Abstract
Bacterial microcompartments (BMCs) are polyhedral metabolic organizers in bacteria that consist of a thin, selectively permeable protein boundary and a set of encapsulated enzymes. Most BMCs participate in the catabolism of specific carbon sources; the carboxysome is the only anabolic BMC and promotes efficient fixation of CO2 by ribulose 1,5-bisphosphate carboxylase/oxygenase (RubisCO) in many autotrophic bacteria. Despite the diverse functions of BMCs, commonalities among them are the oxygen sensitivity of one of the cargo enzymes and the volatility of intermediates that are generated in their interior. The BMC shell prevents the escape of these metabolites, thereby ensures their availability for downstream reactions within the BMC lumen and averts toxic effects on other cell components. In this chapter, we discuss recent advances that have led to new insights into BMC structure, function and cellular interactions.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
1 Introduction
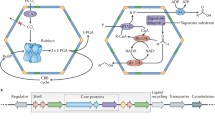
Many bacteria segregate selected metabolic steps into intracellular compartments, termed bacterial microcompartments (BMCs), that are separated from the cytosol by a semipermeable molecular boundary. Functionally analogous to eukaryotic organelles, BMCs, however, lack a lipid bilayer-based permeability barrier and are instead delimited by a thin (2–4 nm) protein shell that determines their polyhedral shape (Fig. 1) and serves to enhance metabolic efficiency of the enzymatic steps within their interior in part through control of metabolite access to and exit from the lumen. Genes for BMC proteins exist in approximately a fifth to a fourth of all sequenced bacterial genomes. Apart from the anabolic carboxysomes of photosynthetic cyanobacteria and some chemoautotrophs, all other known BMCs, often termed metabolosomes, harbour enzymes that aid in the catabolism of specialty carbon sources (Table 1 and reviewed in (Kirst and Kerfeld 2019; Kerfeld et al. 2018)).
The protein shell of BMCs constitutes a permeability barrier for small, volatile metabolites that are formed in their lumen, such as the aldehydes generated in the catabolic BMCs. Limiting their diffusion into the cytosol allows these intermediates to be efficiently processed further within the BMC interior and prevents them from exerting undesirable, often toxic effects on the cell. Likewise, the shell of the only known anabolic BMC, the carboxysome, ensures that CO2, the substrate of ribulose 1,5-bisphosphate carboxylase/oxygenase (RubisCO) that is generated in the lumen by the carboxysomal carbonic anhydrase (CA), remains trapped in the organelle and thereby supports effective fixation by the otherwise catalytically inefficient RubisCO (Dou et al. 2008).
All BMC shells are constructed from the same, structurally conserved basic building modules: three types of small proteins that readily assemble into disc-shaped oligomers of characteristic stoichiometries (Fig. 2). Hexamers of highly abundant pfam00936 proteins (BMC-H) tile into the single layers that form the bulk of the polyhedral BMC facets (Kerfeld et al. 2005; Tanaka et al. 2008; Tsai et al. 2007; Crowley et al. 2008; Mallette and Kimber 2017; Takenoya et al. 2010; Tanaka et al. 2010). Interspersed within the hexamer layer are trimers (Sagermann et al. 2009; Crowley et al. 2010; Heldt et al. 2009; Pang et al. 2011; Tanaka et al. 2010; Takenoya et al. 2010) or stacked dimers of trimers (Cai et al. 2013; Klein et al. 2009; Mallette and Kimber 2017) formed by paralogs (BMC-T) with tandem pfam00936 domains. The vertices of the polyhedral BMC shell are closed by pentamers of pfam03319 proteins (BMC-P) (Tanaka et al. 2008; Cai et al. 2009; Wheatley et al. 2013; Keeling et al. 2014; Mallette and Kimber 2017). Additional shell-associated proteins exist that are BMC type-specific and play a role in assembly and/or function of the organelle (see BMC Superloci).
