Abstract
In the recent literature on multi-label classification, a lot of attention is given to methods that exploit label dependencies. Most of these methods assume that the dependencies are static over the entire instance space. In contrast, here we present an approach that dynamically adapts the label partitions in a multi-label decision tree learning context. In particular, we adapt the recently introduced predictive bi-clustering tree (PBCT) method towards multi-label classification tasks. This way, tree nodes can split the instance-label matrix both in a horizontal and a vertical way. We focus on hierarchical multi-label classification (HMC) tasks, and map the label hierarchy to a feature set over the label space. This feature set is exploited to infer vertical splits, which are regulated by a lookahead strategy in the tree building procedure. We evaluate our proposed method using benchmark datasets. Experiments demonstrate that our proposal (PBCT-HMC) obtained better or competitive results in comparison to its direct competitors, both in terms of predictive performance and model size. Compared to an HMC method that does not produce label partitions though, our method results in larger models on average, while still producing equally large or smaller models in one third of the datasets by creating suitable label partitions.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
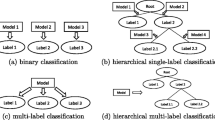
Most of the research on machine learning has investigated traditional classification problems whose classes are mutually exclusive, meaning that one instance may not belong to more than one label (class) simultaneously. Certain applications, however, present more complex learning tasks. In multi-label classification for instance, instances can be associated to multiple labels at the same time.
Originally, two main approaches were used for multi-label classification [1]: local and global approaches. The local approach transforms the multi-label setting to a single-label setting, such that traditional classification algorithms can be applied. Binary relevance and label powerset methods [1] are examples of a local approach. The global approach (also called big bang) adapts classification algorithms, such that they are able to work with the multi-label structure directly. Predictive clustering trees [2] are an example of a global approach. Despite having their respective advantages, the literature does not present a consensus on which strategy is superior. Later, approaches that group labels in different subsets were presented [3,4,5], resulting in methods in-between the global and local ones. That is, certain subsets of labels are believed to be more correlated among themselves than the label set al.together. In the recent years, a lot of attention in the multi-label literature goes to developing methods to optimally handle such correlations hidden in the label space. Many of these methods require ensemble methods [6,7,8] or exploit correlations in a pre-processing step [9,10,11]. In either case, the label dependencies are handled in a static way. In contrast, we hypothesize here that label correlations may differ throughout the instance space. Thus, rather than partitioning the label space as a pre-processing step in the same manner for all instances, models should dynamically detect such correlations, and create suitable partitions during their induction process.
Recently, a method has been introduced in the context of pairwise learning [12], that predicts interactions between two data points by bi-clustering the interaction matrix. Bi-clustering, also called co-clustering or two-way clustering, is the simultaneous clustering of the rows and columns of a matrix. We leverage this idea, which was set in the predictive clustering tree (PCT) framework, to the multi-label classification task, by bi-clustering the label space. This means that the PCT can induce both horizontal and vertical splits in the label matrix, instead of only horizontal ones. The vertical splits allow the model to partition the label set and group labels with a similar interaction pattern. This procedure is performed dynamically during the induction process: at each node to be split, horizontal as well as vertical candidate splits are considered.
Although the bi-clustering method for interaction prediction [12] also employed multi-label classification techniques, the end goal considered here is different, and hence, some adaptations to the method are required. In particular, we generate the vertical splits by incorporating a lookahead strategy in the tree learning procedure. Lookahead [13] is a technique that alleviates the myopia in greedy tree learning algorithms by taking into account the effect that a split has on deeper levels of the tree, when calculating its quality. In order to control computational complexity (which would become a bottleneck when considering all possible \(2^{|L|}\) candidate splits of labels, with L the label set for the node under consideration) and maximize interpretability, we employ a feature-based splitting strategy, just as with the horizontal splits.
