Abstract
Identification of drug-disease associations play an important role for expediting drug development. In comparison with biological experiments for drug repositioning, computational methods may reduce costs and shorten the development cycle. Thus, a number of computational approaches have been proposed for drug repositioning recently. In this study, we develop a novel computational model WGMFDDA to infer potential drug-disease association using weighted graph regularized matrix factorization (WGMF). Firstly, the disease similarity and drug similarity are calculated on the basis of the medical description information of diseases and chemical structures of drugs, respectively. Then, weighted \( K \)-nearest neighbor is implemented to reformulate the drug-disease association adjacency matrix. Finally, the framework of graph regularized matrix factorization is utilized to reveal unknown associations of drug with disease. To evaluate prediction performance of the proposed WGMFDDA method, ten-fold cross-validation is performed on Fdataset. WGMFDDA achieves a high AUC value of 0.939. Experiment results show that the proposed method can be used as an efficient tool in the field of drug-disease association prediction, and can provide valuable information for relevant biomedical research.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
New drug research and development is still a time-consuming, high-risky and tremendously costly process [1,2,3,4]. Although the investment in new drug research and development has been increasing, the number of new drugs approved by the US Food and Drug Administration (FDA) has remained limited in the past few decades [5,6,–7]. Therefore, more and more biomedical researchers and pharmaceutical companies are paying attention to the repositioning for existing drugs, which aims to infer the new therapeutic uses for these drugs [8,9,10,11]. For example, Thalidomide, and Minoxidil, were repositioned as a treatment to insomnia and the androgenic alopecia, respectively [12,13,14,15]. In other words, drug repositioning is actually to infer and discover potential drug-disease associations [16].
Recently, some computational methods have been presented to identify associations of drugs with diseases, such as deep walk embedding [17, 18], rotation forest [19,20,21,22], network analysis [23,24,25], text mining [26, 27] and machine learning [28,29,30,31], etc. Martínez et al. proposed a new approach named DrugNet, which performs disease-drug and drug-disease prioritization by constructing a heterogeneous network of interconnected proteins, drugs and diseases [32]. Wang et al. developed a triple-layer heterogeneous network model called TL-HGBI to infer drug-disease potential associations [33]. The network integrates association data and similarity about targets, drugs and diseases. Luo et al. utilized Bi-Random walk algorithm and comprehensive similarity measures (MBiRW) to infer new indications for existing drugs [34]. In fact, predicting associations of drug with disease can be transformed into a recommendation system problem [35,36,37,38]. Luo et al. developed a drug repositioning recommendation system (DRRS) to identify new indications for a given drug [39]. In this work, we develop a novel computational model WGMFDDA, which utilizes graph regularized matrix factorization to infer the potential associations between drugs and diseases. The experiment results indicate that the performance of WGMFDDA is better than other compared methods.
2 Methods and Materials
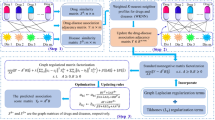
2.1 Method Overview
To predict potential associations of drugs with diseases, the model of WGMFDDA consists of three steps (See Fig. 1): (1) we measure the similarity for drugs and diseases based on the collected dataset; (2) According to the weighted \( {\text{K}} \)-nearest neighbor profiles of drugs and diseases, the drug-disease association adjacency matrix is re-established; (3) the graph Laplacian regularization and Tikhonov (\( {\text{L}}_{2} \)) terms are incorporated into the standard Non-negative matrix factorization (NMF) framework to calculate the drug-disease association scores.
2.2 Dataset
In this study, we obtain the dataset (Fdataset) from Gottlieb et al. [40]. This dataset is used as the gold standard datasets for identifying drug-disease associations, which includes 1933 known associations between 313 diseases and 593 drugs [41, 42]. In order to more conveniently describe the drug-disease associations information, the drug-disease association adjacency matrix \( Y^{n \times m} \) is constructed, where \( n \) and \( m \) are the number of drugs and diseases, respectively. The element \( Y\left( {i,j} \right) = 1 \) if drug \( r_{i} \) associated with disease \( d_{j} \), otherwise \( Y\left( {i,j} \right) = 0 \). The similarities for drugs and diseases are obtained from the Chemical Development Kit (CDK) [43] based on SMILES [44] and MimMiner [45] based on the OMIM [41] database, respectively. In ten-fold cross-validation experiments, all known associations are random divided into ten equal sized subsets, in which the training data set occupies 9/10, and the remaining partition is utilized as the test set.
