Abstract
Glioblastoma, formerly known as glioblastoma multiforme (GBM), is remarkable for its degree of both morphologic and genomic heterogeneity. In children and adults, as defined by the World Health Organization (WHO), GBM is a grade IV neoplasm with a diffusely infiltrative growth pattern populated by cells showing predominately astrocytic differentiation. As discussed in previous chapters, GBM features prominent nuclear atypia and cellular pleomorphism, as well as high tumor cell mitotic activity accompanied by microvascular proliferation and/ or necrosis. As its former name indicates, GBM may include one or many morphologic patterns within a single tumor. In some tumors, this reflects underlying clonal evolution with newly acquired genetic changes in tumor cell subpopulations. Even more variable than GBM morphology is the range of genomic heterogeneity, creating multiple molecular signatures that define tumor behavior, prognosis and treatment response independent of tumor histopathology. Furthermore, while pediatric and adult glioblastoma share many histopathologic similarities, they are unequivocally biologically distinct neoplasms. In many pediatric and adult gliomas, the molecular subgroup is a better predictor of tumor behavior than histologic grade. In particular, WHO grade II or III diffuse astrocytomas lacking morphologic features associated with GBM (microvascular proliferation and necrosis), but with certain defined molecular alterations, should be considered GBM for prognostic and therapeutic purposes due to their expected WHO grade IV-like behavior. The focus of this chapter centers on the genomic heterogeneity and pathobiology of aggressive pediatric and adult diffuse astrocytomas with WHO grade IV behavior independent of morphologic features.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
Introduction
Glioblastoma, formerly known as glioblastoma multiforme (GBM), is remarkable for its degree of both morphologic and genomic heterogeneity. In children and adults, as defined by the World Health Organization (WHO), GBM is a grade IV neoplasm with a diffusely infiltrative growth pattern populated by cells showing predominately astrocytic differentiation [1]. As discussed in previous chapters, GBM features prominent nuclear atypia and cellular pleomorphism, as well as high tumor cell mitotic activity accompanied by microvascular proliferation and/or necrosis. As its former name indicates, GBM may include one or many morphologic patterns within a single tumor. In some tumors, this reflects underlying clonal evolution with newly acquired genetic changes in tumor cell subpopulations [2,3,4,5]. Even more variable than GBM morphology is the range of genomic heterogeneity, creating multiple molecular signatures that define tumor behavior, prognosis and treatment response independent of tumor histopathology. Furthermore, while pediatric and adult glioblastoma share many histopathologic similarities, they are unequivocally biologically distinct neoplasms. In many pediatric and adult gliomas, the molecular subgroup is a better predictor of tumor behavior than histologic grade. In particular, WHO grade II or III diffuse astrocytomas lacking morphologic features associated with GBM (microvascular proliferation and necrosis), but with certain defined molecular alterations, should be considered GBM for prognostic and therapeutic purposes due to their expected WHO grade IV-like behavior. The focus of this chapter centers on the genomic heterogeneity and pathobiology of aggressive pediatric and adult diffuse astrocytomas with WHO grade IV behavior independent of morphologic features.
Pediatric GBM is traditionally grouped with anaplastic astrocytoma and diffuse intrinsic pontine glioma (DIPG) as the high-grade gliomas (HGG). HGG are common in adults, whereas in children low-grade gliomas are more common; nonetheless, HGG are estimated to occur in slightly less than 1 per 100,000 children each year [6]. In adults, HGG commonly arise in the supratentorial structures, while in children GBMs tend to be located in midline structures such as the thalamus, spinal cord and pons. DIPGs are diffusely infiltrative astrocytomas of the pons and many, but not all, fulfill WHO histological criteria for HGG. DIPGs occur nearly exclusively in children with a peak age incidence of 6–8 years, while HGG located in supratentorial structures occur in older children with a peak incidence in adolescence. Overall, pediatric HGG patients have a grim prognosis with less than 5% of GBM patients surviving 5 years after diagnosis while DIPG patients have a median survival that is less than 1 year after diagnosis [7]. Curiously, infants with histologic HGG tend to show better clinical outcomes than older children do and can have a more favorable clinical course than infants with low-grade gliomas, suggesting that infant HGG has unique biological properties [8, 9]. Clearly, there is a pressing need to better understand the biology of these tumors and for effective therapeutic approaches for pediatric GBM.
The diversity of pediatric GBM biology was recognized relatively recently when advanced molecular testing techniques were applied in collaborative studies that amassed large cohorts of GBM. The data generated from these studies has clearly shown that childhood GBM is a distinct disease from adult GBM. Furthermore, these studies conclusively demonstrate that pediatric GBMs develop following unique molecular pathogenetic events, which are different from those that underlie the pathogenesis of their adult counterparts. The advances in genomic and epigenetic profiling have further permitted the characterization of these molecular and cellular differences with precision, expanding our understanding of GBM biology, and resolving distinct subgroups of GBM that arise in children and adults.
The application of molecular profiling to large cohorts of tumor samples from pediatric and adult GBM patients has identified a number of recurrent genomic and epigenetic alterations that subdivide GBM into discrete subgroups, which correlate with various clinical parameters including patient age and tumor location [10, 11]. Moving away from traditional histologic classification of GBM, this chapter will provide an overview of the key features of adult and pediatric GBM based on gene expression profiling, genomic structural variations, copy number alterations (CNA), DNA methylation profiling, and the mutational landscape of single nucleotide variants (SNV). This will be followed by a description of adult and pediatric GBM subgroups that have emerged from these studies.
Gene Expression Profiling
Genome wide gene expression profiling microarray studies analyzing large cohorts of adult and pediatric GBM have been used to characterize differentially expressed genes that successfully identified subgroups within GBM that were not discernable histopathologically. The gene expression profiling studies were initially performed using cohorts of adult GBMs and subsequently the gene expression subgroups were identified in pediatric GBM (Table 9.1). The first gene expression profiling study in GBM identified three subgroups according to the functions of signature genes: proneural, mesenchymal and proliferative, with different patient outcomes including longer survival of patients with proneural tumors [12]. Subsequent gene expression profiling performed on the Cancer Genome Atlas (TCGA) tumor cohort characterized four different groups of GBM in adults: proneural, neural, classical and mesenchymal [13, 14]. The identified gene expression subtypes showed additional correlative molecular features. For instance, the proneural subtype had alterations in PDGFRA or IDH, while NF1 mutations occurred in mesenchymal tumors and EGFR mutations appeared in the classical subgroup. In time, proneural GBMs were distinguished as glioma-CpG island methylator phenotype (G-CIMP)-positive and G-CIMP-negative subtypes based on IDH1 mutation status and DNA methylation pattern. The favorable prognosis of proneural GBM was discovered to apply to the G-CIMP-positive subset, while C-CIMP-negative and mesenchymal tumors have a less favorable prognosis [13].
Gene expression profiling studies of pediatric GBM demonstrated significant differences from adult GBM, suggesting distinct pathogenetic mechanisms are responsible (Table 9.1) [15, 16]. A prominent proportion of pediatric GBMs show enhanced PDGFRA-driven gene expression, which is not surprising given the high frequency of PDGFRA gene amplification in pediatric tumors [10, 16,17,18,19]. Gene expression profiling studies demonstrated similarities between some midline GBMs and DIPG, foreshadowing their common pathogenesis prior to sequencing studies that later showed these tumor types share H3F3A K27M mutations [10, 20, 21].
Genomic Structural Changes and Copy Number Alterations
The genome of pediatric GBM may contain a number of DNA copy number alterations (CNA) and structural alterations, although these changes tend to occur less frequently in childhood than in adult GBM (Table 9.1) [18, 19, 22, 23]. These alterations are variable in degree and range from simple rearrangements to complicated structural anomalies due to chromothripsis [20, 22]. Most pediatric GBM have about five large CNAs along with amplifications and focal deletions, although some cases have fewer and some lack detectable CNAs altogether [16, 19]. Chromosome 1q gain occurs at a higher rate than it does in adult GBM and may be enriched in H3F3A G34-mutated tumors [16]. As more pediatric GBM are studied and technological advances continue, smaller CNAs can be resolved and more complex structural rearrangements and gene fusion events can be identified.
