, generalized-mean (GEM) pooling is applied to the learned features, which is defined as
where \(\mathcal {X}\) and \(\mathbf {f}\) represent the input and output, respectively. When \(p_{k}\) equals to \(\infty \) and 1, the equation denotes the max pooling and average pooling, respectively. Following [14], this work set \(p_{k}\) to 3.
After the GEM operation, the Meta info (age information) is added to the final classification feature vector. Then, a classification and a regression branches are appended as the cross-entropy (\(L_{BCE}\)) and smooth L1 (L1) losses, respectively, to achieve more robust brain tumor classification.
where \(y_{l}\) and \(\hat{y}_{l}\) denote the ground truth and predicted annotations.
For the 3D MRI image classification network, the input image is MRI images with four channels, which correspond to the four modalities. The backbone adopts 3D ResNet, following by global average pooling and fully connected layer to grade the brain tumor. In the MRI-based classification process, the loss function also adopts the cross-entropy. To minimize the loss function, Adam optimization algorithm is used.
The ensemble of the 2D pathology and 3D MRI classification models is the probability average of the two types of networks.
4 Results and Discussion
4.1 Data and Evaluation Metrics
Dataset. The CPM-RadPath 2020 MICCAI challenge provides paired radiology scans and digitized histopathology images of brain gliomas, and image-level ground truth label as well. The goal of CPM2020 is classifying each case into three sub-types: Glioblastoma (grade IV), Oligodendroglioma (grade II or III), and Astrocytoma (grade II or III).
The overall architecture of our glioma grading system. A two-stage classification strategy is applied to both the 2D pathology and 3D MRI images. The glioblastoma with more serious anatomy representation is detected in the first step. Then, in the second step, our algorithm focuses on the classification of astrocytoma and oligodendroglioma.
The detailed 2D CNN network. The backbone includes EfficientNet-B2, EfficientNet-B3, and SE-ResNext101. In the final feature representation, the Meta info (age information) is included. A regression branch with a smooth L1 loss function is added to relieve the overfitting. The classification branch with the cross-entropy loss function is used to complete the classification procedure.
Specifically, for each patient, the provided 3D MRI images comprise four modalities: native (T1), post-contrast T1-weighted (T1Gd), T2-weighted (T2), and T2 Fluid Attenuated Inversion Recovery (T2-FLAIR). All MRI images were co-registered to the same anatomical template and interpolated to the same resolution (1 cubic mm) in three directions. Also, the contest provides one digitized whole slide tissue image for each case, which were scanned at 20x or 40x magnifications. The dataset is divided into 221, 35, 73 cases for training, validation, and testing respectively.
Evaluation Metrics. The algorithmic performance is evaluated from three aspects: F1-Score, Balanced Accuracy, and Cohen’s Kappa. Suppose TP, FP, FN is the number of true positives, false positives, and false negatives respectively. Then the three metrics can be computed as below.
where K is the number of classes, \({p_o}\) is the relative observed agreement among raters, and \({p_e}\) is the hypothetical probability of chance agreement, using the observed data to calculate the probabilities of each observer randomly seeing each category [3]. The aforementioned evaluation metrics are suitable for imbalanced categories, avoiding depending on the proportion of each class.
4.2 Experiments and Discussion
In the training stage, we perform 5-fold cross validation for local validation. Adam optimizer is used with an initial learning rate of 0.001 and decreases by 10 times every 50 epochs. All models are implemented based on the Pytorch framework [11] and trained on a workstation with Intel(R) Xeon(R) CPU E5-2680 v4 2.40 GHz processors and four NVIDIA Tesla P40s (24 GB) installed.
In the inference stage, the multimodal MRI images and the whole-slide pathology images are pre-processed and sent to the classification network respectively. The average predictions of the 5-fold models are employed to get more accurate results. Then, we sum the probabilities of the two networks to obtain the final ensemble prediction.
As the online evaluation performances shown in Table 1, our proposed two-stage coarse-to-fine classification framework contributes to gaining higher accuracy on all three evaluation metrics. What’s more, the classification model of MRI and pathological images can complement each other to obtain more robust and accurate results.
5 Conclusion
This paper proposed a two-stage glioma grading algorithm to classify the brain tumor into three types: astrocytoma, glioblastoma, and oligodendroglioma. The classification algorithm is designed based on the feature representation difference between the severe and lower glioma grades. The more serious glioblastoma grade is separated out in the first stage, and the second stage eliminates the interference of glioblastoma and only focuses on learning the difference between astrocytoma and oligodendroglioma. Our two-stage strategy is applied on the classification networks for the pathology (2D CNN) and MRI images (3D ResNet), respectively. By testing on the validation data, we have achieved state-of-the-art performance by the ensemble of the 2D pathology and 3D MRI images classification networks. In the final submission of this challenge, we omit the 3D MRI image classification network, since the diagnosis based on pathology is adopted as the gold standard in the clinic. In this CPM-RadPath 2020 MICCAI challenge, the number of training samples is limited, which can greatly influence the robustness of designed network. In future work, small sample based deep learning could be developed to build a more general model.