Schematic of a generic BMC shell and its protein components. BMC-H (teal) = pfam00936 protein hexamers forming the BMC facets; BMC-P (yellow) = pfam03319 protein pentamers at the BMC vertices; BMC-T (purple) = tandem pfam00936 protein stacked dimers of trimers interspersed within the BMC-H hexamers. The image was generated by Markus Sutter and Cheryl Kerfeld
Since BMCs have been the subject of several recent reviews (Heinhorst et al. 2014; Kirst and Kerfeld 2019; Kerfeld et al. 2018; Sommer et al. 2017; Yeates et al. 2013), Table 1 provides an overview of BMCs whose functions are either known or have been inferred bioinformatically or through metabolic studies. The remainder of this chapter focuses on recent advances that have significantly enhanced our understanding of BMC structure, function and interaction with other cell components.
2 Shell Structure and Function
Metabolite Transfer
Since the structure of the BMC shell proteins was first elucidated and shell models were established (Kerfeld et al. 2005; Tanaka et al. 2008, 2010), the central pores in the oligomeric assemblies of BMC proteins have been viewed as the most likely candidates for mediating and/or regulating metabolite exchange across the shell. Pore size, geometry and surface charge vary between oligomers of different paralogs and orthologs and thereby provide selectivity for the metabolite(s) that are able to traverse the shell of the various BMC types. Lending indirect support to the proposed role of the pores as conduits for small metabolites are the anions (Tsai et al. 2007; Tanaka et al. 2009; Takenoya et al. 2010) and substrate molecules (Pang et al. 2012) that were shown to occupy the pores in various BMC protein crystals. Furthermore, molecular dynamics simulations of substrate (1,2 propanediol) and product (propionaldehyde) diffusion across the pores of the PduA hexamers of the Pdu (propanediol utilization) BMC predict a higher energy barrier for the toxic aldehyde intermediate than for the substrate (Park et al. 2017).
To address the role of the pores in metabolite transfer across the BMC shell in vivo, Choudhury and colleagues (2015) created a series of Salmonella enterica mutants in which PduA was altered to yield hexamers with occluded or less polar pore openings. These changes affect the diffusion rate of 1,2-propanediol into and/or that of propionaldehyde out of the Pdu BMC. A point mutation that changes the pore lining and renders the PduA pore characteristics similar to those of the Pdu BMC of lactobacilli also allows passage of the substrate glycerol (metabolized in the Pdu BMC of lactoballi but not by that of S. enterica) into the Pdu BMC of the resulting S. enterica mutant. Slininger Lee et al. (2017) expanded these mutant studies by creating a library of 15 PduA pore mutants in which a single pore residue at a key position is replaced with a different amino acid. The authors were able to correlate the resulting mutant growth phenotypes with the characteristics of the substituent amino acid and identified side chain charge and hydrophobicity as crucial determinants for the retention of propionaldehyde in the lumen of the Pdu BMC.
Recombinant, chimeric Pdu BMCs in which the Eut (ethanolamine utilization) BMC shell protein EutM substitutes for PduA appear to be morphologically and compositionally similar to wild-type Pdu BMCs. However, despite their structural similarity, PduA and EutM have different pores: lysine residues line the central pore of PduA hexamers, and glutamine is found in EutM pores. The chimeric Pdu BMC shows improved utilization of 1,2 propanediol, which the authors attribute to more effective retention of luminal metabolites and their increased availability for downstream reactions within the organelle (Slininger Lee et al. 2017).
Similar mutant studies that assess pore permeability have not yet been reported for carboxysomes; however, Mahintchichaichan et al. (Mahinthichaichan et al. 2018) used molecular dynamics simulations and free energy calculations to assess the permeability for HCO3 −, CO2 and O2 of hexamer pores formed by the α- and β-carboxysome shell proteins CsoS1A from Halothiobacillus neapolitanus and CcmK4 from Synechocystis sp. PCC 6803, respectively. For both pore types, they found preferred interactions between the anion and the positively charged pore surface, corroborating the previously observed selective permeability of the carboxysome shell for bicarbonate over CO2 (Dou et al. 2008). Retention of CO2, which is generated from HCO3 − by the carboxysomal CA and released into the lumen, ensures that RubisCO is supplied with ample amounts of its substrate. Likewise, exclusion of O2 from the carboxysome interior is believed to prevent the unproductive conversion of this competing RubisCO substrate.