A number of CNA including gain and losses of chromosomes as well as chromothripsis have been described in adult GBM [13, 24, 25]. Total CNAs are associated with overall prognosis in a number of diffuse glioma subsets [26, 27]. IDH-wildtype GBM have uniformly high total CNA and poor outcomes [27]. Increased numbers of total CNAs in addition to CDK4 amplification and CDKN2A/B homozygous deletion are found in IDH-mutant low grade gliomas that show rapid progression and outcomes similar to IDH-wildtype GBM [26].
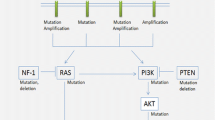
EGFR amplification is an important copy number alteration occurring in approximately 40–50% of adult glioblastomas, mostly primary glioblastomas arising in the fourth decade of life and beyond, and the level of amplification may correlate with patient outcomes [28]. EGFR is positioned on the short arm of chromosome 7 (7p12) and encodes a cell surface receptor tyrosine kinase (EGFR/Erb-1). EGFR is activated following binding of its growth factor ligand to the extracellular domain of EGFR with subsequent phosphorylation of its intracellular tyrosine kinase domain. Activation of EGFR initiates signal transduction of the Ras/MAPK and PI3K/Akt pathways resulting in increased DNA transcription, cellular proliferation, angiogenesis and resistance to apoptosis (Fig. 9.1) [29]. Importantly, EGFR amplification is defined as high level gains of the EGFR gene by validated molecular techniques including fluorescence in-situ hybridization (FISH), next generation sequencing (NGS) and array comparative genomic hybridization (aCGH). Low level gains such as trisomy of chromosome 7 are insufficient for designation as an EGFR amplified tumor. Currently immunohistochemistry for EGFR protein expression is not considered a reproducible test for detection of EGFR amplification.
Summary of key signaling pathways and epigenetic modifiers involved in the pathogenesis of glioblastoma. Multiple genes encoding proteins in the RTK-PI3K-MAPK signaling axis, which are involved in cell growth, proliferation and survival, are affected in GBM. The involved proteins include RTK on the cell membrane and its downstream mediators such as PI3K, RAS and BRAF. The function of negative regulators of this axis such as NF1 and PTEN may be ablated due to gene mutations. BMP signaling is upregulated in a subset of DIPGs. Diverse epigenetic modifiers are important to the pathogenesis of GBM and include direct mutations of histone 3 proteins (K27M, G34R/V) and indirect alterations such as IDH mutations that generate the oncometabolite, 2-hydroxyglutarate, which alters methylation marks on chromatic and in turn, gene expression
Gain of chromosome 7 (+7) and loss of chromosome 10 (−10) are the most common chromosomal aberrations and occur in about 80% of GBMs arising in adults and especially older adults however, these chromosomal changes are far less common in pediatric GBM [13, 24]. As such, GBMs in the receptor tyrosine kinase (RTK) II or classical methylation cluster are significantly more likely to harbor +7+ and −10, while they are distinctly uncommon in proneural GBM and GBM bearing IDH or H3F3A mutations.
Grade II or III IDH-wildtype diffuse astrocytomas with combined loss of whole chromosome 10 and gain of whole chromosome 7 typically exhibit WHO grade IV behavior and overall poor survival. Some studies suggest that partial gains (e.g. +7q or +7p) or partial losses (e.g. -10q or -10p) in diffuse astrocytomas can also predict aggressive clinical behavior, but larger studies are needed to confirm [30,31,32,33,34]. The −10/+7 molecular profile has also been reported, together with BRAF V600E and homozygous loss of CDKN2A/B in pleomorphic xanthroastrocytoma, a potential morphologic mimic of GBM on small biopies, and therefore caution should be used in diagnostically challenging cases [31, 35].
Interchromosomal and intrachromosomal rearrangements are present in most GBMs [36]. In adult GBM certain rearrangements result in the upregulation of growth factor signaling pathways, including EGFR, resulting in the activation of TRK-PI3K-MAPK signaling. The well-known EGFR variant III (EGFRvIII) of adult RTK II GBM occurs following an intrachromosomal deletion that removes exons 2–7 leading to the constitutive activation of EGFR signaling [37]. In adults, intragenic deletions or missense mutations involve exons that encode the extracellular domains of EGFR that are often present on the amplified EGFR allele [13, 38]. EGFR alterations are far less common in pediatric GBM (about 4%) but they seem to characterize an emerging and possible new subtype [9, 39]. This new subtype consists of bithalamic gliomas that characteristically show small in-frame insertions involving exon 20 of EGFR, which encodes the intracellular tyrosine kinase domain, as well as TP53 mutations and unlike unilateral thalamic GBMs, only rare histone H3 mutations [39]. These bithalamic GBM also have a distinct genome methylation profile that shows some overlap with the RTK III methylation subclass of IDH wild-type GBM, a poorly defined group of cerebral pediatric GBMs with EGFR amplification. Nonetheless this suggests that bithalamic tumors may arise due epigenetic mechanisms distinct from other pediatric GBM [39]. Other commonly altered RTKs include a subset of PDGFRA amplified GBMs in adults and children, which show intrachromosomal deletions that generate constitutively active PDGFRA [19, 36, 40,41,42].
Intrachromosomal CNA can be identified in the majority of pediatric HGG and when present in high number, are associated with shorter overall survival, while their absence is associated with a longer overall survival. These data are likely driven by the infant age group, which is known to have a better clinical outcome relative to older children [23, 43,44,45]. GBMs bearing H3F3A K27M have increased numbers of intrachromosomal CNA [23]. The DNA breakpoints of most intrachromosomal structural alterations disrupt the involved gene, leading to a loss of function, which provides a mechanism by which tumor cells can obviate the effects of tumor suppressor genes such as RB1 and NF1, among others [23].
Most chromosomal structural alterations are damaging; however, others may result in a gain of function that promotes gliomagenesis. If the breakpoints come together and align in a single reading frame a novel protein with new function may result and promote gliomagenesis. These so-called gene fusions can be detected using RNA sequencing and are seen in about 50% of childhood GBMs and they are particularly prominent in infant GBMs [22]. A high proportion of fusion genes are potentially targetable therapeutically [8, 22, 23, 45, 46]. It should be noted that many of the discovered gene fusions are not unique to GBM or even limited to HGG [8, 47, 48].
A large international cohort study defined three subgroups of infant GBM and Group 1 was primarily made up of HGG hemispheric tumors bearing fusions of the receptor tyrosine kinase genes ALK, ROS1, NTRK1/2/3 or MET [8]. Rearrangements involving any one of the three NTRK genes may generate fusions with a variety of partner genes in pediatric HGG and are believed to constitutively activate MAPK, PI3K and PKC signaling pathways potentiating cell growth and proliferation and aiding cell survival [22, 47]. MET fusions are less common than NTRK fusions and they also activate MAPK signaling [46].
Gene fusions can also occur in adult gliomas although they are much less common. In frame FGFR3-TACC3 fusions are found in WHO grade II to IV diffuse astrocytomas and these patients may benefit from targeted therapy with FGFR inhibitors [49,50,51]. The FGFR3-TACC3 fusion protein results in a constitutively active tyrosine kinase domain and promotes aneuploidy [52]. These tumors share morphologic similarities including monomorphous ovoid nuclei, nuclear palisading, and thin capillary networks [50]. Identification of these tumors can be challenging without a full molecular panel or directed sequencing; however, immunoreactivity with an FGFR3 antibody shows promise as a robust screening method [50, 53]. The FGFR3-TACC3 fusion gene appears to be mutually exclusive with IDH mutations and EGFR, PDGFR and MET amplification in adult GBM [49, 54].
