Abstract
Pulse transit time (PTT) provides a cuffless method to measure and predict blood pressure, which is essential in long term cardiac activity monitoring. Photoplethysmography (PPG) sensors provide a low-cost and wearable approach to obtain PTT measurements. The current approach to calculating PTT relies on quasi-periodic pulse event extractions based on PPG local signal characteristics. However, due to inherent noise in PPG, especially at uncontrolled settings, this approach leads to significant errors and even missing potential pulse events. In this paper, we propose a novel approach where global features (all samples) of the time-series data are used to develop a machine learning model to extract local pulse events. Specifically, we contribute 1) a new noise resilient machine learning model to extract events from PPG and 2) results from a study showing accuracy over state of the art (e.g. HeartPy) and 3) we show that MLPTT outperforms HeartPy peak detection, especially for noisy photoplethysmography data.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Continuous monitoring of blood pressure over long periods is essential to prevent critical cardiovascular events that can cause irreversible health damages and even death. The classical approach to measuring blood pressure uses pressure cuffs, requiring mechanical apparatuses which are undesired in long-term monitoring. Researchers have proposed alternative methods to measure blood pressure using pulse transit time (PTT), which does not need mechanical cuffs [1,2,3,4,5,6,7]. An extensive review of blood pressure monitoring theory and practice using PTT can be found in [8]. Researchers also found a strong correlation among PTT, the measurement of respiratory effort [9] and the detection of microarousals [9]. Other applications included the indication of cardiovascular changes during obstetric spinal anesthesia [10], myocardial performance [11], respiratory events [12] and hypertension detection [13, 14].
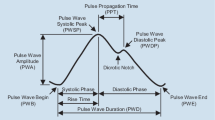
PTT is the duration that it takes for a pulse wave (PW) to travel between two arterial points [13]. It can be measured with different methods, such as arterial probes [15], electrocardiogram (ECG) or contactless photoplethysmography (PPG). ECG electrically records a graph of voltage versus time, which is typically acquired from electrodes attached to the chest or limbs whereas PPG optically records blood volume changes vs time.
While ECG may have suffered from measurement artefacts [16] such as electromagnetic interference or loose leads, it was still considered the gold standard for continuous heart rate monitoring [17]. Despite the high signal quality, its usability was limited by the requirement of leads attached to the skin. In comparison, optical PPG sensors were found to be the least invasive and therefore overall desirable. However, they were also sensitive to sensor pressure and artefacts caused by motion [18]. Pulse transit times can also be measured between different physical locations and temporal parts of the cardiac cycle. The cardiac cycle is a repeating series of pressure changes within the heart of living organisms and was often described to have several peaks, particularly the P wave, QRS complex and T wave [19]. No standardised PTT definition had been established due to various measurement possibilities regarding
-
(i)
different sensor types
-
(ii)
sensor or lead locations and
-
(iii)
temporal reference points.
Van Velzen, et al. identified 43 different methods to determine PTT [20] between an ECG R-peak and a PPG signal [21,22,23,24,25]. A PTT for ECG-PPG measurements is called Pulse Arrival Time (PAT). All reviewed methods [20] used anchor points to calculate the PAT (Fig. 1A).
These anchor points could be the foot (or onset), peak or a percentage (e.g. 50%, 25%, etc.) of the pulse wave. PAT could be calculated by
where n was the individual heartbeat. Many PPG sensor signal quality-related challenges were identified in previous research. Their readings were found to contain artefacts [18, 26] or malformed segments induced by sensor motion or attachment pressure variations [20]. To compensate for undesired signal anomalies, at least 19 of the 43 previously reviewed methods [20] used signal filtering for PTT calculated from anchor points. Although PTT is the duration that it takes for a pulse wave to travel between two arterial points, it was so far considered as the time between two selected points on two curves, with \(PAT_{peak-foot}\) and \(PAT_{peak-peak}\) most widely used. Table 1 provides a summary of previously used anchor points in literature.
