Abstract
Computer-aided diagnosis of polyps has played an important role in advancing the screening capability of computed tomographic colonography (CTC). Texture features, including intensity, gradient, and curvature, are the essential components in differentiation of neoplastic and nonneoplastic polyps. Clinical study has shown that the malignancy rates of polyps increase in correlation to their size. In this chapter, we present a study to investigate the effect of separating polyps based on size on the performance of machine learning. First, the volume of interest of each polyp was extracted and further confirmed by Radiologists. All polyp masses in this study have a diameter size ranging from 6 to 30 mm. Then, we group polyps into three groups based on their sizes: 6–9 mm, 10–30 mm, and a combined group of 6–30 mm. The corresponding malignancy risks of polyps were also recorded. From each polyp volume, we extracted the traditional 14 Haralick texture features plus 16 additional features with a total 30 texture features. Those features were further grouped as descriptors for intensity, gradient, and curvature characteristics. Finally, we employed the Random Forest classifier to differentiate neoplastic and nonneoplastic polyps. The proposed texture feature analysis was studied using 228 polyp masses. We generated a measure of area under the curve (AUC) values of the receiver operating characteristics (ROC) curve. The performance was determined by assessing the sensitivity and specificity values. Experimental results demonstrated that gradient and curvature features were ideal for differentiation of the malignancy risk for medium-sized polyps, whereas intensity feature was better for smaller-sized polyps.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
According to recent statistics published by the American Cancer Association, colorectal cancer is the second leading cause of deaths of men and women combined, with an estimated 101,420 new cases in 2019 (American Cancer [1, 9]). Early detection and removal of polyps significantly reduces the risk of death and screenings are recommended for those 50 or older. However, using clinical optical colonoscopy (OC), to screen the growing population of those over 50 is severely limited by our current resources. Alternatively, computed tomography colonography (CTC) has been a rising noninvasive solution to detect possible cancers [3, 5, 6, 11, 12], decreasing the amount of people being screened under OC. Furthermore, the majority of polyps are found to be nonneoplastic, named hyperplastic, which are abnormal growths with no risk, only taking up valuable resources when removed. Therefore, being able to distinguish between hyperplastic and adenomatous polyps through CTC screenings has become crucial.
In our previous research work, texture feature extraction techniques have been established and modified to improve the accuracy of diagnoses of polyps found through CTC screenings [4, 10]. Those texture features include intensity, gradient, and curvature. Pickhardt et al. suggests that the malignancy rates of polyps increase in correlation to their size [8]. Besides polyp’s malignancy rates, general kernel approach for gradient and curvature texture extraction may also get influenced by the polyp size [7]. Thus, in this study, we design experiments to investigate the effect of separating polyps based on size on the performance of machine learning.
The remainder of this chapter is organized as follows. Section 2 depicts our method for conducting feature analysis in computer-aided diagnosis of colorectal polyps. In Sect. 3, experimental design and results are reported. Finally, discussion of our work and conclusions are given in Sect. 4.
2 Materials and Methods
The aim of this study is to investigate the performance of machine learning on texture features based on different polyp sizes. Figure 1 shows a 6-mm small-sized polyp and a 22-mm medium-sized polyp visualized via two-dimensional (2D) axial image.
2.1 Flowchart of Our Method
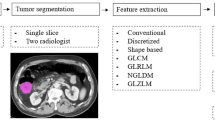
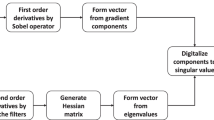
The flowchart of our methods can be summarized in Fig. 2. First, from the original CT image, the volume of interest (VOI) of each polyp was extracted and further confirmed by the radiologists. Second, we applied an extended traditional Haralick model [2] with a total of 30 texture features [4], which were obtained from the VOIs of each polyp. In our study, the texture features of polyps consist of intensity, gradient, and curvature. Third, we performed a receiver operating characteristics (ROC)-based analysis to select the best feature sets. Finally, we employed a machine learning method, the Random Forest (RF) classifier, for feature classification.
2.2 Malignancy Risk of the Polyps
According to histopathology, polyps can be divided into two categories: nonneoplastic and neoplastic. Nonneoplastic polyps are benign polyps, including the subtypes of hyperplasic, mucosal polyp, and inflammatory, etc. On the other hand, neoplastic are malignancy risky, including serrated adenoma, tubular adenoma, tubulovillous adenoma, adenocarcinoma, etc. In this study, we labeled the nonneoplastic polyp as 0 and neoplastic polyps as 1, indicating their benign or malignancy risk accordingly.
2.3 Data Preparation
The dataset used in this study consists of a total number of 228 polyp masses found through a CTC database. Those polyp masses were confirmed by OC. The spatial resolution of the CT image is 0.7 by 0.7 by 1.0 mm3. All the polyp masses have a diameter size ranging from 6 to 30 mm. Polyps were grouped into three groups based on their sizes: 6–9 mm, 10–30 mm, and a combined group of 6–30 mm. For a fair performance comparison, we have a balanced polyp pool of 114 polyps in each size group. Within each size group, all of the datasets had 57 benign polyps and 57 malignant polyps.