Not all of the identified gene fusions involve receptor tyrosine kinase genes. BCL6 corepressor protein is encoded by BCOR and mediates transcriptional silencing of genes epigenetically via its interactions with histone deacetylases and the polycomb repressive complex [55, 56]. BCOR alterations are known to occur in other types of brain tumors and in some tumor types arising outside of the central nervous system (CNS) [57, 58]. BCOR fusions are uncommon and not unique to HGG, but are associated with aggressive clinical behavior [59, 60]. While most reported fusions result in a gain-of-function such as enhanced kinase activity, BCOR fusions may drive gliomagenesis by leading to a loss of function in BCOR and its fusion partner [59, 60]. This would be consistent with a tumor suppressor effect as described with BCOR nonsense, frameshift, deletions and splice site mutations encountered in histone H3-mutated gliomas [9, 22, 56].
Focal copy number alterations in pediatric GBM overlap with those occurring in adult GBM and include deletions in PTEN, RB1, CDKN2C, NF1 and TP53 as well as amplifications of CDK4, CDK6, PDGFRA, MET and MDM2 among others [61]. PDGFRA and either MYC or MYCN amplifications are more frequent in pediatric than adult GBMs [10, 19, 62]. PDGFRA amplifications are prominent in DIPG, particularly in DIPG with H3 mutations and are enriched in radiation-induced gliomas [10, 17, 18, 63]. The H3 K27-mutant midline gliomas show a high frequency of PDGFRA amplifications and are enriched for proneural gene expression signature [10]. In DIPG PDGFRA amplifications are associated with a dismal prognosis and resistance to therapy [17, 62]. MYCN amplifications are seen in a subgroup of DIPGs that have hypermethylated genomes [64].
DNA Methylation Profiling
Hypermethylation of gene promoter regions is an epigenetic mechanism of gene silencing that impacts the expression of many types of genes including tumor suppressors and others that are important in cell cycle regulation and additional key functions of neoplastic cells. The most widely used application of DNA methylation testing concerns assessing the methylation status of the MGMT promoter, which regulates the expression of O6-methylguanine methyltransferase. MGMT promoter methylation silences gene expression and limits the ability of the cell to repair DNA damage induced by alkylating agent chemotherapy, particularly temozolomide. MGMT promoter methylation is a useful biomarker in adult GBM patients and is responsible in part to the improved outcome in IDH-mutant tumors, but its predictive capacity in pediatric patients is unclear [65,66,67].
DNA microarray technology facilitated the evaluation of the pediatric GBM methylome in an unbiased, genome-wide fashion and led to the delineation of tumor subgroups that differed from those identified in adult GBM patients. Early studies of the GBM methylome in adults discerned a group in the proneural gene expression class with hypermethylation at many loci and coexisting IDH1 alterations, i.e. the so-called G-CIMP-positive tumors [68]. A more comprehensive genome-wide DNA methylation analysis of a large cohort of pediatric and adult GBMs demonstrated that, when correlated with gene expression profiles, mutational status, and DNA copy number alterations, GBMs cluster into six distinct groups known as IDH, K27, G34, receptor tyrosine kinase (RTK) I and II, and mesenchymal that also align with clinicopathologic parameters including patient age and tumor location (Table 9.1) [10]. These GBM methylation groups correlated relatively well with the groups identified by expression profiling, underscoring the importance of epigenetic mechanisms in the pathogenesis of GBM [14].
A subset of GBM in the RTK I cluster arose in some pediatric patients and included PDGFRA amplified, G-CIMP-negative, proneural GBMs [10, 14]. Tumors in the IDH cluster were G-CIMP-positive, proneural gliomas that primarily arose in young adults; however, some adolescents were also affected (median age 40 years; range 13–71 years). GBMs clustering in the mesenchymal methylation group arose in patients with a wide range of ages that included elderly patients and showed a mesenchymal pattern of gene expression as well as PTEN and NF1 mutations. The RTK II cluster showed classical gene expression profile, chromosome 7 loss and chromosome 10 gain as well as EGFR alterations and had a median age 58 years with no pediatric GBM patients.
Two of the methylation clusters, K27 and G34, preferentially occurred in the pediatric population and were tightly correlated with the presence of H3F3A mutations at positions K27 and G34 [10]. H3F3A encodes the replication-independent histone, H3.3, which primarily binds transcriptionally activated genes and telomeres. H3.3 is often methylated at or near the mutated residues, which affects DNA methylation likely by altering DNA accessibility, so H3F3A mutations can account for the global changes in methylation noted in these GBM subgroups. The two main recurrent H3F3A mutations are only seven amino acid residues apart in the H3.3 protein, yet they result in significant differences in terms of methylation profile, tumor location in the central nervous system and clinical setting. Tumors in the K27 cluster predominately arise in the midline central nervous system structures including the thalami and pons of children with a median age of about 10 years, while G34 tumors tend to occur in the cerebral hemispheres of adolescents with a median age of 18 years [10, 20, 69]. IDH1 and H3F3A mutations are mutually exclusive between individual GBMs, yet it seems that each favors tumor development by impairing the normal differentiation program of progenitor cells using a pathogenetic mechanism with some overlapping features [70]. Mutant IDH1 protein produces 2-hydroxyglutarate, an oncometabolite, which is responsible for the increased global methylation pattern of CIMP-positive, proneural GBM. Increased intracellular 2-hydroxyglutarate concentrations increases H3 K27 methylation, inhibiting progenitor cell differentiation by increasing the expression of stem cell markers while decreasing the expression of differentiation-related markers. In a related fashion, H3.3 bearing mutations at G34 favors tumor formation in part, by reducing the expression of the important developmental transcription factor, OLIG2, due to hypermethylation of its gene locus [10].
The K27 and G34 mutations are associated with different methylation patterns [10]. Despite the increased methylation of the OLIG2 locus, tumors in the G34 subgroup show global hypomethylation across the genome, which is prominent in nonpromoter regions, including subtelomeric zones near the ends of chromosomes, suggesting that the loss of methylation in subtelomeric areas may have a role in the alternative lengthening of telomeres observed in tumors with H3F3A G34 mutations [10, 20]. The marked differences in methylation signatures correlate with tumor location and clinical course, suggesting that the mechanisms by which these epigenetic alterations lead to tumor formation are critical to understand and will likely be a focus of intense study for years to come.
Approximately half of pediatric GBMs have recurrent somatic mutations in histone H3 genes or in IDH1/2. The remaining GBMs are a diverse group; however, genome-wide molecular profiling studies are starting to delineate new subgroups, identify potential previously unrecognized therapeutic targets, and also reveal prognostic information in these tumors [67, 71]. A cohort of H3-/IDH-wild type pediatric GBM was comprehensively studied using an integrated approach that included genome-wide DNA methylation, targeted mutation detection and CNA and identified three molecular subtypes with different genomic and epigenetic signatures and clinical behavior. These were designated as MYCN, enriched for MYCN amplification, RTK 1, enriched for PDGFRA amplification and RTK 2, enriched for EGFR amplification [67]. The MYCN subtype of H3-/IDH- wild type tumors is the most aggressive with a survival period that is similar to that of H3 K27M mutant tumors [10, 67, 69]. These tumors tend to arise outside of the brainstem, have high level MYCN amplification, which often co-exist with amplification of Inhibitor of DNA Binding 2 (ID2) and recurrent TP53 mutations [67, 72]. The RTK 1 subtype of H3-/IDH- wild type tumors frequently bear PDGRFA amplification and a paucity of other typical GBM cytogenetic alterations [67]. The RTK I tumors have an intermediate prognosis. H3-/IDH- wild type tumors of the RTK 2 subtype differed from adult GBM despite having EGFR amplification in common. Tumors in this subtype showed an overall 5 year survival at close to 50% and the methylation profile differed from all adult GBM variants [67].
Korshunov et al. studied a large cohort of pediatric GBMs using genome wide DNA methylation and candidate gene screening, which revealed that a subset of histologically high-grade tumors showed a methylation profile similar to low-grade glioma or pleomorphic xanthoastrocytoma [71]. These tumors had a favorable prognosis and often had BRAF V600E mutations with chromosome 9p21 loss leading to homozygous deletion of the CDKN2A tumor suppressor locus [71]. BRAF V600E is common in PXA, but its frequency is unknown in genuine GBM and the histologic features of PXA, especially anaplastic PXA, overlap with GBM, so molecular testing will likely be required to distinguish these entities with confidence [73, 74].