In this paper, we apply the proposed idea to the task of hierarchical multi-label classification (HMC), i.e., problems where each data instance can be associated to multiple paths of a hierarchy defined over the label set. Such a hierarchy reflects a general-to-specific structure of the labels, and predictions must conform with the hierarchy constraint, that is, if a given label is predicted, all its ancestor labels must be predicted as well. One example is the task of image classification into a topic hierarchy, where an image may be classified as belonging to the paths Nature \(\rightarrow \) Tree \(\rightarrow \) Pine and Nature \(\rightarrow \) Snow.
To evaluate our proposed algorithm, we have performed experiments using 24 benchmark datasets from the domain of functional genomics, email classification and medical X-Ray images. We have compared our approach (which we call PBCT-HMC) to three other approaches: 1) Regular PCTs for HMC (Clus-HMC) [14]; 2) Predictive bi-clustering trees, as originally proposed to interaction prediction tasks [12] and later also applied to multi-label classification tasks [15] (PBCT); and 3) A static approach where label subsets are defined upfront according to the subtrees in the label hierarchy (Clus-Subtree).
The results show that PBCT-HMC outperforms PBCT and Clus-Subtree in terms of predictive performance and model size. Compared to Clus-HMC, while the predictive performance is comparable, our model sizes are larger on average. Nevertheless, for 8 out of 24 datasets, PBCT-HMC yields equally or more interpretable models than Clus-HMC by creating suitable label partitions.
The remainder of this paper is organized as follows: Sect. 2 discusses recent methods from the multi-label classification literature that also focus on exploiting label correlations. Section 3 presents our approach. Section 4 exposes the empirical comparison, with a discussion of the results. Finally, Sect. 5 provides a conclusion of this work and future work directions.
2 Related Work
Many multi-label studies have focused on exploiting label relationships, an important aspect for good multi-label predictors. In many of these studies, label relationships are defined by the problem domain, such as in hierarchical multi-label classification (HMC), where a taxonomy organizes the classes involved.
Predictive clustering trees for HMC were investigated in [14]. This work proposes a global method, named Clus-HMC, which induces a single decision tree predicting all the hierarchical labels at once, and shows that it outperforms local variants. The method was later extended to an ensemble approach [16], boosting the performance at the cost of reduced interpretability.
HMC has also been addressed by neural network based methods. In [17], the proposed method associates one multi-layer perceptron (MLP) to each hierarchical level, each MLP being responsible for the predictions in its associated level. The predictions at one level are used to complement the feature vectors of the instances used to train the neural network associated to the next level. Novel architectures for HMC tasks have been proposed in [18], where local and global loss functions are optimized for discovery of local hierarchical class-relationships and global information, being able to penalize violations of the hierarchy.
In [19], the authors propose a component to the output layers of neural networks that is applicable to HMC tasks. It makes use of prior knowledge on the output domain to map the hierarchical structure to the network topology. The so-called adjacency wrapping matrix incorporates the domain-knowledge directly into the learning process of neural networks.
In the absence of explicit label relationships, many studies have focused on extracting relationships from the label vectors of the instances, in order to exploit them during training. In [3], a probabilistic method is presented, that enforces discovery of deterministic relationships among labels in a multi-label model. The study focused on two main relationships: pairwise positive entailment and exclusion. Such relationships are represented as a deterministic Bayesian network.
A label co-occurence graph is created in [5]. Then, community detection methods are applied on the graph, as an alternative to the random partitioning performed by the RAkEL method [20]. A label powerset approach is used on the created label subsets. The results of the community detection methods on label co-occurence graphs outperformed the results obtained by using random partitioning. Another study [10] proposes to modify multi-label datasets by creating new features that represent correlations, in order to exploit instance correlations in the classification task. The features take into account distances between instances in the feature space.
Although the previous methods aim at extracting label relationships from a class hierarchy or label vectors, these relationships are considered to be fixed throughout the dataset. Recently, ensemble methods have been proposed, that allow different relations (modeled as label partitions) in each individual model.
In [7], an ensemble of PCTs is proposed, that consider random output subspaces to multi-label classification: each tree of the model uses a subset of the label space, thereby combining problem transformation and algorithm adaptation approaches into one method. The same approach was also proposed for multi-target regression [8]. Also, this work proposed a new prediction aggregation function that provides best results when used with extremely randomized PCTs. A recent study [21] proposed to use evolutionary algorithms for multi-label ensemble optimization. It selects diverse multi-label classifiers, each focusing on a label subset, in order to build an ensemble which takes into account data attributes, such as label relationship and label imbalance degree.