Collectively, theoretical and experimental evidence lends strong support to BMC-H shell pores serving as the main conduits for small metabolites between BMC lumen and the exterior. Furthermore, pore properties are specific for the substrates, intermediates and products that are relevant for a particular BMC type, and pore characteristics can be genetically engineered without compromising the structural integrity of the BMC shell. This flexibility opens up the possibility of designing and engineering specific passages for small metabolites that could enhance and/or broaden the range of BMC functions (Cai et al. 2015b; Kirst and Kerfeld 2019; Kerfeld et al. 2018).
In addition to the abundant BMC-H proteins that form the majority of the single-layer shell facets, the trimers and stacked dimers of trimers of BMC-T proteins are also integral components of the BMC shell (Roberts et al. 2012; Cai et al. 2013). Their pores are generally larger than those of the BMC-H hexamers and have unique geometric and charge properties. The double-layered dimers of trimers seen in some BMC-T protein crystals contain an internal channel or nanocompartment with either one or both pores in the closed conformation (Klein et al. 2009; Mallette and Kimber 2017; Cai et al. 2013; Larsson et al. 2017), suggesting that these proteins may have the ability to bind, temporarily store and gate the transfer of larger metabolites. However, to date experimental evidence for substrate traffic though BMC-T proteins is lacking. Considering their very low abundance in the carboxysome shell (Roberts et al. 2012), at least for the carboxysome a role in substrate transfer does not seem likely.
BMC-T protein assemblies might control entry and/or exit of larger regulatory metabolites that affect the reactions within the BMC and serve to align processes in the BMC lumen with the metabolic status of the cell, or the BMC-T pores may serve as gated conduits to replenish larger cofactors (e.g. NAD(P)H, ATP, cobalamin) (Mallette and Kimber 2017; Larsson et al. 2017). The EutL trimers crystallize with either a closed (Sagermann et al. 2009; Takenoya et al. 2010) or an open (Tanaka et al. 2010) central pore. Exposure of closed-pore EutL crystals to Zn2+ induces a conformational change to the open-pore state (Takenoya et al. 2010) that is suggestive of a gating mechanism. The observation that cobalamin, a required cofactor for the catabolism of 1,2-propanediol and ethanolamine in Pdu and Eut BMCs, respectively, binds to Clostridium perfringens EutL assemblies lends support to the proposed role of BMC-T proteins in cofactor replenishment (Thompson et al. 2014). However, the binding of only one ligand per trimer towards the periphery, rather than in the pore, casts doubt on the physiological significance of this association and points to the need for in vivo experiments that can help elucidate the contributions of EutL and other BMC-T proteins to BMC function.
Electron and Proton Permeability
The electron and proton impermeability of the lipid bilayers that bound eukaryotic organelles begs the question whether BMC protein shells display a similar selectivity. Several observations support the possibility that electrons or redox-active metabolites may be able to cross the BMC boundary.
Several shell protein oligomers of catabolic BMCs are associated with FeS clusters (Crowley et al. 2010; Parsons et al. 2008; Pang et al. 2011; Thompson et al. 2014; Ferlez et al. 2019). The BMC-T protein PduT of the Pdu BMC and the BMC-H protein GrpU found in several glycyl radical propanediol (Grp) BMCs have spectral properties that indicate the presence of a 4Fe-4S cluster, a cofactor in 1,2 propanediol metabolism. Modelling results predict a location of the ligand in the central pore. Although the exact mode of action of the redox moiety has not been experimentally ascertained, likely scenarios are a shuttling of electrons or of the entire 4Fe-4S cluster across the BMC shell to support cofactor regeneration within the BMC lumen (Crowley et al. 2010; Parsons et al. 2008; Pang et al. 2011; Thompson et al. 2014).