References
Banerjee, S., Mitra, S., Masulli, F., Rovetta, S.: Glioma classification using deep radiomics. SN Comput. Sci. 1(4), 1–14 (2020)
Chan, H.-W., Weng, Y.-T., Huang, T.-Y.: Automatic classification of brain tumor types with the MRI scans and histopathology images. In: Crimi, A., Bakas, S. (eds.) BrainLes 2019. LNCS, vol. 11993, pp. 353–359. Springer, Cham (2020). https://doi.org/10.1007/978-3-030-46643-5_35
Cohen, J.: A coefficient of agreement for nominal scales. Educ. Psychol. Measur. 20(1), 37–46 (1960)
Decuyper, M., Bonte, S., Van Holen, R.: Binary glioma grading: radiomics versus pre-trained CNN features. In: Frangi, A.F., Schnabel, J.A., Davatzikos, C., Alberola-López, C., Fichtinger, G. (eds.) MICCAI 2018. LNCS, vol. 11072, pp. 498–505. Springer, Cham (2018). https://doi.org/10.1007/978-3-030-00931-1_57
Decuyper, M., Holen, R.V.: Fully automatic binary glioma grading based on pre-therapy MRI using 3D convolutional neural networks (2019)
Fusun, C.E., Zeynep, F., Ilhami, K., Ugur, T., Esin, O.I.: Machine-learning in grading of gliomas based on multi-parametric magnetic resonance imaging at 3t. Comput. Biol. Med. 99, 154–160 (2018)
Ge, C., Qu, Q., Gu, I.Y.H., Jakola, A.S.: 3D multi-scale convolutional networks for glioma grading using MR images. In: 2018 25th IEEE International Conference on Image Processing (ICIP), pp. 141–145. IEEE (2018)
Goodenberger, M.K.L., Jenkins, R.B.: Genetics of adult glioma. Cancer Genet. 205(12), 613–621 (2012)
Ismael, S.A.A., Mohammed, A., Hefny, H.: An enhanced deep learning approach for brain cancer MRI images classification using residual networks. Artif. Intell. Med. 102, 101779 (2020)
Ma, X., Jia, F.: Brain tumor classification with multimodal MR and pathology images. In: Crimi, A., Bakas, S. (eds.) BrainLes 2019. LNCS, vol. 11993, pp. 343–352. Springer, Cham (2020). https://doi.org/10.1007/978-3-030-46643-5_34
Paszke, A., et al.: Automatic differentiation in pytorch (2017)
Pei, L., Vidyaratne, L., Hsu, W.-W., Rahman, M.M., Iftekharuddin, K.M.: Brain tumor classification using 3D convolutional neural network. In: Crimi, A., Bakas, S. (eds.) BrainLes 2019. LNCS, vol. 11993, pp. 335–342. Springer, Cham (2020). https://doi.org/10.1007/978-3-030-46643-5_33
Pintelas, E., Liaskos, M., Livieris, I.E., Kotsiantis, S., Pintelas, P.: Explainable machine learning framework for image classification problems: case study on glioma cancer prediction. J. Imaging 6(6), 37–50 (2020)
Radenović, F., Tolias, G., Chum, O.: Fine-tuning CNN image retrieval with no human annotation. IEEE Trans. Pattern Anal. Mach. Intell. 41(7), 1655–1668 (2018)
Su, C., et al.: Radiomics based on multicontrast MRI can precisely differentiate among glioma subtypes and predict tumour-proliferative behaviour. Eur. Radiol. 29(4), 1986–1996 (2019)
Wang, X., et al.: Machine learning models for multiparametric glioma grading with quantitative result interpretations. Frontiers Neurosci. 12, 1–10 (2019)
Xue, Y., et al.: Brain tumor classification with tumor segmentations and a dual path residual convolutional neural network from MRI and pathology images. In: Crimi, A., Bakas, S. (eds.) BrainLes 2019. LNCS, vol. 11993, pp. 360–367. Springer, Cham (2020). https://doi.org/10.1007/978-3-030-46643-5_36
Yang, Y., et al.: Glioma grading on conventional MR images: a deep learning study with transfer learning. Frontiers Neurosci. 12, 1–10 (2018)
Zhang, Z., et al.: Deep convolutional radiomic features on diffusion tensor images for classification of glioma grades. J. Digit. Imaging 33(4), 826–837 (2020). https://doi.org/10.1007/s10278-020-00322-4
Acknowledgement
This research was funded by the National Natural Science Foundation of China, grant number 61571314.
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 Springer Nature Switzerland AG
About this paper
Cite this paper
Wang, X., Yang, S., Wu, X. (2021). Automatic Glioma Grading Based on Two-Stage Networks by Integrating Pathology and MRI Images. In: Crimi, A., Bakas, S. (eds) Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries. BrainLes 2020. Lecture Notes in Computer Science(), vol 12659. Springer, Cham. https://doi.org/10.1007/978-3-030-72087-2_40
Download citation
DOI: https://doi.org/10.1007/978-3-030-72087-2_40
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-72086-5
Online ISBN: 978-3-030-72087-2
eBook Packages: Computer ScienceComputer Science (R0)