Differently from these approaches, in the current paper we present a method that learns a single (i.e., interpretable) tree model, that allows different label partitions in different parts of the instance space.
3 Predictive Bi-clustering Trees for HMC
3.1 Background
A predictive clustering tree (PCT) is a generalization of a decision tree, where each node corresponds to a cluster. Depending on the information contained in the leaf nodes, PCTs can be used for different learning tasks, including clustering, classification (single or multi-label) and regression (single or multi-target). They are implemented in the Clus softwareFootnote 1. PCTs are built in a top-down approach, meaning that the root node corresponds to the complete training set, which is recursively partitioned in each split. The heuristic used to select tests to include in the tree is the reduction of intra-cluster label variance, i.e., each split aims to maximize the inter-cluster label variance. In the multi-label classification setting, the labels are represented by a binary label vector, and the variance of a set of instances S is defined as:
where d is the distance between each instance’s label vector \(\mathbf{h_{i}}\) and the mean label vector from the set, \(\overline{\mathbf{h}}\).
In HMC tasks, it is convenient to consider that the similarity of higher levels of the hierarchy are more important than the similarity of lower levels. In order to implement such idea, the HMC implementation of PCTs in Clus (Clus-HMC) uses a weighted Euclidean distance in the function d above [14]. The class weight w decreases exponentially with the depth of the class in the hierarchy. The weight w of a class c, is given as:
A predictive bi-clustering tree (PBCT) is a PCT that is capable to predict interactions between two node sets in a bipartite network [12]. Interaction prediction is found in many applications, such as predicting drug-protein interactions, user-product interactions, etc. The network can be described by an adjacency matrix where each row and each column corresponds to a node, and each cell receives the value 1 if its corresponding nodes are connected, and 0 otherwise (Fig. 1). Each row and each column is also associated to a feature vector (e.g., chemical characteristics for drugs and structural properties for proteins in a drug-protein interaction network).
As PCTs, the PBCT is also built in a top-down approach. That means that in each iteration, a test is applied to one of the features. The test is chosen considering both sets of features for row and column nodes (H and V, as shown in Fig. 1), based on a heuristic and a stop criterion. The heuristic is the same as for multi-label classification (minimize intra-cluster variance of the label vectors), in the sense that for a horizontal split, row-wise label vectors are considered, while for a vertical split, column-wise label vectors.
3.2 Proposed Method: PBCT-HMC
In order to leverage PBCTs to the HMC domain, we apply the PBCT construction procedure to the label matrix of the HMC problem. Figure 2 provides an example, where for the label matrix shown in the top left box, there is no proper horizontal split, but if a vertical split is performed first, then it is possible to find two horizontal splits afterwards, resulting in four pure bi-clusters.
While the PBCT method has been applied to multi-label classification tasks before [15], it was designed for a different task (interaction prediction), and thus, we argue that some additions and adaptations to the algorithm described above are necessary in order to make the method fully accommodated to the HMC context. Our proposed approach is further denoted as PBCT-HMC. We introduce the following notation. \(\mathrm {H}\) and \(\mathrm {V}\), the two node sets from before, now respectively denote the set of instances and the set of labels. Each of these sets is composed by a (usually real-valued) feature matrix (\(\mathrm {H}^\mathrm {F}\) and \(\mathrm {V}^\mathrm {F}\)) and a (binary valued) target matrix (\(\mathrm {H}^\mathrm {T}\) and \(\mathrm {V}^\mathrm {T}\)), such that \(\mathrm {H}^\mathrm {T}\) is equivalent to transposing \(\mathrm {V}^\mathrm {T}\). Vectors are denoted in boldface and we use the notation \(\mathbf{w}[Z]\) to denote the subvector of \(\mathbf{w}\), restricted to the components defined by Z.