2.3 Reformulate the Drug-Disease Association Adjacency Matrix
Let \( R = \left\{ {r_{1} ,r_{2} , \cdots ,r_{n} } \right\} \) and \( D = \left\{ {d_{1} ,d_{2} , \cdots ,d_{m} } \right\} \) are the set of \( n \) drugs and \( m \) diseases. \( Y\left( {r_{i} } \right) = \left( {Y_{i1} ,Y_{i2} , \cdots ,Y_{im} } \right) \) and \( Y\left( {d_{j} } \right) = \left( {Y_{1j} ,Y_{2j} , \cdots ,Y_{nj} } \right) \) are the \( i{\text{th}} \) row vector and \( j{\text{th}} \) column vector of matrix \( Y \), respectively. \( Y\left( {r_{i} } \right) \) and \( Y\left( {d_{j} } \right) \) denote the interaction profiles of drugs and diseases, respectively. Since many drug-disease pairs with unknown associations (i.e. the value of these elements in \( Y \) is zero) may be potential true associations, this will affect prediction performance. In order to assign associated likelihood scores to drug-disease pairs with unknown associations, weighted \( K \)-nearest neighbor (WKNN) is implemented to calculate new interaction profiles of drugs and diseases [38, 46].
For each drug \( r_{p} \) (or disease \( d_{q} \)), the novel interaction profile can be calculated as follows:
or
\( a \in \left[ {0,\,1} \right] \) denotes a decay term. \( S^{R} \) and \( S^{D} \) are the similarity matrices for drugs and diseases, respectively.
Subsequently, we define the updated association adjacency matrix \( Y \) as follows:
where
2.4 WGMFDDA
The standard Nonnegative matrix factorization (NMF) aims to find two low-rank Nonnegative matrices whose product as more as possible to approximation to the original matrix [36, 47,48,49]. \( Y \cong A^{T} B\left( {k \le { \hbox{min} }\left( {n,m} \right)} \right) \), \( A \in R^{k \times n} \) and \( B \in R^{k \times m} \). To avoid overfitting, the graph Laplacian regularization and Tikhonov (\( L_{2} \)) terms are introduced into the standard NMF model. The objective function of WGMFDDA can be constructed as follows:
where \( \left\| \cdot \right\|_{F} \) denotes the Frobenius norm. \( \lambda \) and \( \beta \) are the regularization parameters. \( a_{j} \) and \( b_{j} \) are \( j{\text{th}} \) column of matrices \( A \) and \( B \), respectively. \( S^{R*} \) and \( S^{D*} \) denote the sparse similarity matrices for drugs and diseases, respectively.
According to the spectral graph theory, the \( p \)-nearest neighbor graph can preserve the intrinsic geometrical structure of the original data [46]. Therefore, \( p \)-nearest neighbors is utilized to construct the graphs \( S^{R*} \) and \( S^{D*} \). The details are as follows:
where \( N_{p} \left( {r_{i} } \right) \) and \( N_{p} \left( {r_{j} } \right) \) denote the sets of \( p \)-nearest neighbors of \( r_{i} \) and \( r_{j} \) respectively. Then, we define the sparse matrix \( S^{R*} \) of drug as follows:
Similarly, the sparse matrix \( S^{D*} \) of disease can be expressed as follows:
The Eq. (5) can be written as:
Here, \( L_{r} = D_{r} - S^{R*} \) and \( L_{d} = D_{d} - S^{D*} \) are the graph Laplacian matrices for \( S^{R*} \) and \( S^{D*} \), respectively. \( D_{r} \left( {i,i} \right) = \sum\nolimits_{p} {S_{ip}^{R*} } \) and \( D_{d} \left( {j,j} \right) = \sum\limits_{q} {S_{jq}^{D*} } \) are diagonal matrices, \( Tr\left( \cdot \right) \) denotes the trace of matrix.