Single Nucleotide Variants and Deregulated Cancer Cell Signaling Pathways
The 2016 update of the WHO Classification of CNS tumors subclassifies diffuse astrocytomas including GBM by IDH mutation status. This distinction principally pertains to adults and a subset of older adolescents as pediatric gliomas are very rarely IDH-driven [61, 75, 76]. As with essentially all GBM, IDH-wildtype and IDH-mutant tumors are not distinguishable on morphology despite their distinct biologic differences. IDH-wildtype GBM are significantly more common with poorer overall survival [77,78,79]. IDH-mutant glioblastomas account for 10% or fewer of all GBM [1, 80]. IDH1 and IDH2-mutant GBM are associated with younger age at presentation, DNA hypermethylation phenotype, and overall better outcome compared to IDH-wildtype GBM [78, 80]. Despite a better overall prognosis, IDH-mutant GBM are designated WHO grade IV as most patients develop tumor progression and die of their disease.
Patient outcomes are highly variable among IDH-mutant GBM suggesting genomic heterogeneity among IDH-mutant tumors [81]. Furthermore, histologic grading of IDH-mutant tumors and assessment of mitotic activity are not good predictors of overall patient outcomes [80, 82]. Analysis of large cohorts of IDH-mutant GBM has revealed a strong association between patient outcome and distinct copy number alterations, particularly homozygous deletion of CDKN2A/B [30, 80, 84,85,85]. As a result, the Consortium to Inform Molecular and Practical Approaches to CNS Tumor Taxonomy (cIMPACT-NOW) issued recommendations to reflect the current understanding of IDH-mutant tumors. The recommended integrated diagnosis for IDH-mutant diffuse astrocytomas with either necrosis, microvascular proliferation, or CDKN2A/B homozygous deletion is “astrocytoma, IDH-mutant, grade 4” [86, 87]. CDKN2A/B homozygous deletions are also found in approximately 60% of IDH-wildtype GBM but do not carry the same prognostic significance.
Sequencing technology has evolved and an ever-increasing number of recurrent somatic SNV have been identified, sometimes at low frequency, and occasionally in genes that were not expected to be involved in gliomagenesis or even in neoplasia. Adult and pediatric GBM share common SNV in a number of genes, although some are unique to one patient population or the other [61]. Nonetheless, the gene mutations encountered in pediatric GBM perturb many of the same cancer cell pathways and functions that are altered in adult GBM, which include RTK-RAS-PI3K pathway, p53 function, cell cycle control and epigenetic regulation [10, 20, 88].
The genetic hallmarks of adult IDH-wildtype and IDH-mutant tumors are described in previous chapters and will only be briefly mentioned here. Common somatic mutations in adult IDH-wildtype GBM include TERT, PTEN, EGFR, TP53, NF1, PIC3CA, and RB1, while ATRX mutations are very rare [1, 89, 90]. TP53 and ATRX mutations are found in 70–80% of IDH-mutant GBM while EGFR and PTEN mutations are exceedingly rare [89, 90]. H3F3A K27M mutations are uncommon in adults but do occur in older patients over, and immunohistochemistry or other testing should be considered in diffusely infiltrating midline gliomas presenting at any age (Fig. 9.2) [91, 92].
Mutations in the promoter region of telomerase reverse transcriptase (TERT) gene are frequent in IDH-wildtype GBM and less frequent among IDH-mutant GBM but do occur [89, 90]. Telomerase, a reverse transcriptase that maintains telomere length, is inactive in mature somatic cells in adulthood but activated in glioma cells via defined TERT promoter mutations. Mutations in the TERT promoter result in upregulation of telomerase complex activity and elongation of telomere length leading to uncontrolled cell proliferation. The most common mutations in the TERT promoter, C228T and C250T are located upstream of the TERT start site. Recent studies have elucidated the prognostic significance of TERT promoter mutations by demonstrating patients with WHO grade II and III IDH-wildtype gliomas often follow a similar clinical course as IDH-wildtype WHO grade IV GBM [93]. As such, the cIMPACT-NOW working group has recommended an integrated diagnosis of “diffuse glioma, WHO grade II or III, with molecular features of glioblastoma”. This recommended terminology refers to diffuse astrocytomas of any grade with TERT promoter mutations, EGFR amplification, and/ or chromosome +7/−10. Long-term follow-up studies have subsequently validated this recommendation and conclude diffuse astrocytomas with TERT promoter mutations are as clinically aggressive as WHO grade IV tumors [93]. It is important to note that TERT promoter mutations are not limited to IDH-wildtype or IDH-mutant GBM. TERT promoter mutations are very common in adult oligodendrogliomas and occasionally found in pleomorphic xanthoastrocytoma, ependymoma, and low-grade glioneuronal tumors [94, 95]. TERT promoter mutations frequently occur together with EGFR amplification or chromosome −10/+7. TERT promoter mutations are rare in the pediatric population [96].
As mentioned, the RTK EGFR, commonly mutated to potentiate cell growth and proliferation signaling in adult GBM is rare in childhood GBM. PDGFRA is the most commonly mutated RTK in children and is seen in about 30% of all childhood HGGs [16, 42, 63]. In addition, pediatric GBMs may activate the RTK-RAS-PI3K pathway via MET amplification or by activating mutations in other RTKs, such as FGFR1 [22, 97]. Inhibiting RTK is an attractive treatment strategy; however, the degree of intratumoral heterogeneity within a given pediatric GBM can be considerable, suggesting that a given tumor would have a population of drug resistant cells prior to initiating therapy, since not all of the cells in the tumor may contain the therapeutic target [18].
GBM cells can also activate RTK-RAS-PI3K signaling downstream of RTK. BRAF binds RAS, transducing the cell growth signal of the RTK-RAS-PI3K pathway. Amino acid position 600 of BRAF is a hotspot where the amino acid, valine is substituted for a glutamic acid, due to a point mutation in the BRAF gene. BRAF V600E mutations are present in a subset of pediatric and adult HGG [20, 73, 98]. PI3K is activated in pediatric GBM by either gain of function mutations in PIK3CA, which encodes the catalytic subunit of PI3K or by deregulating mutations in PIK3R1, which encodes the regulatory subunit of PI3K [97]. In adults, PI3K activity may be upregulated following biallelic inactivation of the tumor suppressor, PTEN but this is uncommon in pediatric HGG [24, 88].
Loss of cell cycle control is a key event in pediatric GBM pathogenesis. Approximately half of all pediatric HGGs bear mutations in TP53, which encodes p53, a critical tumor suppressor that regulates cell division and survival (apoptosis) as well as senescence [20, 22]. Proteins of the cyclin-CDK complex phosphorylate RB at the G1 checkpoint, which is a key cell cycle regulatory step that commits a cell to synthesize DNA and divide. Amplifications of CDK4, CDK6 or the cyclin genes, CCND1, CCND2 or CCDN3 have been identified in pediatric HGG and have potential to allow a tumor cell to divide by overcoming the checkpoint [18]. Homozygous deletion of CDKN2A, which encodes the tumor suppressor proteins, p14 and p16, occurs in about a quarter of non-brainstem HGGs [61]. CDK4 and CDK6 normally induce the cell to divide by promoting the completion of the cell cycle. The p16 protein normally binds CDK4 and CDK6, blocking their ability to stimulate cell proliferation. The p14 protein normally binds p53, protecting it from degradation. Therefore, CDKN2A deletions can profoundly deregulate the cell cycle; losing the effect of p14 on p53 effectively removes a brake on the cell cycle, while losing the effect of p16 on CDK4 and CDK6 is akin to depressing a cell cycle accelerator. Curiously, unlike in adults, RB1 biallelic loss of function mutations are uncommon childhood GBM, although about 30% of tumors show chromosome 13q loss, which contains the RB1 locus [20, 22].
Perhaps the most unexpected finding to come from genome-wide sequencing studies of large cohorts of pediatric GBM and DIPG was the prominence of SNV in H3 histone family 3A (H3F3A) and histone cluster 1, H3b (HIST1H3B) genes, encoding H3.3 and H3.1, respectively [20, 22, 99]. H3F3A and HIST1H3B mutations are extremely rare adult GBM and likely contribute to the significant differences in HGG pathogenesis in adults and children and the previously discussed differences in the tumor methylome between these patients.