Feature Representation of Label Hierarchy. In order to represent the feature matrix \(\mathrm {V}^\mathrm {F}\), we propose the following simple mapping of the corresponding label hierarchy. We provide a single categorical feature, that denotes for each label l the name of the label just below the root, that is on the path from l to the root. A toy hierarchy is shown in Fig. 3, along with its representation. The proposed mapping takes into account the structural properties of the label hierarchy, it guarantees to produce predictions that fulfil the hierarchy constraint (i.e., the prediction probability for a child label can not exceed that for its parent label), and has the advantages to be small (which is beneficial for the lookahead approach - see further), very simple to model and to be domain independent.
Tree Induction and Split Heuristic. As in PBCT, tree induction is performed in a top-down manner. Any given node k of the tree can be associated to a bi-cluster defined by a pair \((H_k,V_k)\) with \(H_k \subseteq H^T\) and \(V_k \subseteq V^T\). The subsets \(H_k\) and \(V_k\) can be obtained by following the path of split nodes all the way from the root until node k. The root node is associated to (H, V).
In order to split node k, we first go through each feature in \(\mathrm {H}^\mathrm {F}\), in order to choose the best horizontal test. We apply the variance reduction heuristic, given by Eq. 1, to \(\mathrm {H}^\mathrm {T}\), in order to evaluate the quality of each split. In doing so, we restrict the label vectors \(\mathbf{h_i}\) and \(\mathbf{\overline{h}}\) to those components that are in \(V_k\). As we are dealing with a hierarchical task, we take into account the weights for each class, as exposed in Eq. 2. This results in the following variance definition:
and, consequently, in the following heuristic function for horizontal splits:
In Eq. 4, L and R refer to the left and right child nodes that are created for node k, after applying horizontal split s.
In a regular PBCT [12], the same procedure would be applied to the features in \(\mathrm {V}^\mathrm {F}\), after which the best overall test would be chosen to split the node. However, since the goal here is to perform HMC, we are not interested in the variance reduction over \(\mathrm {V}^\mathrm {T}\). Instead of measuring the quality of a vertical split directly, we need to make sure that the choice of a vertical split will indeed benefit the instance space partitioning, since in the end we want to make predictions for new (unseen) rows. To do so, the algorithm goes through each possible test from \(\mathrm {V}^\mathrm {F}\), defined by taking a subset of its feature valuesFootnote 2, and uses a lookahead approach [13] (illustrated in Fig. 4): For each test, the vertical split is simulated, as well as the next horizontal split (if any) in both resulting child nodes \((H_k,V_{kL})\) and \((H_k,V_{kR})\) of \((H_k,V_k)\). Thus, the heuristic function for vertical splits is based on the best value of the subsequently applied heuristic for horizontal splits. For computational reasons, we only perform a lookahead of depth one. This results in the following function to evaluate the quality of a vertical split s:
Splits \(s_L\) and \(s_R\) are chosen to maximize the values of the horizontal heuristic in the left and right child nodes of node k, respectively. The split s that is chosen to split the node k, is the one giving the overall maximal value for Eq. 4 or Eq. 5.
Before applying the split, an F-test is used to check if the variance reduction induced by the split is statistically significant. If the reduction is not significant, a leaf node is created instead. Otherwise, the test is included in the tree, the subsets of \(H_k\) or \(V_k\) are created to form new bi-clusters and the induction is recursively called until a stopping criterion, e.g. the minimal number of instances in a leaf, is reached. When a vertical split is included, the two subsequent horizontal splits are also included, i.e., a vertical split yields six instead of two new nodes, four of them being nodes to consider for splitting in the next iteration.
Each leaf receives a prototype vector, which corresponds to the vector of classwise (i.e., rowwise) averages. Figure 5(a) shows a small example of a tree resulting from our approach, using the label hierarchy from Fig. 3.
(a) Illustration of an induced PBCT-HMC tree. H\(_{n}\) is the nth feature from \(H^F\), and \(V_{1}\) is the (single) categorical feature from \(V^F\). (b) Illustration of the prediction procedure for the constructed PBCT-HMC tree. The two bi-clusters where the new instance arrives are colored in gray. The prototypes from these bi-clusters are then copied and concatenated into the prediction vector.