Machine learning and particularly neural networks had shown great potential at extracting spatial information from data [27]. We hypothesized that while filtering had refined PTT measurements, accuracy could be improved further by not considering an individual point on the curve but the shift of the shape of the curve using all points. We therefore propose Machine Learned Pulse Transit Time (MLPTT), which was trained on a sliding frame to detect curve properties relative to a virtual anchor point (Fig. 1B). The proposed PAT measurement could be defined with the following equation
where n was an individual heartbeat. PAT is a particular type of PTT and was used to validate the proposed method by comparing the predicted PAT with PAT calculated by HeartPy, a toolkit that was designed to handle (noisy) PPG data [28, 29]. While the advantages of PAT are that R-peaks of the cardiovascular QRS complex can be detected with numerous algorithms [30] such as Pan-Tompkins [31], it requires conductive electrodes attached to the skin in selected locations and is, therefore, more disruptive than PPG, which measures optically. Gao et al. compared the pulse transit time estimates of PAT and PPG-PTT with invasive I-PTT using arterial probes as a reference [32]. They concluded that PPG-PTT correlated well with all blood pressure levels. To validate MLPTT, the PAT of three BIDMC PPG and Respiration Dataset [33] patients was calculated with HeartPy and MLPTT. Both methods provided independent results from each other, the overall validation workflow is shown in Fig. 2.
Contribution: We propose a machine learning based PTT calculation method and show it is more accurate than the existing signal processing approaches. The specific contributions can be summarized as follows:
-
a)
We propose a new noise resilient machine learning model to extract events from PPG and demonstrate it particularly on PAT extraction.
-
b)
Results from a study showing accuracy over state of the art (i.e. HeartPy) with statistical significance.
-
c)
We show that MLPTT outperforms HeartPy peak detection especially for noisy photoplethysmogram data.
-
d)
We discuss and evaluate the results.
2 Design
Pulse transit time was previously measured between two anchor points on ECG, PPT or arterial probe pressure signals [32]. Since all these signals are quasi-periodic, we hypothesized that PTT can be considered signal phase shift per heartbeat period. While phase shift can be calculated between the same anchor point on different signals, we propose to use machine learning to consider all points on the signal to estimate phase shift and therefore PTT.
2.1 Machine Learned Pulse Transit Time
The idea to consider all points on the signal was implemented in MLPTT. MLPTT consisted of a sequence of 4 processes: frame segmentation, waveform binary classifier, frame segmentation, anchor point classifier (Fig. 3: 3, 4, 7, 8).
MLPTT Frame Segmentation for Binary Classification. PPG and ECG data were loaded from a dataset (Fig. 3: 1, 2). To avoid discontinuities in the time series data, 70% training and 30% test ratio were used for all patients instead of k-fold verification. The first two MLPTT processes (Fig. 3: 3 and 4) aimed at finding a periodic pattern in the PPG signal. This was achieved by using the known periods from ECG signals R-peaks, detected with the Pan-Tompkins algorithm (Fig. 3: 5 and [31]), as virtual anchor point labels. Frames of 60 samples each were created by shifting the frame in increments of one sample. In the case of the BIDMC dataset sampled 125 Hz, one sample was 8ms long. Each frame was labelled respective containing a virtual anchor or not.
Waveform Binary Classifier. The segmented and by anchor points labelled frames were subsequently used as training input for the ML waveform binary classifier (Fig. 4A: 4).
The goal of this step was to train a classifier that could detect quasi-periodic waveforms in longer PPG time series which contained one virtual anchor. This could also be considered a heart rate classification based on PPG data, with the detected consecutive virtual anchors manifested as one heartbeat each. Settings for Python Scikit-learn 0.22.1 KNN, SVC, Gaussian process, decision tree, random forest, MLP, AdaBoost and Gaussian process Naïve Bayes classifiers were grid searched with Gaussian Naïve Bayes achieving the highest classification accuracy with default settings. The trained classifier model was then used for binary prediction of all frames, either containing a virtual anchor or not.
Frame Segmentation for Anchor Point Classification. The frames predicted to contain virtual anchors were then automatically selected for a second classifier (Fig. 3: 7). Frames predicted to contain no virtual anchors were not evaluated further.