In order to optimize the objective function in Eq. (9), the corresponding Lagrange function \( {\mathcal{H}}_{f} \) is defined as:
In which, \( \varPhi = \left\{ {\phi_{ki} } \right\} \) and \( \varPsi = \left\{ {\psi_{kj} } \right\} \) are Lagrange multipliers that constrain \( a_{ki} \ge 0 \) and \( b_{kj} \ge 0 \), respectively. We calculate \( \frac{{\partial {\mathcal{H}}_{f} }}{\partial A} \) and \( \frac{{\partial {\mathcal{H}}_{f} }}{\partial B} \) as follows:
After using Karush–Kuhn–Tucker (KKT) conditions \( \phi_{ki} a_{ki} = 0 \) and \( \psi_{kj} b_{kj} = 0 \), the updating rules can be obtained as follows:
The predicted drug-disease association matrix is obtained by \( Y^{*} = A^{T} B \). Generally, the larger the element value in predicted matrix \( Y^{*} \), the more likely the drug is related to the corresponding disease.
3 Experimental Results
In this study, the model of WGMFDDA has six parameters that determine by grid search. The ROC curve and AUC value are widely used to evaluate the predictor [50,51,52,53,54]. WGMFDDA produces best AUC values when \( P = 5 \), \( K = 5 \), \( a = 0.5 \), \( k = 160 \), \( \lambda = 1 \) and \( \beta = 0.02 \). We implement ten-fold cross-validation (CV) experiments on the Fdataset and compare it with the previous methods: DrugNet [32], HGBI [33], MBiRW [34] and DDRS [39]. To implement 10-CV experiment, all known drug-disease associations in Fdataset are random divided into ten equal sized subsets. the training data set occupies 9/10, while the remaining partition is utilized as the test set. As shown in Fig. 2 and Table 1, WGMFDDA achieves the AUC value of 0.939, while DrugNet, HGBI, MBiRW and DDRS are 0.778, 0.829, 0.917and 0.930, respectively. This result shows that compared with DDRS, MBiRW, HGBI and DrugNet, WGMFDDA obtains the best performance.
4 Conclusions
The purpose of drug repositioning is to discover new indications for existing drugs. Compared to traditional drug development, drug repositioning can reduce risk, save time and costs. In this work, we present a new prediction approach, WGMFDDA, based on weighted graph regularized matrix factorization. The proposed method casts the problem of inferring the associations between drugs and diseases into a matrix factorization problem in recommendation system. The main contribution of our method is that a preprocessing step is performed before matrix factorization to reformulate the drug-disease association adjacency matrix. In ten-fold cross-validation, experiment results indicate that our proposed model outperforms other compared methods.