Stop Criterion. The smaller the prototype vector gets (after vertical splits), the easier it becomes to find a statistically significant subsequent horizontal split, using a fixed significance level \(l_0\). This results in the tree making more splits than necessary and becoming overfitted. For that reason, we apply a correction to the F-test significance level: when checking the significance of a split in a node k defined by (\(H_k\),\(V_k\)), we use \(l = l_0 \times (|V_k|/|V|)\) as significance level. Thus, the significance criterion becomes stricter, as the target vectors become smaller.
Making Predictions. After the tree is built, it is possible to enter a test set and get the predicted vector of probabilities for each test instance. To do this, each test instance is sorted down the tree, starting with the root node. Whenever a horizontal split is encountered, one of the child nodes is followed, according to the outcome of the test. Whenever a vertical split is encountered, however, both child nodes are followed, since a prediction is required for all labels. As such, the test instance may end up in multiple leaf nodes. The final prediction is constructed by concatenating the prototypes from these leaf nodes. Figure 5(b) illustrates the prediction procedure.
Pseudocode. Our proposed PBCT-HMC algorithm is presented in Algorithm 1.

4 Experiments and Discussion
4.1 Datasets
We used 24 datasets in our experiments, from the domain of functional genomics, image and email classification. Twenty datasets contain proteins from the Saccharomyces cerevisiae (yeast, 16 datasets) or Arabidopsis thaliana (4 datasets, with suffix ‘_ara’) genomeFootnote 3. The datasets differ mainly in how the protein features are represented, e.g., using sequence, cellcycle, or expression information. The label hierarchy employed is the Functional Catalogue (FunCat), a hierarchically structured classification system that enables the functional description of proteins from almost any organism (prokaryotes, fungi, plants and animals). The FunCat annotation scheme covers functions like cellular transport, metabolism and protein activity regulation [22]. The yeast datasets come in 2 versions: the 2007 version datasetsFootnote 4 were used in the original Clus-HMC publication [14] and the 2018 version datasetsFootnote 5 come from a recent update of the class labels [23]. The datasets ImgClef07a and ImgClef07d contain annotations of medical X-Ray images, with the attributes being descriptors obtained from EHD diagrams [24]. The Diatoms dataset contains descriptors of image data from diatoms, unicellular algae found in water and humid places [25]. Finally, the Enron dataset specifies a taxonomy for the classification of a corpus of emails [26]Footnote 6.
Table 1 summarizes the datasets used, presenting the number of instances in each subset (Train, Valid and Test), the number of attributes of each type (Categorical and Numerical), and the number of labels per level of the hierarchy.
4.2 Comparison Methods and Evaluation Measures
We compare PBCT-HMC to its three competitor methods, all set in the PCT framework. Although a lot of recent work on HMC is based on neural networks [17,18,19], here, next to predictive performance, we also want to assess the interpretability of the models. To do that, we compare the following methods:
-
Clus-HMC: Original PCT models proposed for HMC tasks [14]Footnote 7 (Sect. 3.1);
-
PBCT: Original bi-clustering PCT proposed for interaction prediction tasks [12], as described in Sect. 3.1, which has also been applied to multi-label classification tasks before [15];
-
Clus-Subtree: A static approach where one Clus-HMC model is built for each subtree of the root of the hierarchy;
-
PBCT-HMC: Our proposed methodFootnote 8;
In order to have a fair comparison, we have re-implemented the PBCT method, which was originally implemented in Python, in Clus. For the label feature space, it uses the same mapping of the label hierarchy as PBCT-HMC. The main difference is the split heuristic, which uses rowwise and columnwise variance reduction, without class weights.
The Clus-Subtree method was included to compare against a static variant of our proposed method. In this case, the vertical splits are made upfront: for each subtree of the label hierarchy root, a Clus-HMC model is constructed. Afterwards, all prediction vectors are concatenated. For each model, all available training instances are used, thus including the instances with an all-zeroes label vector (negative instances).