Multilabel Anchor Position Classifier. As the final step of the proposed MLPTT method, the anchor position in the selected frames was predicted. This classifier was trained on ECG derived peaks for PPG training data. In addition to the classifiers evaluated for the binary classification, a 5-layer sequential TensorFlow model was developed. The model consisted of 120 neurons for the input layer and 60 for the output. After grid searching hyperparameters for this model, it outperformed the baseline Scikit-learn MLP mode and was used for the prediction results in the following chapters. The trained MLPTT was then used to predict \(PAT(MLPTT)_{peak-virtual\_anchor}\) by predicting the virtual anchors for test data extracted from the BIDMC dataset, which were then downsampled to one anchor per period.
2.2 HeartPy Pulse Transit Time
HeartPy is a toolkit designed to handle noisy PPG data and was used to detect peaks in PPG signals. For three out of four tested PAT definitions, Rajala et al. reported the smallest relative error for \(PAT_{peak-peak}\) [34] and this definition was found to be the second most widely used in a literature review of 43 published papers [20]. Therefore, \(PAT(HeartPy)_{peak-peak}\) was calculated by HeartPy. As ECG reference, the same Pan-Tompkins detected peaks were used as for MLPTT. PPG peaks were detected with HeartPy using following settings: \(sample\_rate = 125\), \(hampel\_correct = False\), \(high\_precision\_fs=1000\).
3 Analysis
In this section, we show how the proposed method was compared to the established toolkit.
3.1 Methods
For PTT and PAT, there is no ground truth readily available. We therefore initially compared MLPTT PAT with HeartPy PAT as a reference, to disprove the null hypothesis that their correlation is not statistically relevant. PAT was used instead of PPG PTT because ECG R-peak detection accuracy was found to be higher than for PPG, ECG R-peak detection has been standard practice and for some populations, comparatively better signal to noise ratios had been observed [35] as well as lower morphological variance [36]. Because our eventual goal was to measure PTT and therefore phase shift between signals, we considered only the AC component of the time series PTT estimations for comparison. Accordingly, the PAT of 196–240 ECG-detected heartbeats was calculated for 5 patients with HeartPy and MLPTT.
We then disproved that there is no statistically significant correlation between both curves. If the following relationship was true
where \(hb_k\) were the heartbeats of each patient from the beginning of the test data (\(k=0\)) to the last heartbeat (\(k=n\)), MLPTT PAT and HeartPy PAT were not correlated. If they were linearly correlated, plotting \(x =MLPTT PAT(hb_k)\) and \(y = HeartPy PAT(hb_k)\) would have formed a straight line. The correlation was to be proven with Pearson correlation and its respective p-value <0.05.
4 Results
We measured performance by showing a linear relation between our proposed method PAT results and HeartPy PAT results and plotted resulting PAT curves plotted on top of each other with the respective difference filled (Fig. 5).
Out of 5 tested patients (BIDMC patients 2, 6, 8, 42, 53), HeartPy failed to detect exactly one PPG peak per ECG period for patients 2 and 53, in some instances detecting 0 and in some instances 2. These patients were removed and only patients 6, 8 and 42 used for comparison. The Pearson correlation between HeartPy PAT and MLPTT PAT was the following (Table 2).
F1 score and RMSE were calculated between the ECG signals R-peaks and the MLPTT detected virtual anchor from PPG. The predicted R-peak from PPG data was used as a reference to benchmark the stability of the method, but it should not be considered as ground truth, which is indeterminable for PPG with current methods and can only be approximated intravenously [32]. For patients 2–42 the F1 score was in the range of 0.15–0.30 with RMSE extending between 3.83 and 8.01 samples (Table 3). Patient 53 showed the lowest accuracy with an F1 score of 0.04 and a RMSE of 15.66 samples. Support was the number of analysed samples in the time series, 11661 to 13278 samples for patient 2–42 and 9014 for patient 53.
4.1 BIDMC Patient 6 Drilldown
These were the MLPTT subprocess results sampled from one out of the 3/5 BIDMC patients of whom HeartPy detected all PPG peaks.
HeartPy and MLPTT PAT Patient 6 Pearson Correlation. A scatterplot with HeartPy calculated PAT on the x-axis and MLPTT PAT on the y-axis was produced (Fig. 6). A histogram on top of each axis showed the respective PAT distribution. The Pearson correlation coefficient p was listed for all tested patients in Table 2.