References
Li, J., Zheng, S., Chen, B., Butte, A.J., Swamidass, S.J., Lu, Z.: A survey of current trends in computational drug repositioning. Briefings Bioinform. 17, 2–12 (2016)
Huang, Y.-A., Hu, P., Chan, K.C., You, Z.-H.: Graph convolution for predicting associations between miRNA and drug resistance. Bioinformatics 36, 851–858 (2020)
Chen, Z.-H., You, Z.-H., Guo, Z.-H., Yi, H.-C., Luo, G.-X., Wang, Y.-B.: Prediction of drug-target interactions from multi-molecular network based on deep walk embedding model. Front. Bioeng. Biotechnol. 8, 338 (2020)
Wang, L., You, Z.-H., Li, L.-P., Yan, X., Zhang, W.: Incorporating chemical sub-structures and protein evolutionary information for inferring drug-target interactions. Sci. Rep. 10, 1–11 (2020)
Kinch, M.S., Griesenauer, R.H.: 2017 in review: FDA approvals of new molecular entities. Drug Discovery Today 23, 1469–1473 (2018)
Wang, L., et al.: Identification of potential drug–targets by combining evolutionary information extracted from frequency profiles and molecular topological structures. Chem. Biol. Drug Des. (2019)
Jiang, H.-J., You, Z.-H., Huang, Y.-A.: Predicting drug − disease associations via sigmoid kernel-based convolutional neural networks. J. Transl. Med. 17, 382 (2019)
Hurle, M., Yang, L., Xie, Q., Rajpal, D., Sanseau, P., Agarwal, P.: Computational drug repositioning: from data to therapeutics. Clin. Pharmacol. Ther. 93, 335–341 (2013)
Huang, Y.-A., You, Z.-H., Chen, X.: A systematic prediction of drug-target interactions using molecular fingerprints and protein sequences. Curr. Protein Pept. Sci. 19, 468–478 (2018)
Wang, L., You, Z.-H., Chen, X., Yan, X., Liu, G., Zhang, W.: Rfdt: a rotation forest-based predictor for predicting drug-target interactions using drug structure and protein sequence information. Curr. Protein Pept. Sci. 19, 445–454 (2018)
Li, Y., Huang, Y.-A., You, Z.-H., Li, L.-P., Wang, Z.: Drug-target interaction prediction based on drug fingerprint information and protein sequence. Molecules 24, 2999 (2019)
Graul, A.I., et al.: The year’s new drugs & biologics-2009. Drug News Perspect 23, 7–36 (2010)
Sardana, D., Zhu, C., Zhang, M., Gudivada, R.C., Yang, L., Jegga, A.G.: Drug repositioning for orphan diseases. Briefings Bioinform. 12, 346–356 (2011)
Zhang, S., Zhu, Y., You, Z., Wu, X.: Fusion of superpixel, expectation maximization and PHOG for recognizing cucumber diseases. Comput. Electron. Agric. 140, 338–347 (2017)
Li, Z., et al.: In silico prediction of drug-target interaction networks based on drug chemical structure and protein sequences. Sci. Rep. 7, 1–13 (2017)
Zheng, K., You, Z.-H., Wang, L., Zhou, Y., Li, L.-P., Li, Z.-W.: Dbmda: a unified embedding for sequence-based mirna similarity measure with applications to predict and validate mirna-disease associations. Mol. Therapy-Nucleic Acids 19, 602–611 (2020)
Guo, Z., Yi, H., You, Z.: Construction and comprehensive analysis of a molecular association network via lncRNA–miRNA–disease–drug–protein graph. Cells 8, 866 (2019)
Chen, Z., You, Z., Zhang, W., Wang, Y., Cheng, L., Alghazzawi, D.: Global vectors representation of protein sequences and its application for predicting self-interacting proteins with multi-grained cascade forest model. Genes 10, 924 (2019)
You, Z.-H., Chan, K.C., Hu, P.: Predicting protein-protein interactions from primary protein sequences using a novel multi-scale local feature representation scheme and the random forest. PLoS ONE 10, e0125811 (2015)
Guo, Z., You, Z., Wang, Y., Yi, H., Chen, Z.: A learning-based method for LncRNA-disease association identification combing similarity information and rotation forest. iScience 19, 786–795 (2019)
Wang, L., et al.: Using two-dimensional principal component analysis and rotation forest for prediction of protein-protein interactions. Sci. Rep. 8, 1–10 (2018)