Histone H3 mutations in GBM occur at two amino acid positions resulting in the lysine at position 27 being replaced by methionine (K27M) or the glycine at position 34 being replaced by either arginine (G34R) or valine (G34V) [20, 22]. Ten identical genes encode histone 3.1, but HIST1H3B is most commonly mutated in GBM [20, 22]. Nucleosomes are disrupted during the cell cycle and the cell synthesizes H3.3 in all phases of the cell cycle to replace lost histones [100, 101]. The role of H3.1 and H3.2 is to package newly synthesized DNA and these histones are synthesized during S phase of the cell cycle [100, 101].
K27M mutations in H3F3A or HIST1H3B are both encountered in DIPG and other midline gliomas that arise in the spinal cord of thalamus [10, 20, 22, 69, 99]. H3.3 K27M bearing tumors have an age of onset at about 6 years and a median survival period of about 12 months [22, 64, 102,103,104]. H3.3 K27M can be identified in about two-thirds of DIPGs and in two-thirds of midline line HGGs arising outside of the brainstem [9]. Curiously, co-segregating alterations differ by tumor location with PDGFRA alterations tending to occur in the pons and FGFR alterations occurring in the thalamus [9, 102]. A meta-analysis showed H3.3 K27M tumors located in the pons are associated with CCND2 amplification, while non-brainstem midline tumors have amplification of CDK4 [9]. A complex rearrangement in H3.3 K27M tumors was found resulting in amplification at 17p11.2 increasing TOP3A copy number and expression and the loss of the distal aspect of 17p, which contains the TP53 locus [9]. TOP3A is a topoisomerase with roles in homologous recombination and alternative lengthening of telomeres and in H3.3 K27M DIPG TOP3A alterations are mutually exclusive with ATRX mutations, providing another means gliomas can use to activate alternative lengthening of telomeres [105].
HIST1H3B K27M mutations are associated with a younger age of onset and a slightly longer survival period [22, 64, 102, 103]. H3.1 K27M tumors are restricted to the pons. DIPGs harboring a HIST1H3B mutation also often have coexisting somatic activating mutations in ACVR1 suggesting that BMP signaling is important to their pathogenesis [22, 64, 102, 103]. ACVR1 mutations are exclusive to DIPG, predominate in females and enriched in tumors bearing histone 3.1 gene mutations [22]. ACVR1 mutations phosphorylate and activate SMAD proteins, increasing the expression of downstream effectors including the ID protein family members ID1 and ID2 [64, 102, 103]. Histone H3 and ACVR1 mutations seem to work cooperatively to enhance the downstream effect on ID proteins [64]. H3.1 K27M tumors are enriched for mutations that activate the PI3K pathway, including PIK3CA and PIK3R1 and are associated with BCOR mutations [9].
G34 mutations do not occur in HIST1H3B and GBMs bearing H3F3A G34 mutations have distinct clinical profiles. In contrast to tumors with K27M, G34 tumors arise in the peripheral aspect of the cerebral hemispheres and not in midline structures. In addition, G34 tumors tend to occur in older children with an onset at about 13 years and patients survive longer (about 24 months) [22, 64, 102,103,104]. The differences in the timing of diagnosis and the survival interval likely stem from the differences in tumor location with brainstem and midline tumors presenting earlier than hemispheric tumors and affording little opportunity for maximal safe surgical resection, which may be achieved in hemispheric tumors. G34R/V and K27M mutations predominate in children but can be identified in adult GBMs, with the latter seen in the majority of young adults with thalamic tumors [10, 106]. H3.3 G34R/V mutations co-segregate with ATRX and TP53 mutations and are the only pediatric GBM subgroup to commonly show MGMT promoter methylation [71]. A meta-analysis showed frequent loss of chromosomal arms at 3q, 4q, 5q and 18q, with loss of 4q31.3 leading to loss of FBXW7, a candidate tumor suppressor [9, 107]. FBXW7 is part of the SCF-like ubiquitin ligase complex that targets MYC and MYCN for proteasomal destruction, so the loss of FBXW7 in H3.3 G34R/V tumors would enhance the life span of MYC proteins [108]. MYCN expression is also increased in H3.3 G34R/V tumors due to the effects of H3 K36me3, making MYNC a critically important driver of this subgroup [109].
K27M and G34R/V have structural consequences that impact histone function in gene transcription. These amino acids occur in the amino-terminal tail of the histone H3 protein and the post-translation modification of the tail region has regulatory roles on gene transcription and chromatin compaction and structure that are important in cell differentiation [101, 110, 111]. The post-translational modifications of the tail region are diverse and include acetylation, methylation and ubiquitylation of lysine residues, phosphorylation of serine of threonine residues and methylation of arginine residues. A wide scope of enzymes catalyze these changes and include writers which add and erasers which remove these moieties, while readers are effector proteins that bind to chromatin according to the pattern of moieties on the histone tail, thereby regulating the location of the transcriptional complexes to chromatin [101, 112]. These readers, writers and erasers are increasingly recognized as critical to the pathogenesis of many different types of cancer arising in diverse tissues [113, 114].
The role histone mutations play in gliomagenesis is not entirely understood. The lysine that is abrogated due to the K27M mutation is normally either methylated or acetylated. When K27 is trimethylated (K27me3) it interacts with the polycomb repressive complex 2 (PCR2) to selectively repress gene transcription. The K27M mutation effectively removes the ability of this key methylation site on the histone tail leading to general hypomethylation followed by an upregulation in gene expression with PRC2 target gene derepression, which together potentiate gliomagenesis [21, 101, 115]. The impact of the K27M mutation is likely enormous to tumor cell biology. Immunostaining shows that the H3 K27M mutant protein is often detectable in nearly 100% of tumor cells, which suggests that tumor cells bearing H3 K27M are favorably selected within the overall tumor cell population [69, 99]. Furthermore, H3 K27M protein expression makes up a small fraction of the total H3 protein in the tumor cell, yet these tumors essentially show a total loss of K27me3, indicating a trans-dominant-negative effect across all isoforms of wild type H3 occurs in the tumor cell [97, 116]. K27 seems to hold a critical place in pediatric gliomagenesis because mutations in the writers or erasers of each post-translationally modified lysine residue of the histone tail has been identified except for K27, which is only impacted by direct mutation [20, 22].
H3 bearing G34R/V mutations also impair epigenetic regulation of gene expression but the mechanism is different from that of K27M. The G34 residue does not undergo a direct post-translational modification it is, however, near the lysine residue at position 36 (K36), which is directly modified. In support of the importance of K36 methylation, SETD2, the methyltransferase that writes the methyl marks on K36, is mutated in about 10% of pediatric GBMs [22, 102, 104]. K36 trimethylation activates gene transcription, and also impacts alternative splicing and DNA repair [97]. G34R is associated with reduced K36 trimethylation on one mutant allele so the dominant effect over total cell histone H3 as seen in the presence of K27M, does not occur [97, 115]. Experiments show that histone 3 bearing G34V binds genes associated with stem cell maintenance and cortical development with the oncogene, MYCN, showing significant G34 binding and increased expression [97, 109]. The presence of MYNC amplification and G34R/V are mutually exclusive [109]. The differential impact of G34R and G34V substitution on gliomagenesis are not fully characterized, although the arginine in G34R has the potential to undergo post-translational modifications, suggesting that there may be differences [97, 117].
Mutations in other chromatin regulatory genes besides histone genes have been identified in pediatric GBM including members of the MLL (writers), KDM (erasers) and CHD (chromatin remodelers) gene families [20, 22]. ATRX and DAXX encode histone chaperones that load histone H3.3 to regions of heterochromatin located at telomeres and co-segregate with H3 G34R/and they are mutated in up to 20% of pediatric HGG [20, 22, 102]. Tumors with H3F3A and ATRX or DAXX mutations show telomerase independent alternative lengthening of telomeres [20]. TERT promoter mutations are rare in pediatric GBMs but are present in most adult GBMs and in adults they are mutually exclusive to ATRX mutations [22, 118].