Waveform Binary Classifier. The waveform binary classifier achieved at an overall anchor prediction accuracy of 97% for 17977 frames of 60 samples each. Precision, recall, f-1 score and support were listed in Table 4. Precision was the ratio of correctly positively predicted virtual anchors to all positive virtual anchors (\(Precision = TP/TP+FP\), where TP was True Positive and FP was False Positive). The calculated precision was 0.92 for frames without anchors and 0.99 for frames with anchors. Recall, the ratio of correctly predicted virtual anchors to all virtual anchors in the class was 0.99 for frames without anchor and 0.95 for frames containing a virtual anchor (\(Recall = TP/TP+FN\), where TP was True Positive and FN False Negative). Support was the number of samples in the respective class (Table 4).
Multilabel Anchor Position Classifier. The multilabel anchor position classifier achieved an overall prediction accuracy of 30% (Table 3), with most predictions scattered closely around a confusion matrix diagonal.
5 Discussion
PTT is an important measure that can be used in many clinical applications and many researchers tried to measure it precisely. The goal of this study was to assess if the accuracy of established PAT and PTT methods that relied on signal filtering and fixed specific points can be matched and potentially exceeded. We addressed this challenge by creating a novel approach that used machine learning to find quasi-periodic patterns in PPG signals based on a series of samples.
For five tested BIDMC dataset patients, HeartPy did not detect one PPG peak for every ECG peak. MLPTT was more robust and correctly classified the presence of a quasi-periodic waveform in 97% of all frames for patient 6. Since the frame was shifted in increments of one sample through the entire signal, the classifier would predict 125 frames for one heartbeat at a heart rate of 60bpm and a sample rate 125 Hz. We expected the increased accuracy to be driven by at least three factors, the first was MLPTT’s ability to learn malformed PPG shapes of any form as long as they were contained in the training sequence. This could be particularly helpful for PPG-PPG PTT where it is desired to measure the signal phase shift accurately. The second factor was that previous PTT measurements were dependent on high signal quality at a specific location during each period. For example when measuring the peak-foot PAT, a pronounced foot would have to occur in the PPG signal which could be corrupted by motion artefacts or other noise. If the peak was still in its true position for a period with a corrupted foot, HeartPy would not benefit while MLPTT could learn and still predict the correct virtual anchor point. The third expected reason was that MLPTT was stepping through the entire signal in frames of 1 sample interval. Therefore, the multilabel classifier had the chance to predict every virtual anchor point for the number of intervals in one frame, 60 times in our implementation.
The calculated HeartPy- and MLPTT PAT correlated with statistical significance for in total 637 tested heartbeats of BIDMC patients 6, 8 and 42. Although PAT for patients 2 and 53 could not be compared due to HeartPy’s PPG peak detection inconsistencies compared to ECG, the MLPTT PAT prediction did not show any significant variations in the AC component that could have been caused by anchor point misdetection. MLPTT showed a pronounced confusion matrix diagonal for the prediction of virtual anchors. Overall, MLPTT correlated with HeartPy with statistical significance for the tested dataset, required no adjustments for individual patients and showed more robust PAT measurements for patients with noisy measurements.
5.1 Limitations
Despite the strong Pearson correlation with HeartPy PAT measurements, the method and current implementation are not without limitations. The behaviour for medically relevant outliers was not tested and patients’ background was not investigated. Some BIDMC signals also showed apparent sawtooth and potentially sine type noise. Lin et al. reported a sawtooth pattern in a different PPG dataset [37], which indicates that this type of noise may be prevalent in PPG recordings of medical equipment. Furthermore, despite our attempts to find the ideal parameters for HeartPy, HeartPy could offer additional built-in signal filters that we did not use. These might have improved HeartPy performance further. No custom filtering was applied for MLPTT for different patients.