3 Results
We investigated the performance of the algorithm on the 6–9 mm, 10–30 mm, and 6–30 mm groups. The studied features include intensity, gradient, curvature, all combined features. We generated a measure of area under the curve (AUC) values of ROC curve. The performance was determined by assessing the sensitivity and specificity values. The averaged AUC information is illustrated in Table 1. Figure 3 shows the averaged ROC curve for the intensity feature. Figure 4 shows the averaged ROC curve for the gradient feature. Figure 5 shows the averaged ROC curve for the curvature feature. And Fig. 6 shows the averaged ROC curve for all combined features.
4 Discussion and Conclusions
Experimental results demonstrated that gradient and curvature were ideal distinguishing features for medium-sized polyps, whereas intensity was better for smaller-sized polyps. Due to their negligible proportions, the curvature and gradient features were flawed for the 6–9 mm polyps during the experiment. The opposite is true for the medium-sized polyp group.
When examining all features, the AUC value of the 10–30 mm polyps was greater than the AUC value of the 6–30 mm polyps, suggesting that separating the small- and medium-sized polyps would be beneficial in identifying medium-sized polyps. However, the AUC value of the 6–9 mm polyps was lower than that of the 6–30 mm polyps, suggesting that this separation may not be ideal for identifying smaller polyps. The AUC value for all polyps is greater than that of the individual group of smaller polyps because it is averaged out by the higher performance of the identification of the 10–30 mm polyps. Furthermore, the smaller polyps have less identifiable features. This study shall facilitate computer-aided diagnosis of polyps to achieve high performance by taking into account the contributions of different features among different polyp sizes.
References
American Cancer Society, Cancer facts & figures 2019. Atlanta (2019)
R.M. Haralick, K. Shanmugam, I. Dinstein, Textural features for image classification. IEEE Trans. Syst. Man Cybern. 3(6), 610–621 (1973)
L. Hong, A. Kaufman, Y. Wei, et al., 3D virtual colonoscopy. IEEE Biomedical Visualization Symposium (IEEE CS Press, Los Alamitos, 1995), pp. 26–32
Y. Hu, Z. Liang, B. Song, et al., Texture feature extraction and analysis for polyp differentiation via computed tomography colonography. IEEE Trans. Med. Imaging 35(6), 1522–1531 (2016)
Z. Liang, VC: An alternative approach to examination of the entire colon. INNERVISION 16, 40–44 (2001)
Z. Liang, R. Richards, Virtual colonoscopy v.s. optical colonoscopy. Expert Opin. Med. Diagn J 4(2), 149–158 (2010)
J. Liu, S. Kabadi, R.V. Uitert, et al., Improved computer-aided detection of small polyps in CT colonography using interpolation for curvature estimation. Med. Phys. 38(7), 4276–4284 (2011)
P.J. Pickhardt, K.S. Hain, D.H. Kim, et al., Low rates of cancer or high-grade dysplasia in colorectal polyps collected from computed tomography colonography screening. Clin. Gastroenterol. Hepatol. 8, 610–615 (2010)
R.L. Siegel, K.D. Miller, A. Jemal, Cancer statistics, 2019. CA Cancer J. Clin. 69(1), 7–34 (2019)
B. Song, G. Zhang, H. Lu, et al., Volumetric texture features from higher-order images for diagnosis of colon lesions via CTC. Int. J. Comput. Assist. Radiol. Surg. 9(6), 1021–1032 (2014)
D. Vining, D. Gelfand, R. Bechtold et al., Technical feasibility of colon imaging with helical CT and virtual reality. Annual Meeting of American Roentgen Ray Society, New Orleans, pp. 104 (1994)
H. Zhu, Y. Fan, H. Lu, et al., Improved curvature estimation for computer-aided detection of colonic polyps in CT colonography. Acad. Radiol. 18(8), 1024–1034 (2011)
Acknowledgments
This work was partially supported by NIH grant #CA206171 of the National Cancer Institute and the Computer Science and Informatics Research Experience Program at Stony Brook University.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 Springer Nature Switzerland AG
About this paper
Cite this paper
Choi, Y., Wei, A., Wang, D., Liang, D., Zhang, S., Pomeroy, M. (2021). An Investigation of Texture Features Based on Polyp Size for Computer-Aided Diagnosis of Colonic Polyps. In: Arabnia, H.R., Deligiannidis, L., Shouno, H., Tinetti, F.G., Tran, QN. (eds) Advances in Computer Vision and Computational Biology. Transactions on Computational Science and Computational Intelligence. Springer, Cham. https://doi.org/10.1007/978-3-030-71051-4_66
Download citation
DOI: https://doi.org/10.1007/978-3-030-71051-4_66
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-71050-7
Online ISBN: 978-3-030-71051-4
eBook Packages: Computer ScienceComputer Science (R0)