In accordance with the HMC literature [14, 18, 19], we adopted the Pooled Area Under Precision-Recall Curve (Pooled AUPRC) as the evaluation measure. It is the micro-averaged area under the label-wise precision-recall curves, generated by using threshold values (steps of 0.02) ranging from 0 to 1.
We have optimized the F-test significance level parameter (\(l_0\)), a parameter used in all compared methods, using the following values: 0.001, 0.005, 0.01, 0.05, 0.1 and 0.125. For each of these values, a model is built using only the training subset, and evaluated using the validation dataset. The value associated to the best performance is selected. Finally, the optimized model is built using the train and validation datasets together, and results are reported using the test dataset. For the parameters minimum of examples perf leaf, we have fixed the value to 5.
4.3 Results and Discussion
Table 2 shows the results regarding the Pooled AUPRC for each dataset and each approach. The best results are highlighted. Looking at this table, we can notice that Clus-HMC and PBCT-HMC presented superior results when compared to Clus-Subtree and PBCT. In all datasets, apart from 3 exceptions, either Clus-HMC or PBCT-HMC was superior. When compared between themselves, Clus-HMC and PBCT-HMC yielded very similar results. Although on average, Clus-HMC has a slight advantage, in 7 cases PBCT-HMC outperformed or resulted in the same performance as Clus-HMC. In Fig. 6, we present a critical distance diagram following a Friedman and Nemenyi test, as proposed by [27], comparing the Pooled AUPRC of all methods. The Friedman test resulted in a p-value = 5.441e–07. PBCT-HMC and Clus-HMC present a statistically significant difference when compared to PBCT, but not between themselves. Likewise, PBCT and Clus-Subtree do not present a statistically significant difference. Although PBCT-HMC did not present a statistically significant difference compared to Clus-Subtree, their average ranks are at the limit of the critical difference.
The contrasting performance between PBCT-HMC and PBCT is associated to the lookahead procedure and the F-test significance level adjustment. PBCT, which targets interaction prediction and aims to maximize also column-wise variance reduction, performs vertical splits prematurely, leading to suboptimal splits. As for the F-test, not adjusting its significance level can result in overfitting.
As for model size (Table 3), we see that PBCT-HMC results in models that are 4 and 2 times smaller on average compared to Clus-Subtree and PBCT, respectively. Thus, compared to the methods that also partition the label space, our method can be expected to yield more interpretable models. This finding is expected for Clus-Subtree since it contains multiple models, and thus its size consists of the summed size of all models. As for PBCT, we suspect that the models may be overfitted due to their larger size, but inferior performance. In contrast, when compared to Clus-HMC, on average, our models are twice as large. This can be explained by the fact that Clus-HMC only introduces horizontal splits and that each vertical split in PBCT-HMC results in six new nodes compared to only two nodes for horizontal splits, as explained before. Since at prediction time, the vertical nodes are not used semantically, in the sense that their split condition does not need to be checked for the test instances (they follow the paths to both child nodes), and hence, they do not add to the prediction time, we also report the number of horizontal splits for the bi-clustering-based methods (reported as PBCT_HOR and PBCT-HMC_HOR).
Although Clus-HMC induced smaller models on average, we see that PBCT-HMC, specially when considering only the horizontal nodes, yielded considerably smaller models for some datasets. Nevertheless, our results raise the question under which conditions label partitions should be considered to exploit correlations.
Finally, Table 4 exposes the induction time for each experiment. Clus-HMC presented lower induction times overall due to its simplicity. On the other hand, Clus-Subtree demands most time since it builds multiple models per dataset. PBCT-HMC comes in third, after PBCT, due to the lookahead strategy. The induction time depends on the number of categorical feature values of the dataset V, meaning the hierarchy size has a direct effect on the induction time. However, as the table shows, the run time is still feasible.
5 Conclusions
In this work, we have proposed a predictive bi-clustering tree method for hierarchical multi-label classification (HMC). Opposed to traditional approaches for HMC, our method, named PBCT-HMC, automatically partitions the label space during its induction process. To achieve this, a lookahead approach is incorporated in the tree construction, such that a label space partition is introduced if it leads to a better instance space partition in a deeper level of the tree.