Theoretical limitations included that the tested implementation with a frame length of 0...60 samples allowed for a maximum of 125bpm heart rate. Exceeding this heart rate would have led to more than one period per frame, which the implementation was not designed to handle in its first revision. This could be addressed in further revisions. Another theoretical limitation were edge cases in which the virtual anchor point is at either edge of the frame. It can be expected that these frames were more difficult to classify, which resulted in a slight decrease in classification accuracy for the outermost classes. This could be mitigated in further revisions by not considering those frames and only processing frames with a predicted minimum distance from the outer frame limits. Foremost, although both tested methods showed a statistically significant correlation, there were no readily available ground truth measurements for PAT and PTT. Invasive arterial probe measurements were found to produce the smallest errors for blood pressure prediction [32] and can be considered to be more reliable due to measuring the arterial pressure directly. Testing PPG-PTT against I-PTT would be more meaningful, but no dataset was available at the time of writing. We recognize the risks that incorrect PTT measurements may cause if used in clinical applications. All PTT measurements based on the new method should be revalidated against methods such as I-PTT with statistical significance before clinical deployment.
6 Conclusion
The key contribution of this paper is to provide a novel approach where global features (all samples) of the time-series data are used to develop a machine learning model to extract local pulse events. We evaluated the performance of MLPTT for more than 50000 samples of a reference dataset and validated the performance in comparison to a reference method for over 1000 heartbeats of 5 patients. The analyses show that MLPTT copes significantly better with inherently noisy PPG data than the reference method. The proposed technique is suitable for analysis of other medical recordings and for application in many other domains that rely on time series data.
References
Geddes, L.A., Voelz, M.H., Babbs, C.F., Bourland, J.D., Tacker, W.A.: Pulse transit time as an indicator of arterial blood pressure. Psychophysiology 18(1), 71–74 (1981). https://doi.org/10.1111/j.1469-8986.1981.tb01545.x. ISSN 0048-5772
Poon, C.C.Y., Zhang, Y.T.: Cuff-less and noninvasive measurements of arterial blood pressure by pulse transit time. In: 2005 IEEE Engineering in Medicine and Biology 27th Annual Conference, pp. 5877–5880 (2005). https://doi.org/10.1109/IEMBS.2005.1615827
Lane, J.D., Greenstadt, L., Shapiro, D., Rubinstein, E.: Pulse transit time and blood pressure: an intensive analysis. Psychophysiology 20(1), 45–49 (1983). https://doi.org/10.1111/j.1469-8986.1983.tb00899.x. ISSN 0048-5772
Pitson, D.J., Stradling, J.R.: Value of beat-to-beat blood pressure changes, detected by pulse transit time, in the management of the obstructive sleep apnoea/hypopnoea syndrome. Eur. Respir. J. 12(3), 685–692 (1998)
Gesche, H., Grosskurth, D., Küchler, G., Patzak, A.: Continuous blood pressure measurement by using the pulse transit time: comparison to a cuff-based method. Eur. J. Appl. Physiol. 112, 309–315 (2011). https://doi.org/10.1007/s00421-011-1983-3
Ripoll, V.R., Vellido, A.: Blood pressure assessment with differential pulse transit time and deep learning: a proof of concept. Kidney Dis. 5(1), 23–27 (2019). https://doi.org/10.1159/000493478
Sklarsky, A., Garvin, N.M., Pawelczyk, J.A.: Limitations of pulse transit time to estimate blood pressure. FASEB J. 33(1 supplement), 562.13 (2019). https://doi.org/10.1096/fasebj.2019.33.1_supplement.562.13