Future Directions and Conclusion
The amount of information regarding the origin and drivers of GBM has vastly expanded in the past several years. It is now well understood that molecular features better predict tumor behavior and patient outcomes compared to traditional histologic classification, especially on small or non-representative tumor biopsies. Many of the recently recognized molecular subgroups will undoubtedly find their way into future revisions of the WHO classification. Despite our increasing understanding of distinct glioma molecular signatures, it is important to emphasize that a combined approach of morphologic evaluation and molecular characterization improves diagnostic classification for glioblastoma [119].
In this timeframe, we have also learned that pediatric and adult GBMs are significantly different biologically. Adult and pediatric GBM are truly a group of diseases that arise due to unique pathogenetic mechanisms, but how can this new information be applied in patient care to improve outcomes for this dreaded, essentially fatal disease? First, we cannot expect that data from clinical trials of adults GBM patients will properly inform trials in pediatric GBM patients. This approach was used in the past and as a result, trials were largely ineffective with only slight improvements in clinical outcome. Clearly, these biological differences in adult and pediatric disease cannot be overlooked. Second, we now realize that each pediatric GBM subgroup and some adult subgroups appear to have distinct cellular origins and oncogenic drivers, which may be therapeutically targetable [120, 121]. It is hoped that tumors targeted with such specificity would lead to more effective therapies and perhaps fewer treatment-related complications. We now realize that past clinical trials, which enrolled an unselected patient population and failed to take GBM subgroups into account, were underpowered to detect subgroup-specific efficacy [120]. Taking tumor subgroup into account when tailoring therapeutic approach is critical as clinical behavior is more accurately predicted by tumor biology, which is reflected in genetic and epigenetic alterations rather than tumor grade or various clinical parameters [120].
BRAF V600E mutations, while not unique to GBM, are known to activate MAPK signaling and BRAF inhibiting drugs such as the MEK-inhibitors and BRAF V600E-specific inhibitors have shown success [122]. RTK gene fusions are now recognized as key drivers of tumor biology in a significant subset of GBMs, particularly those arising in infants and these alterations are potential therapeutic targets [8, 123,124,125]. DIPG and midline GBM with histone 3 mutations may be treatable with drugs that are epigenetic modifiers with activity toward histone demethylases and deacetylases [126, 127]. IDH mutations are rare in pediatric GBM and likely represent the tail end of the peak incidence in adulthood. As such patients with these tumors may be better off enrolling on separate strata of adult clinical trials [120].
It is hoped that as our knowledge of the molecular alterations driving tumor biology grows, new molecular targets will continue to emerge and innovative agents can be designed to allow even more refined subgroup specific therapies and trials with the goal of improving patient outcome.
References
Louis DN, et al. WHO classification of tumours of the central nervous system. Revised 4th ed. Lyon: International Agency for Research on Cancer; 2016.
Fujisawa H, et al. Acquisition of the glioblastoma phenotype during astrocytoma progression is associated with loss of heterozygosity on 10q25-qter. Am J Pathol. 1999;155(2):387–94.
Georgescu M-M, Olar A. Genetic and histologic spatiotemporal evolution of recurrent, multifocal, multicentric and metastatic glioblastoma. Acta Neuropathol Commun. 2020;8(1):10.
Consortium TG. Glioma through the looking GLASS: molecular evolution of diffuse gliomas and the Glioma Longitudinal Analysis Consortium. Neuro Oncol. 2018;20(7):873–84.
Riehmer V, et al. Genomic profiling reveals distinctive molecular relapse patterns in IDH1/2 wild-type glioblastoma. Genes Chromosom Cancer. 2014;53(7):589–605.
Ostrom QT, et al. CBTRUS Statistical Report: Primary brain and other central nervous system tumors diagnosed in the United States in 2012–2016. Neuro Oncol. 2019;21(Supplement_5):v1–v100.
Stupp R, et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10(5):459–66.
Guerreiro Stucklin AS, et al. Alterations in ALK/ROS1/NTRK/MET drive a group of infantile hemispheric gliomas. Nat Commun. 2019;10(1):4343.
Mackay A, et al. Integrated molecular meta-analysis of 1,000 pediatric high-grade and diffuse intrinsic pontine glioma. Cancer Cell. 2017;32(4):520–537 e5.
Sturm D, et al. Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell. 2012;22(4):425–37.
Jones DTW, et al. Molecular characteristics and therapeutic vulnerabilities across paediatric solid tumours. Nat Rev Cancer. 2019;19(8):420–38.
Phillips HS, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9(3):157–73.
Brennan CW, et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155(2):462–77.
Verhaak RG, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110.
Faury D, et al. Molecular profiling identifies prognostic subgroups of pediatric glioblastoma and shows increased YB-1 expression in tumors. J Clin Oncol. 2007;25(10):1196–208.
Paugh BS, et al. Integrated molecular genetic profiling of pediatric high-grade gliomas reveals key differences with the adult disease. J Clin Oncol. 2010;28(18):3061–8.
Puget S, et al. Mesenchymal transition and PDGFRA amplification/mutation are key distinct oncogenic events in pediatric diffuse intrinsic pontine gliomas. PLoS One. 2012;7(2):e30313.
Paugh BS, et al. Genome-wide analyses identify recurrent amplifications of receptor tyrosine kinases and cell-cycle regulatory genes in diffuse intrinsic pontine glioma. J Clin Oncol. 2011;29(30):3999–4006.
Bax DA, et al. A distinct spectrum of copy number aberrations in pediatric high-grade gliomas. Clin Cancer Res. 2010;16(13):3368–77.
Schwartzentruber J, et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012;482(7384):226–31.
Bender S, et al. Reduced H3K27me3 and DNA hypomethylation are major drivers of gene expression in K27M mutant pediatric high-grade gliomas. Cancer Cell. 2013;24(5):660–72.
Wu G, et al. The genomic landscape of diffuse intrinsic pontine glioma and pediatric non-brainstem high-grade glioma. Nat Genet. 2014;46(5):444–50.
Carvalho D, et al. The prognostic role of intragenic copy number breakpoints and identification of novel fusion genes in paediatric high grade glioma. Acta Neuropathol Commun. 2014;2:23.
Cancer Genome Atlas Research, N. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061–8.
Malhotra A, et al. Breakpoint profiling of 64 cancer genomes reveals numerous complex rearrangements spawned by homology-independent mechanisms. Genome Res. 2013;23(5):762–76.
Richardson TE, et al. Genetic and epigenetic features of rapidly progressing IDH-mutant astrocytomas. J Neuropathol Exp Neurol. 2018;77(7):542–8.
Mirchia K, et al. Total copy number variation as a prognostic factor in adult astrocytoma subtypes. Acta Neuropathol Commun. 2019;7(1):92.
Hobbs J, et al. Paradoxical relationship between the degree of EGFR amplification and outcome in glioblastomas. Am J Surg Pathol. 2012;36(8):1186–93.
An Z, et al. Epidermal growth factor receptor and EGFRvIII in glioblastoma: signaling pathways and targeted therapies. Oncogene. 2018;37(12):1561–75.
Aoki K, et al. Prognostic relevance of genetic alterations in diffuse lower-grade gliomas. Neuro Oncol. 2018;20:66–77.
Stichel D, et al. Distribution of EGFR amplification, combined chromosome 7 gain and chromosome 10 loss, and TERT promoter mutation in brain tumors and their potential for the reclassification of IDHwt astrocytoma to glioblastoma. Acta Neuropathol. 2018;136(5):793–803.
Tabouret E, et al. Prognostic impact of the 2016 WHO classification of diffuse gliomas in the French POLA cohort. Acta Neuropathol. 2016;132(4):625–34.
Weller M, et al. Molecular classification of diffuse cerebral WHO grade II/III gliomas using genome- and transcriptome-wide profiling improves stratification of prognostically distinct patient groups. Acta Neuropathol. 2015;129(5):679–93.
Wijnenga MMJ, et al. Molecular and clinical heterogeneity of adult diffuse low-grade IDH wild-type gliomas: assessment of TERT promoter mutation and chromosome 7 and 10 copy number status allows superior prognostic stratification. Acta Neuropathol. 2017;134(6):957–9.