Penrod and Roth (2006) had proposed that the maintenance of a pH differential between BMC interior and its surroundings could constitute a potential mechanism by which all BMCs are able to enhance the metabolic reactions that take place in their lumen. A lumenal pH that is lower than that of the cytosol would promote catabolism of volatile metabolites in catabolic BMCs and, in the carboxysome, would favour the conversion of bicarbonate to CO2 by the carboxysomal CA. However, work by Menon et al. (Menon et al. 2010) convincingly showed that the α-carboxysome shell is permeable to protons. Because the shells of the various BMCs share identical design principles, this property is thought to apply to all BMC types.
Orientation of Shell Protein Oligomers
The first published model of a BMC shell (Tanaka et al. 2008) predicted that the hexamers of the abundant α-carboxysome shell proteins are all oriented in the same direction; however, the model was unable to resolve the relative orientations of their concave and convex sides. In the CsoS1A hexamer, only the pore portion that faces the concave side is positively charged; the one on the convex side has a negative surface potential and presents a higher energy barrier for entry of HCO3 − into the funnel leading towards the narrowest pore diameter (Tanaka et al. 2008). Based on those characteristics, the authors predicted a shell protein orientation in which the concave side of the hexamer faces outward and facilitates the transfer of HCO3 − into the carboxysome interior. However, size and composition heterogeneity of naturally occurring BMCs have rendered them refractory to crystallization, and to date, the orientation of their shell proteins has not been resolved experimentally.
Using a combination of genes that encode BMC-H, BMC-T and BMC-P shell proteins from Haliangium ochraceum, Sutter et al. (2017) recombinantly expressed uniformly sized synthetic BMCs in E. coli. Their crystal structure corroborates the previously predicted orientation of the carboxysomal protein pentamers (Tanaka et al. 2008) that seal the icosahedral carboxysome shell at the vertices of its flat facets (Cai et al. 2009). Most importantly, the synthetic BMC structure revealed that the concave surface of the hexameric protein assemblies that form the BMC shell facets faces the cytosol. This finding has important implications for the structure and metabolite transfer properties of all BMC types.
BMC-H Heterohexamers
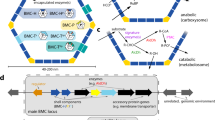
The large majority of gene clusters and operons that encode the various BMC types contain multiple paralogs of BMC-H and BMC-P shell proteins (Axen et al. 2014), raising questions about functional redundancy or complementarity between individual paralogs. Informed by structural characterization of homo-oligomeric crystals of these proteins (Kerfeld et al. 2005; Tanaka et al. 2008; Tsai et al. 2007, 2009; Mallette and Kimber 2017; Cai et al. 2015b), current models of BMC shells are based solely on homo-oligomeric shell proteins. However, the existence of hetero-oligomers in BMC shells has long been considered to be a means by which bacteria could modulate pore properties, broaden the range of metabolites that can cross the shell, and/or regulate small-molecule traffic into and out of the organelle.
Sommer et al. (2019) investigated two BMC-H shell protein paralogs of β-carboxysomes, CcmK3 and CcmK4. Their genes always co-occur in a satellite locus distant from the main ccm gene cluster in the ~90% of cyanobacterial genomes in which they exist (Sommer et al. 2017), which suggests that they are not redundant but functionally connected. Mutant studies in Synechococcus elongatus PCC 7942 and Synechocystis sp. PCC 6803 revealed that CcmK4 is essential for efficient photoautotrophic growth in air (Zhang et al. 2004; Sommer et al. 2019). Both proteins are carboxysome components, but unlike CcmK4 (Kerfeld et al. 2005; Cai et al. 2015b), recombinant CcmK3 does not form homohexamers (Sommer et al. 2019). However, subjecting differentially tagged CcmK protein combinations to sequential affinity purification yielded stable CcmK3/CcmK4 heterohexamers for the orthologs from Halothece PCC 7418 and S. elongatus PCC 7942. Using a similar approach, Garcia-Alles and colleagues (2019) isolated tagged recombinant Synechocystis sp. PCC 6803 CcmK3/CcmK4, as well as CcmK1/CcmK2 heterohexamers. Interestingly, the CcmK3/CcmK4 hexamers also assemble into dodecamers in a pH-dependent manner (Sommer et al. 2019). Because of the poor conservation of residues lining the hetero-hexamer edges and the at-best very low abundance of CcmK3 in purified β-carboxysomes, Sommer et al. (2019) proposed that heterohexamers might be transiently associating with the outer shell surface, forming concave-to-concave side dodecamers with existing hexamers in the shell facets and changing their permeability properties temporarily.