Mukkamala, R., et al.: Toward ubiquitous blood pressure monitoring via pulse transit time: theory and practice. IEEE Trans. Biomed. Eng. 62(8), 1879–1901 (2015). https://doi.org/10.1109/tbme.2015.2441951. ISSN 0018-9294
Smith, R.P., Argod, J., Pépin, J.-L., Lévy, P.A.: Pulse transit time: an appraisal of potential clinical applications. Thorax 54(5), 452–457 (1999). https://doi.org/10.1136/thx.54.5.452
Sharwood-Smith, G., Bruce, J., Drummond, G.: Assessment of pulse transit time to indicate cardiovascular changes during obstetric spinal anaesthesia. BJA Br. J. Anaesth. 96(1), 100–105 (2005). https://doi.org/10.1093/bja/aei266. ISSN 0007-0912
Obrist, P.A., Light, K.C., McCubbin, J.A., Hutcheson, J.S., Hoffer, J.L.: Pulse transit time: relationship to blood pressure and myocardial performance. Psychophysiology 16(3), 292–301 (1979). https://doi.org/10.1111/j.1469-8986.1979.tb02993.x. ISSN 0048-5772
Pèpin, J.-L., et al.: Pulse transit time improves detection of sleep respiratory events and microarousals in children. CHEST 127(3), 722–730 (2005). https://doi.org/10.1378/chest.127.3.722. ISSN 0012-3692
García, M.T.G., et al.: Can pulse transit time be useful for detecting hypertension in patients in a sleep unit? Archivos de Bronconeumología (English Ed.) 50(7), 278–284 (2014). https://doi.org/10.1016/j.arbr.2014.05.001. ISSN 15792129
Elgendi, M., et al.: The use of photoplethysmography for assessing hypertension. NPJ Digital Med. 2(1), 60 (2019). https://doi.org/10.1038/s41746-019-0136-7. ISSN 2398-6352
Nabeel, P.M., Joseph, J., Sivaprakasam, M.: Arterial compliance probe for local blood pulse wave velocity measurement, vol. 2015 (2015). https://doi.org/10.1109/EMBC.2015.7319689
Samaniego, N.C., Morris, F., Brady, W.J.: Electrocardiographic artefact mimicking arrhythmic change on the ECG. Emerg. Med. J. 20(4), 356–357 (2003). https://doi.org/10.1136/emj.20.4.356
Anton, O., Fernandez, R., Rendon-Morales, E., Aviles-EspinosaÂ, R., Jordan, H., Rabe, H.: Heart rate monitoring in newborn babies: a systematic review. Neonatology 116(3) (2019). https://doi.org/10.1159/000499675
Pollreisz, D., TaheriNejad, N.: Detection and removal of motion artifacts in PPG signals. Mob. Netw. Appl (2019). https://doi.org/10.1007/s11036-019-01323-6. ISSN 1572-8153
Ponnle, A., Ogundepo, O.: Development of a computer-aided application for analyzing ECG signals and detection of cardiac arrhythmia using back propagation neural network-part I: model development. Int. J. Appl. Inf. Syst. 9 (2015). https://doi.org/10.5120/ijais15-451378
van Velzen, M.H.N., Loeve, A.J., Niehof, S.P., Mik, E.G.: Increasing accuracy of pulse transit time measurements by automated elimination of distorted photoplethysmography waves. Med. Biol. Eng. Comput. 55(11), 1989–2000 (2017). https://doi.org/10.1007/s11517-017-1642-x
Kortekaas, M., Niehof, S., Van Velzen, M., Galvin, E., Huygen, F., Stolker, R.: Pulse transit time as a quick predictor of a successful axillary brachial plexus block. Acta Anaesthesiologica Scandinavica 56, 1228–1233 (2012). https://doi.org/10.1111/j.1399-6576.2012.02746.x
Foo, J.Y.A., et al.: Effects of poorly perfused peripheries on derived transit time parameters of the lower and upper limbs. Generic (2008). https://doi.org/10.1515/BMT.2008.023
Foo, J.Y.A., Lim, C.S.: Difference in pulse transit time between populations: a comparison between Caucasian and Chinese children in Australia. J. Med. Eng. Technol. 32(2), 162–166 (2008). https://doi.org/10.1080/03091900600632694. ISSN 0309-1902
Foo, J.Y.A., Lim, C.S.: Difference in pulse transit time between populations: a comparison between Caucasian and Chinese children in Australia. Generic (2007). https://doi.org/10.1515/BMT.2007.043
Foo, J.Y.A.: Normality of upper and lower peripheral pulse transit time of normotensive and hypertensive children. J. Clin. Monit. Comput. 21, 243–248 (2007). https://doi.org/10.1007/s10877-007-9080-1