Vaubel RA, et al. Recurrent copy number alterations in low-grade and anaplastic pleomorphic xanthoastrocytoma with and without BRAF V600E mutation. Brain Pathol. 2018;28(2):172–82.
Zheng S, et al. A survey of intragenic breakpoints in glioblastoma identifies a distinct subset associated with poor survival. Genes Dev. 2013;27(13):1462–72.
Biernat W, et al. Predominant expression of mutant EGFR (EGFRvIII) is rare in primary glioblastomas. Brain Pathol. 2004;14(2):131–6.
Wong AJ, et al. Structural alterations of the epidermal growth factor receptor gene in human gliomas. Proc Natl Acad Sci U S A. 1992;89(7):2965–9.
Mondal G, et al. Pediatric bithalamic gliomas have a distinct epigenetic signature and frequent EGFR exon 20 insertions resulting in potential sensitivity to targeted kinase inhibition. Acta Neuropathol. 2020;139(6):1071–88.
Cho J, et al. Glioblastoma-derived epidermal growth factor receptor carboxyl-terminal deletion mutants are transforming and are sensitive to EGFR-directed therapies. Cancer Res. 2011;71(24):7587–96.
Ozawa T, et al. PDGFRA gene rearrangements are frequent genetic events in PDGFRA-amplified glioblastomas. Genes Dev. 2010;24(19):2205–18.
Paugh BS, et al. Novel oncogenic PDGFRA mutations in pediatric high-grade gliomas. Cancer Res. 2013;73(20):6219–29.
Duffner PK, et al. Treatment of infants with malignant gliomas: the Pediatric Oncology Group experience. J Neuro-Oncol. 1996;28(2–3):245–56.
Wu W, et al. Joint NCCTG and NABTC prognostic factors analysis for high-grade recurrent glioma. Neuro Oncol. 2010;12(2):164–72.
Clarke M, et al. Infant high-grade gliomas comprise multiple subgroups characterized by novel targetable gene fusions and favorable outcomes. Cancer Discov. 2020;10(7):942–63.
International Cancer Genome Consortium PedBrain Tumor, P. Recurrent MET fusion genes represent a drug target in pediatric glioblastoma. Nat Med. 2016;22(11):1314–20.
Torre M, et al. Molecular and clinicopathologic features of gliomas harboring NTRK fusions. Acta Neuropathol Commun. 2020;8(1):107.
Jones DT, et al. Recurrent somatic alterations of FGFR1 and NTRK2 in pilocytic astrocytoma. Nat Genet. 2013;45(8):927–32.
Di Stefano AL, et al. Detection, characterization, and inhibition of FGFR-TACC fusions in IDH wild-type glioma. Clin Cancer Res. 2015;21(14):3307–17.
Bielle F, et al. Diffuse gliomas with FGFR3-TACC3 fusion have characteristic histopathological and molecular features. Brain Pathol. 2018;28(5):674–83.
Lasorella A, Sanson M, Iavarone A. FGFR-TACC gene fusions in human glioma. Neuro Oncol. 2017;19(4):475–83.
Costa R, et al. FGFR3-TACC3 fusion in solid tumors: mini review. Oncotarget. 2016;7(34):55924–38.
Granberg KJ, et al. Strong FGFR3 staining is a marker for FGFR3 fusions in diffuse gliomas. Neuro Oncol. 2017;19(9):1206–16.
Parker BC, et al. The tumorigenic FGFR3-TACC3 gene fusion escapes miR-99a regulation in glioblastoma. J Clin Invest. 2013;123(2):855–65.
Huynh KD, et al. BCoR, a novel corepressor involved in BCL-6 repression. Genes Dev. 2000;14(14):1810–23.
Astolfi A, et al. BCOR involvement in cancer. Epigenomics. 2019;11(7):835–55.
Sturm D, et al. New brain tumor entities emerge from molecular classification of CNS-PNETs. Cell. 2016;164(5):1060–72.
Roy A, et al. Recurrent internal tandem duplications of BCOR in clear cell sarcoma of the kidney. Nat Commun. 2015;6:8891.
Torre M, et al. Recurrent EP300-BCOR fusions in pediatric gliomas with distinct clinicopathologic features. J Neuropathol Exp Neurol. 2019;78(4):305–14.
Pisapia DJ, et al. Fusions involving BCOR and CREBBP are rare events in infiltrating glioma. Acta Neuropathol Commun. 2020;8(1):80.
Sturm D, et al. Paediatric and adult glioblastoma: multiform (epi)genomic culprits emerge. Nat Rev Cancer. 2014;14(2):92–107.
Phillips JJ, et al. PDGFRA amplification is common in pediatric and adult high-grade astrocytomas and identifies a poor prognostic group in IDH1 mutant glioblastoma. Brain Pathol. 2013;23(5):565–73.
Zarghooni M, et al. Whole-genome profiling of pediatric diffuse intrinsic pontine gliomas highlights platelet-derived growth factor receptor alpha and poly (ADP-ribose) polymerase as potential therapeutic targets. J Clin Oncol. 2010;28(8):1337–44.
Buczkowicz P, et al. Genomic analysis of diffuse intrinsic pontine gliomas identifies three molecular subgroups and recurrent activating ACVR1 mutations. Nat Genet. 2014;46(5):451–6.
Donson AM, et al. MGMT promoter methylation correlates with survival benefit and sensitivity to temozolomide in pediatric glioblastoma. Pediatr Blood Cancer. 2007;48(4):403–7.
Lee JY, et al. MGMT promoter gene methylation in pediatric glioblastoma: analysis using MS-MLPA. Childs Nerv Syst. 2011;27(11):1877–83.
Korshunov A, et al. H3-/IDH-wild type pediatric glioblastoma is comprised of molecularly and prognostically distinct subtypes with associated oncogenic drivers. Acta Neuropathol. 2017;134(3):507–16.
Noushmehr H, et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17(5):510–22.
Khuong-Quang DA, et al. K27M mutation in histone H3.3 defines clinically and biologically distinct subgroups of pediatric diffuse intrinsic pontine gliomas. Acta Neuropathol. 2012;124(3):439–47.
Lu C, et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 2012;483(7390):474–8.
Korshunov A, et al. Integrated analysis of pediatric glioblastoma reveals a subset of biologically favorable tumors with associated molecular prognostic markers. Acta Neuropathol. 2015;129(5):669–78.
Tauziède-Espariat A, et al. The pediatric supratentorial MYCN-amplified high-grade gliomas methylation class presents the same radiological, histopathological and molecular features as their pontine counterparts. Acta Neuropathol Commun. 2020;8(1):104.
Schindler G, et al. Analysis of BRAF V600E mutation in 1,320 nervous system tumors reveals high mutation frequencies in pleomorphic xanthoastrocytoma, ganglioglioma and extra-cerebellar pilocytic astrocytoma. Acta Neuropathol. 2011;121(3):397–405.
Ida CM, et al. Pleomorphic xanthoastrocytoma: natural history and long-term follow-up. Brain Pathol. 2015;25(5):575–86.
Louis DN, et al. The 2016 World Health Organization Classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131:803.
Pollack IF, et al. IDH1 mutations are common in malignant gliomas arising in adolescents: a report from the Children’s Oncology Group. Childs Nerv Syst. 2011;27(1):87–94.
Nobusawa S, et al. IDH1 mutations as molecular signature and predictive factor of secondary glioblastomas. Clin Cancer Res. 2009;15(19):6002–7.
Yan H, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360(8):765–73.
Hartmann C, et al. Long-term survival in primary glioblastoma with versus without isocitrate dehydrogenase mutations. Clin Cancer Res. 2013;19(18):5146–57.
Korshunov A, et al. Integrated molecular characterization of IDH-mutant glioblastomas. Neuropathol Appl Neurobiol. 2019;45:108–18.
Gerber NK, et al. Transcriptional diversity of long-term glioblastoma survivors. Neuro Oncol. 2014;16(9):1186–95.
Yoda RA, et al. Mitotic index thresholds do not predict clinical outcome for IDH-mutant astrocytoma. J Neuropathol Exp Neurol. 2019;78(11):1002–10.