Our experiments demonstrated that PBCT-HMC obtained better or competitive predictive performances compared to three related methods, yielding the second smallest models overall. Specially in comparison with the original PBCT and the static method Clus-Subtree, PBCT-HMC performed better in the large majority of the datasets, achieving a great interpretability gain. Still, our results put into question the advantage of considering label partitions to exploit label correlations in multi-label classification, a topic that receives a lot of attention. Further research is needed to identify which type of models can benefit from it.
Future extensions of our work might address more challenging applications, e.g., datasets with a label hierarchy structured as a directed acyclic graph, and the application to non-hierarchical multi-label classification tasks.
Notes
- 1.
- 2.
In our implementation, we consider a greedy generation of the subsets.
- 3.
Available at https://dtai.cs.kuleuven.be/clus/hmc-ens/.
- 4.
Available at https://dtai.cs.kuleuven.be/clus/hmcdatasets/.
- 5.
Available at https://itec.kuleuven-kulak.be/supportingmaterial.
- 6.
Available at http://kt.ijs.si/DragiKocev/PhD/resources/doku.php.
- 7.
Available at https://dtai.cs.kuleuven.be/clus.
- 8.
Available at https://github.com/biomal/Clus-PBCT-HMC.
References
Tsoumakas, G., Katakis, I.: Multi-label classification: an overview. Int. J. Data Warehouse. Min. (IJDWM) 3(3), 1–13 (2007)
Blockeel, H., Raedt, L.D., Ramon, J.: Top-down induction of clustering trees. In: Proceedings of the Fifteenth International Conference on Machine Learning, ICML 1998, pp. 55–63 (1998)
Papagiannopoulou, C., Tsoumakas, G., Tsamardinos, I.: Discovering and exploiting deterministic label relationships in multi-label learning. In: Proceedings of the 21th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, pp. 915–924. Association for Computing Machinery (2015)
Madjarov, G., Gjorgjevikj, D., Dimitrovski, I., Džeroski, S.: The use of data-derived label hierarchies in multi-label classification. J. Intell. Inf. Syst. 47(1), 57–90 (2016)
Szymanski, P., Kajdanowicz, T., Kersting, K.: How is a data-driven approach better than random choice in label space division for multi-label classification? CoRR (2016)
Joly, A., Geurts, P., Wehenkel, L.: Random forests with random projections of the output space for high dimensional multi-label classification. In: Machine Learning and Knowledge Discovery in Databases, pp. 607–622 (2014)
Breskvar, M., Kocev, D., Džeroski, S.: Multi-label classification using random label subset selections. In: Discovery Science (2017)
Breskvar, M., Kocev, D., Džeroski, S.: Ensembles for multi-target regression with random output selections. Mach. Learn. 107(11), 1673–1709 (2018). https://doi.org/10.1007/s10994-018-5744-y
Prati, R.C., de França, F.O.: Extending features for multilabel classification with swarm biclustering. In: IEEE Congress on Evolutionary Computation, pp. 2964–2971 (2013)
de Abreu, I.B.M., Mantovani, R.G., Cerri, R.: Incorporating instance correlations in multi-label classification via label-space. In: International Joint Conference on Neural Networks (IJCNN), pp. 581–588 (2017)
Feng, L., An, B., He, S.: Collaboration based multi-label learning. In: Proceedings of the AAAI Conference on Artificial Intelligence, vol. 33, pp. 3550–3557 (2019)
Pliakos, K., Geurts, P., Vens, C.: Global multi-output decision trees forinteraction prediction. Mach. Learn. 107(8), 1257–1281 (2018). https://doi.org/10.1007/s10994-018-5700-x