Double-layered Shell Components
Clearly, ultrastructural evidence has established a BMC shell thickness of 2–4 nm, which corresponds to the thickness of many BMC-H hexamers and strongly supports the single-layer nature of the shell facets (reviewed in (Heinhorst et al. 2014)). However, the possibility that some BMC-H hexamers assemble into double-layered dodecamers under certain conditions has been raised (Sommer et al. 2019; Samborska and Kimber 2012). The pH-dependent stacking of CcmK3/CcmK4 heterohexamers and the stacked dimers of trimers formed by some BMC-T proteins suggest that localized double-layered assemblies may exist transiently in response to changing intracellular conditions or as permanent shell features.
3 BMC Superloci
Aside from the major shell and cargo proteins of the various bacterial microcompartments (Cannon and Shively 1983; Havemann and Bobik 2003; Axen et al. 2014), additional proteins are associated with the organelles and/or are relevant to organelle function, regulation and/or integration of BMC-associated enzymatic steps with the metabolic state of the cell. The genes for these proteins often cluster in the vicinity of the main locus that encodes shell and cargo proteins but can also be located at greater distance. Collectively, these genes constitute what has been referred to as the BMC superlocus (Roberts et al. 2012; Kirst and Kerfeld 2019; Kerfeld et al. 2018; Axen et al. 2014; Pitts et al. 2012).
Structures and functions of shell and lumen proteins are well known for most BMCs, have been covered in several reviews (Heinhorst et al. 2014; Kerfeld et al. 2010, 2018; Yeates et al. 2013; Kirst and Kerfeld 2019) and are therefore not mentioned here. Proteins encoded by the accessory genes in the various BMC superloci play a role in the assembly of a functional organelle, are components of membrane transporters, regulatory components, or may participate in BMC positioning and partitioning (Pitts et al. 2012, Mallette and Kimber 2017). Since aside from their identification by bioinformatics approaches (Axen et al. 2014) not much is known about the role of accessory proteins in the superloci of the diverse catabolic BMC, this section focuses on selected proteins of α- and β-carboxysome superloci.
Pcd-like Protein
The gene for the Pterin-4a-carbinolamine dehydratase (Pcd)-like protein is present in the genome of all α-carboxysome-forming bacteria (Roberts et al. 2012) and encodes a small polypeptide that is structurally very similar to bona fide Pcd but lacks enzymatic activity (Wheatley et al. 2014). Co-expression studies in E. coli with combinations of Pcd-like protein, RubisCO and GroESL, suggest that the Pcd-like protein functions as a RubisCO assembly chaperone in the heterologous host (Wheatley et al. 2014). Recombinant carboxysomes expressed in E. coli from the H. neapolitanus cso operon that lacks the terminal pcd gene, on the other hand, contain enzymatically active RubisCO (Bonacci et al. 2012). Preliminary studies in our lab of a H. neapolitanus pcd knockout mutant (D. Del Valle, unpublished) indicate that the Pcd-like protein affects the ability of H. neapolitanus to grow efficiently at ambient CO2 levels. However, carboxysomes purified from mutant cells are very similar, if not identical, to wild-type organelles in appearance and composition, and the CO2 fixation kinetics of their carboxysomal RubisCOs are near identical. The exact role of the Pcd-like protein in α-carboxysome assembly and/or function in vivo remains to be elucidated, and it is still unclear whether the protein is associated with the organelle.