Singha Roy, M., Gupta, R., Chandra, J.K., Das Sharma, K., Talukdar, A.: Improving photoplethysmographic measurements under motion artifacts using artificial neural network for personal healthcare. IEEE Trans. Instrum. Meas. 67(12), 2820–2829 (2018). https://doi.org/10.1109/TIM.2018.2829488. ISSN 1557-9662
Mehrgardt, P., Zandavi, S.M., Poon, S.K., Kim, J., Markoulli, M., Khushi, M.: U-Net segmented adjacent angle detection (USAAD) for automatic analysis of corneal nerve structures. Data 5(2) (2020). https://doi.org/10.3390/data5020037
van Gent, P., Farah, H., Nes, N., Arem, B.: Analysing noisy driver physiology real-time using off-the-shelf sensors: heart rate analysis software from the taking the fast lane project (2018). https://doi.org/10.13140/RG.2.2.24895.56485
van Gent, P., Farah, H., Nes, N., Arem, B.: Heart rate analysis for human factors: development and validation of an open source toolkit for noisy naturalistic heart rate data (2018)
Kohler, B., Hennig, C., Orglmeister, R.: The principles of software QRS detection. IEEE Eng. Med. Biol. Mag. 21(1), 42–57 (2002). https://doi.org/10.1109/51.993193. ISSN 1937-4186
Pan, J., Tompkins, W.J.: A real-time QRS detection algorithm. IEEE Trans. Biomed. Eng. BME–32(3), 230–236 (1985). https://doi.org/10.1109/TBME.1985.325532
Gao, M., Bari Olivier, N., Mukkamala, R.: Comparison of noninvasive pulse transit time estimates as markers of blood pressure using invasive pulse transit time measurements as a reference. Physiol. Rep. 4(10), e12768 (2016). https://doi.org/10.14814/phy2.12768. ISSN 2051-817X
Pimentel, M.A.F., et al.: Toward a robust estimation of respiratory rate from pulse oximeters. IEEE Trans. Biomed. Eng. 64(8), 1914–1923 (2017). https://doi.org/10.1109/TBME.2016.2613124. ISSN 1558-2531
Rajala, S., Ahmaniemi, T., Lindholm, H., Taipalus, T.: Pulse arrival time (PAT) measurement based on arm ECG and finger PPG signals - comparison of PPG feature detection methods for PAT calculation, vol. 2017 (2017). https://doi.org/10.1109/EMBC.2017.8036809
Becker, D.: Fundamentals of electrocardiography interpretation. Anesth. Prog. 53, 53–63 (2006). https://doi.org/10.2344/0003-3006(2006)53[53:FOEI]2.0.CO;2. quiz 64
Orphanidou, C., Bonnici, T., Charlton, P., Clifton, D., Vallance, D., Tarassenko, L.: Signal-quality indices for the electrocardiogram and photoplethysmogram: derivation and applications to wireless monitoring. IEEE J. Biomed. Health Inform. 19(3), 832–838 (2015). https://doi.org/10.1109/JBHI.2014.2338351. ISSN 2168-2208
Lin, Y.-T., Lo, Y.-L., Lin, C.-Y., Frasch, M.G., Wu, H.-T.: Unexpected sawtooth artifact in beat-to-beat pulse transit time measured from patient monitor data. PLOS ONE 14(9), e0221319 (2019). https://doi.org/10.1371/journal.pone.0221319
Acknowledgements
This work was supported by the University of Sydney Cardiovascular Initiative funding. Dr Withana is the recipient of an Australian Research Council Discovery Early Career Award (DE200100479) funded by the Australian Government.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 Springer Nature Switzerland AG
About this paper
Cite this paper
Mehrgardt, P., Khushi, M., Withana, A., Poon, S. (2020). Machine Learned Pulse Transit Time (MLPTT) Measurements from Photoplethysmography. In: Yang, H., Pasupa, K., Leung, A.CS., Kwok, J.T., Chan, J.H., King, I. (eds) Neural Information Processing. ICONIP 2020. Lecture Notes in Computer Science(), vol 12534. Springer, Cham. https://doi.org/10.1007/978-3-030-63836-8_5
Download citation
DOI: https://doi.org/10.1007/978-3-030-63836-8_5
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-63835-1
Online ISBN: 978-3-030-63836-8
eBook Packages: Computer ScienceComputer Science (R0)