Elomaa, T., Malinen, T.: On lookahead heuristics in decision tree learning. In: Zhong, N., Raś, Z.W., Tsumoto, S., Suzuki, E. (eds.) ISMIS 2003. LNCS (LNAI), vol. 2871, pp. 445–453. Springer, Heidelberg (2003). https://doi.org/10.1007/978-3-540-39592-8_63
Vens, C., Struyf, J., Schietgat, L., Džeroski, S., Blockeel, H.: Decision trees for hierarchical multi-label classification. Mach. Learn. 73(2), 185 (2008)
Pliakos, K., Vens, C., Tsoumakas, G.: Predicting drug-target interactions with multi-label classification and label partitioning. In: IEEE-ACM Transactions On Computational Biology And Bioinformatics, pp. 1–11 (2019)
Schietgat, L., Vens, C., Struyf, J., Blockeel, H., Kocev, D., Dzeroski, S.: Predicting gene function using hierarchical multi-label decision tree ensembles. BMC Bioinf. 11, 2 (2010)
Cerri, R., Barros, R.C., de Carvalho, A.C., Jin, Y.: Reduction strategies for hierarchical multi-label classification in protein function prediction. BMC Bioinform. 17(1), 373 (2016) https://doi.org/10.1186/s12859-016-1232-1
Wehrmann, J., Cerri, R., Barros, R.: Hierarchical multi-label classification networks. In: Proceedings of the 35th International Conference on Machine Learning, vol. 80, pp. 5075–5084 (2018)
Masera, L., Blanzieri, E.: Awx: an integrated approach to hierarchical-multilabel classification. In: Machine Learning and Knowledge Discovery in Databases, pp. 322–336 (2019)
Tsoumakas, G., Katakis, I., Vlahavas, I.: Random k-labelsets for multi-label classification. IEEE Trans. Knowl. Data Eng. 23 1079–1089 (2011)
Moyano, J., Gibaja, E., Cios, K., Ventura, S.: Combining multi-labelclassifiers based on projections of the output space using evolutionary algorithms. Knowl.-Based Syst. 196, 105770 (2020)
Ruepp, A, Zollner, A.M.D.: The funcat, a functional annotation scheme for systematic classification of proteins from whole genomes. Nucleic Acids Res. 32(18), 5539–5545 (2004)
Nakano, F.K., Lietaert, M., Vens, C.: Machine learning for discovering missing or wrong protein function annotations. BMC Bioinf. 20(1), 485 (2019)
Dimitrovski. I., Kocev, D., Loskovska, S., Džeroski, S.: Hierchical annotation of medical images. In: Proceedings of the 11th International Multiconference - Information Society IS 200. IJS, Ljubljana, pp. 174–181 (2008)
Dimitrovski, I., Kocev, D., Loskovska, S., Džeroski, S.: Hierarchical classification of diatom images using ensembles of predictive clustering trees. Ecol. Inf. 7(1), 19–29 (2012)
Klimt, B., Yang, Y.: The Enron corpus: a new dataset for email classification research. In: Boulicaut, J.-F., Esposito, F., Giannotti, F., Pedreschi, D. (eds.) ECML 2004. LNCS (LNAI), vol. 3201, pp. 217–226. Springer, Heidelberg (2004). https://doi.org/10.1007/978-3-540-30115-8_22
Demšar, J.: Statistical comparisons of classifiers over multiple data sets. J. Mach. Learn. Res. 7, 1–30 (2006)
Acknowledgments
We acknowledge Sao Paulo Research Foundation (FAPESP grants #2017/13218-5 and #2016/25078-0) and Research Fund Flanders (FWO) for financial support.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 Springer Nature Switzerland AG
About this paper
Cite this paper
Santos, B.Z., Nakano, F.K., Cerri, R., Vens, C. (2021). Predictive Bi-clustering Trees for Hierarchical Multi-label Classification. In: Hutter, F., Kersting, K., Lijffijt, J., Valera, I. (eds) Machine Learning and Knowledge Discovery in Databases. ECML PKDD 2020. Lecture Notes in Computer Science(), vol 12459. Springer, Cham. https://doi.org/10.1007/978-3-030-67664-3_42
Download citation
DOI: https://doi.org/10.1007/978-3-030-67664-3_42
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-67663-6
Online ISBN: 978-3-030-67664-3
eBook Packages: Computer ScienceComputer Science (R0)