You, Z., Li, X., Chan, K.C.C.: An improved sequence-based prediction protocol for protein-protein interactions using amino acids substitution matrix and rotation forest ensemble classifiers. Neurocomputing 228, 277–282 (2017)
Oh, M., Ahn, J., Yoon, Y.: A network-based classification model for deriving novel drug-disease associations and assessing their molecular actions. PLoS ONE 9, e111668 (2014)
Zheng, K., You, Z.-H., Li, J.-Q., Wang, L., Guo, Z.-H., Huang, Y.-A.: iCDA-CGR: identification of circRNA-disease associations based on Chaos Game Representation. PLoS Comput. Biol. 16, e1007872 (2020)
Yi, H.-C., You, Z.-H., Guo, Z.-H.: Construction and analysis of molecular association network by combining behavior representation and node attributes. Front. Genet. 10, 1106 (2019)
Yang, H., Spasic, I., Keane, J.A., Nenadic, G.: A text mining approach to the prediction of disease status from clinical discharge summaries. J. Am. Med. Inform. Assoc. 16, 596–600 (2009)
Chen, Z.-H., Li, L.-P., He, Z., Zhou, J.-R., Li, Y., Wong, L.: An improved deep forest model for predicting self-interacting proteins from protein sequence using wavelet transformation. Front. Genet. 10, 90 (2019)
Li, L., Wang, Y., You, Z., Li, Y., An, J.: PCLPred: a bioinformatics method for predicting protein-protein interactions by combining relevance vector machine model with low-rank matrix approximation. Int. J. Mol. Sci. 19, 1029 (2018)
Li, S., You, Z.-H., Guo, H., Luo, X., Zhao, Z.-Q.: Inverse-free extreme learning machine with optimal information updating. IEEE Trans. Cybern. 46, 1229–1241 (2015)
Zheng, K., You, Z.-H., Wang, L., Zhou, Y., Li, L.-P., Li, Z.-W.: MLMDA: a machine learning approach to predict and validate MicroRNA–disease associations by integrating of heterogenous information sources. J. Transl. Med. 17, 260 (2019)
Yi, H.-C., You, Z.-H., Wang, M.-N., Guo, Z.-H., Wang, Y.-B., Zhou, J.-R.: RPI-SE: a stacking ensemble learning framework for ncRNA-protein interactions prediction using sequence information. BMC Bioinform. 21, 60 (2020)
Martinez, V., Navarro, C., Cano, C., Fajardo, W., Blanco, A.: DrugNet: network-based drug–disease prioritization by integrating heterogeneous data. Artif. Intell. Med. 63, 41–49 (2015)
Wang, W., Yang, S., Zhang, X., Li, J.: Drug repositioning by integrating target information through a heterogeneous network model. Bioinformatics 30, 2923–2930 (2014)
Luo, H., Wang, J., Li, M., Luo, J., Peng, X., Wu, F.-X., Pan, Y.: Drug repositioning based on comprehensive similarity measures and bi-random walk algorithm. Bioinformatics 32, 2664–2671 (2016)
You, Z., Wang, L., Chen, X., Zhang, S., Li, X., Yan, G., Li, Z.: PRMDA: personalized recommendation-based MiRNA-disease association prediction. Oncotarget 8, 85568–85583 (2017)
Wang, M.-N., You, Z.-H., Li, L.-P., Wong, L., Chen, Z.-H., Gan, C.-Z.: GNMFLMI: graph regularized nonnegative matrix factorization for predicting LncRNA-MiRNA interactions. IEEE Access 8, 37578–37588 (2020)
Huang, Y., You, Z., Chen, X., Huang, Z., Zhang, S., Yan, G.: Prediction of microbe-disease association from the integration of neighbor and graph with collaborative recommendation model. J. Transl. Med. 15, 1–11 (2017)
Wang, M.-N., You, Z.-H., Wang, L., Li, L.-P., Zheng, K.: LDGRNMF: LncRNA-disease associations prediction based on graph regularized non-negative matrix factorization. Neurocomputing (2020)
Luo, H., Li, M., Wang, S., Liu, Q., Li, Y., Wang, J.: Computational drug repositioning using low-rank matrix approximation and randomized algorithms. Bioinformatics 34, 1904–1912 (2018)
Gottlieb, A., Stein, G.Y., Ruppin, E., Sharan, R.: PREDICT: a method for inferring novel drug indications with application to personalized medicine. Mol. Syst. Biol. 7, 496 (2011)