Appay R, et al. CDKN2A homozygous deletion is a strong adverse prognosis factor in diffuse malignant IDH-mutant gliomas. Neuro Oncol. 2019;21:1519.
Cimino PJ, Holland EC. Targeted copy number analysis outperforms histological grading in predicting patient survival for WHO grade II/III IDH-mutant astrocytomas. Neuro Oncol. 2019;21(6):819.
Perry A, et al. CDKN2A loss is associated with shortened survival in infiltrating astrocytomas but not oligodendrogliomas or mixed oligoastrocytomas. Neuro Oncol. 2014;16(suppl_3):iii1–iii22.
Louis DN, et al. cIMPACT-NOW update 6: new entity and diagnostic principle recommendations of the cIMPACT-Utrecht meeting on future CNS tumor classification and grading. Brain Pathol. 2020;30(4):844–856. https://doi.org/10.1111/bpa.12832. Epub 2020 Apr 19. PMID: 32307792.
Brat DJ, et al. cIMPACT-NOW update 5: recommended grading criteria and terminologies for IDH-mutant astrocytomas. Acta Neuropathol. 2020;139(3):603–8.
Parsons DW, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321(5897):1807–12.
Nonoguchi N, et al. TERT promoter mutations in primary and secondary glioblastomas. Acta Neuropathol. 2013;126(6):931–7.
Liu XY, et al. Frequent ATRX mutations and loss of expression in adult diffuse astrocytic tumors carrying IDH1/IDH2 and TP53 mutations. Acta Neuropathol. 2012;124(5):615–25.
Solomon DA, et al. Diffuse midline gliomas with histone H3-K27M mutation: a series of 47 cases assessing the spectrum of morphologic variation and associated genetic alterations. Brain Pathol. 2016;26(5):569–80.
Meyronet D, et al. Characteristics of H3 K27M-mutant gliomas in adults. Neuro Oncol. 2017;19(8):1127–34.
Tesileanu CMS, et al. Survival of diffuse astrocytic glioma, IDH1/2 wildtype, with molecular features of glioblastoma, WHO grade IV: a confirmation of the cIMPACT-NOW criteria. Neuro Oncol. 2020;22(4):515–23.
Lee Y, et al. The frequency and prognostic effect of TERT promoter mutation in diffuse gliomas. Acta Neuropathol Commun. 2017;5(1):62.
Batista R, et al. The prognostic impact of TERT promoter mutations in glioblastomas is modified by the rs2853669 single nucleotide polymorphism. Int J Cancer. 2016;139(2):414–23.
Koelsche C, et al. Distribution of TERT promoter mutations in pediatric and adult tumors of the nervous system. Acta Neuropathol. 2013;126(6):907–15.
Jones C, Baker SJ. Unique genetic and epigenetic mechanisms driving paediatric diffuse high-grade glioma. Nat Rev Cancer. 2014;14(10):651–61.
Nicolaides TP, et al. Targeted therapy for BRAFV600E malignant astrocytoma. Clin Cancer Res. 2011;17(24):7595–604.
Wu G, et al. Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat Genet. 2012;44(3):251–3.
Skene PJ, Henikoff S. Histone variants in pluripotency and disease. Development. 2013;140(12):2513–24.
Wan YCE, Liu J, Chan KM. Histone H3 mutations in cancer. Curr Pharmacol Rep. 2018;4(4):292–300.
Fontebasso AM, et al. Recurrent somatic mutations in ACVR1 in pediatric midline high-grade astrocytoma. Nat Genet. 2014;46(5):462–6.
Taylor KR, et al. Recurrent activating ACVR1 mutations in diffuse intrinsic pontine glioma. Nat Genet. 2014;46(5):457–61.
Fontebasso AM, et al. Mutations in SETD2 and genes affecting histone H3K36 methylation target hemispheric high-grade gliomas. Acta Neuropathol. 2013;125(5):659–69.
Temime-Smaali N, et al. The G-quadruplex ligand telomestatin impairs binding of topoisomerase IIIalpha to G-quadruplex-forming oligonucleotides and uncaps telomeres in ALT cells. PLoS One. 2009;4(9):e6919.
Aihara K, et al. H3F3A K27M mutations in thalamic gliomas from young adult patients. Neuro Oncol. 2014;16(1):140–6.
Davis RJ, Welcker M, Clurman BE. Tumor suppression by the Fbw7 ubiquitin ligase: mechanisms and opportunities. Cancer Cell. 2014;26(4):455–64.
Welcker M, et al. The Fbw7 tumor suppressor regulates glycogen synthase kinase 3 phosphorylation-dependent c-Myc protein degradation. Proc Natl Acad Sci U S A. 2004;101(24):9085–90.
Bjerke L, et al. Histone H3.3. mutations drive pediatric glioblastoma through upregulation of MYCN. Cancer Discov. 2013;3(5):512–9.
Li M, Liu GH, Izpisua Belmonte JC. Navigating the epigenetic landscape of pluripotent stem cells. Nat Rev Mol Cell Biol. 2012;13(8):524–35.
Hirabayashi Y, Gotoh Y. Epigenetic control of neural precursor cell fate during development. Nat Rev Neurosci. 2010;11(6):377–88.
Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293(5532):1074–80.
Klonou A, et al. Chromatin remodeling defects in pediatric brain tumors. Ann Transl Med. 2018;6(12):248.
Plass C, et al. Mutations in regulators of the epigenome and their connections to global chromatin patterns in cancer. Nat Rev Genet. 2013;14(11):765–80.
Lewis PW, et al. Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science. 2013;340(6134):857–61.
Chan KM, et al. The histone H3.3K27M mutation in pediatric glioma reprograms H3K27 methylation and gene expression. Genes Dev. 2013;27(9):985–90.
Di Lorenzo A, Bedford MT. Histone arginine methylation. FEBS Lett. 2011;585(13):2024–31.
Killela PJ, et al. TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. Proc Natl Acad Sci U S A. 2013;110(15):6021–6.
Kam KL, et al. Is next-generation sequencing alone sufficient to reliably diagnose gliomas? J Neuropathol Exp Neurol. 2020;79(7):763–6.
Jones C, et al. Pediatric high-grade glioma: biologically and clinically in need of new thinking. Neuro Oncol. 2017;19(2):153–61.
Coleman C, et al. Pediatric hemispheric high-grade glioma: targeting the future. Cancer Metastasis Rev. 2020;39(1):245–60.
Toll SA, et al. Sustained response of three pediatric BRAF(V600E) mutated high-grade gliomas to combined BRAF and MEK inhibitor therapy. Oncotarget. 2019;10(4):551–7.
Laetsch TW, et al. Larotrectinib for paediatric solid tumours harbouring NTRK gene fusions: phase 1 results from a multicentre, open-label, phase 1/2 study. Lancet Oncol. 2018;19(5):705–14.
Drilon A, et al. Efficacy of larotrectinib in TRK fusion–positive cancers in adults and children. N Engl J Med. 2018;378(8):731–9.
Ziegler DS, et al. Brief Report: Potent clinical and radiological response to larotrectinib in TRK fusion-driven high-grade glioma. Br J Cancer. 2018;119(6):693–6.
Grasso CS, et al. Functionally defined therapeutic targets in diffuse intrinsic pontine glioma. Nat Med. 2015;21(6):555–9.
Hashizume R, et al. Pharmacologic inhibition of histone demethylation as a therapy for pediatric brainstem glioma. Nat Med. 2014;20(12):1394–6.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this chapter
Cite this chapter
Pierson, C.R., Thomas, D.L. (2021). Genomic Heterogeneity of Aggressive Pediatric and Adult Diffuse Astrocytomas. In: Otero, J.J., Becker, A.P. (eds) Precision Molecular Pathology of Glioblastoma. Molecular Pathology Library. Springer, Cham. https://doi.org/10.1007/978-3-030-69170-7_9
Download citation
DOI: https://doi.org/10.1007/978-3-030-69170-7_9
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-69169-1
Online ISBN: 978-3-030-69170-7
eBook Packages: MedicineMedicine (R0)