Hamosh, A., Scott, A.F., Amberger, J.S., Bocchini, C.A., McKusick, V.A.: Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 33, D514–D517 (2005)
Wishart, D.S., et al.: DrugBank: a comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res. 34, D668–D672 (2006)
Steinbeck, C., Han, Y., Kuhn, S., Horlacher, O., Luttmann, E., Willighagen, E.: The Chemistry Development Kit (CDK): an open-source Java library for chemo-and bioinformatics. J. Chem. Inf. Comput. Sci. 43, 493–500 (2003)
Weininge, D.: SMILES, a chemical language and information system. 1. Introduction to methodology and encoding rules. J. Chem. Inf. Comput. Sci. 28, 31–36 (1988)
Van Driel, M.A., Bruggeman, J., Vriend, G., Brunner, H.G., Leunissen, J.A.: A text-mining analysis of the human phenome. Eur. J. Hum. Genet. 14, 535–542 (2006)
Xiao, Q., Luo, J., Liang, C., Cai, J., Ding, P.: A graph regularized non-negative matrix factorization method for identifying microRNA-disease associations. Bioinformatics 34, 239–248 (2018)
Huang, Y.-A., You, Z.-H., Li, X., Chen, X., Hu, P., Li, S., Luo, X.: Construction of reliable protein–protein interaction networks using weighted sparse representation based classifier with pseudo substitution matrix representation features. Neurocomputing 218, 131–138 (2016)
Chen, Z.-H., You, Z.-H., Li, L.-P., Wang, Y.-B., Wong, L., Yi, H.-C.: Prediction of self-interacting proteins from protein sequence information based on random projection model and fast fourier transform. Int. J. Mol. Sci. 20, 930 (2019)
Ji, B.-Y., You, Z.-H., Cheng, L., Zhou, J.-R., Alghazzawi, D., Li, L.-P.: Predicting miRNA-disease association from heterogeneous information network with GraRep embedding model. Sci. Rep. 10, 1–12 (2020)
Chen, Z.-H., You, Z.-H., Li, L.-P., Wang, Y.-B., Qiu, Y., Hu, P.-W.: Identification of self-interacting proteins by integrating random projection classifier and finite impulse response filter. BMC Genom. 20, 1–10 (2019)
Chen, X., Yan, C.C., Zhang, X., You, Z.-H.: Long non-coding RNAs and complex diseases: from experimental results to computational models. Briefings Bioinform. 18, 558–576 (2017)
Jiao, Y., Du, P.: Performance measures in evaluating machine learning based bioinformatics predictors for classifications. Quant. Biol. 4(4), 320–330 (2016). https://doi.org/10.1007/s40484-016-0081-2
Chen, X., Huang, Y.-A., You, Z.-H., Yan, G.-Y., Wang, X.-S.: A novel approach based on KATZ measure to predict associations of human microbiota with non-infectious diseases. Bioinformatics 33, 733–739 (2017)
You, Z.-H., Huang, Z.-A., Zhu, Z., Yan, G.-Y., Li, Z.-W., Wen, Z., Chen, X.: PBMDA: a novel and effective path-based computational model for miRNA-disease association prediction. PLoS Comput. Biol. 13, e1005455 (2017)
Acknowledgement
This work was supported in part by the NSFC Excellent Young Scholars Program, under Grants 61722212, in part by the Science and Technology Project of Jiangxi Provincial Department of Education, under Grants GJJ180830, GJJ190834.
Competing Interests
The authors declare that they have no competing interests.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 Springer Nature Switzerland AG
About this paper
Cite this paper
Wang, MN., You, ZH., Li, LP., Chen, ZH., Xie, XJ. (2020). WGMFDDA: A Novel Weighted-Based Graph Regularized Matrix Factorization for Predicting Drug-Disease Associations. In: Huang, DS., Premaratne, P. (eds) Intelligent Computing Methodologies. ICIC 2020. Lecture Notes in Computer Science(), vol 12465. Springer, Cham. https://doi.org/10.1007/978-3-030-60796-8_47
Download citation
DOI: https://doi.org/10.1007/978-3-030-60796-8_47
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-60795-1
Online ISBN: 978-3-030-60796-8
eBook Packages: Computer ScienceComputer Science (R0)