CbbO and CbbQ
When present in α-carboxysome-forming bacteria, the genes for the putative RubisCO activase CbbQ and its interaction partner, the Van Willebrand Factor A domain-containing protein CbbO, occur together. Sutter et al. (2015) determined the structure and activity of H. neapolitanus CbbQ. The protein, which belongs to the MoxR-type AAA+ ATPase family, forms a hexamer that interacts with one copy of CbbO and has ATPase activity. The CbbQ protein is tightly associated with the shell of H. neapolitanus carboxysomes but does not seem to play a crucial structural or functional role in the organelle. Carboxysomes purified from a cbbQ knockout mutant are structurally and functionally indistinguishable from wild-type organelles, and the mutant does not require elevated CO2 levels for efficient growth.
Although annotated as a putative RubisCO activase, unlike its orthologs from Acidithiobacillus ferrooxidans that stimulate reactivation of inhibited non-carboxysomal form I and form II RubisCO, respectively (Tsai et al. 2015), the CbbQ/O complex encoded in the H. neapolitanus α-carboxysome superlocus does not appear to function as an activase of the carboxysomal RubisCO (Sutter et al. 2015). However, it is possible that the carboxysomal CbbQ/O is required to re-activate carboxysomal RubisCO under certain metabolic conditions that have yet to be identified.
CbbX
The gene for the putative RubisCO activase CbbX (Mueller-Cajar et al. 2011) is part of the superlocus in many α-carboxysome-forming cyanobacteria (Roberts et al. 2012) but absent from the genomes of chemoautotrophs. The CbbX polypeptide also belongs to the AAA+ superfamily. The ortholog from Rhodobacter sphaeroides has been shown to reactivate inhibited red-type RubisCO of that organism (Mueller-Cajar et al. 2011), but the role of CbbX in assembly and/or activation of the cyanobacterial Form IA carboxysomal RubisCO remains to be addressed experimentally.
RbcX
The rbcX gene of most β-carboxysome-forming cyanobacteria is either co-localized with the genes for the carboxysomal Form IB RubisCO (rbcL, rbcS) (Tabita et al. 2007, 2008) in a small operon or is part of the ccmKLN gene cluster (Huang et al. 2019). Onizuka et al. (2004) observed a marked reduction in the amount of RubisCO subunits present and in RubisCO activity in a Synechococcus sp. PCC 7002 frameshift mutant that produces partially inactive RbcX protein. Direct stimulation of RubisCO assembly by RbcX, however, has so far only been shown for recombinant Form IB RubisCO expressed in the heterologous host E. coli (Onizuka et al. 2004; Li and Tabita 1997).
Notable exceptions to the colocalization of the genes encoding RubisCO and its presumed chaperone are S. elongatus PCC 7942 and S. elongatus PCC 6301; in these cyanobacteria, the rbcX gene is located at a considerable distance from rbcL and rbcS. Interruption (Emlyn-Jones et al. 2006b) or deletion (Huang et al. 2019) of rbcX in S. elongatus PCC 7942 does not affect cell growth and RubisCO activity. However, Huang et al. (2019) recently showed that the complete absence of RbcX has a marked effect on the amount of RubisCO present and on size, number and intracellular positioning of carboxysomes. Since the majority of fluorescently labelled RubisCO and RbcX colocalizes to carboxysomes, the collective evidence suggests that, rather than promoting RubisCO assembly, RbcX instead plays a role in carboxysome assembly and distribution within the S. elongatus PCC 7942 cell. Proteomic analysis of purified S. elongatus PCC 7942 carboxysomes (Faulkner et al. 2017) did not reveal any RbcX protein, which suggests that the RubisCO activase is present in the organelle in very small amounts, or that its association with the β-carboxysome is transient.
McdA and McdB
Two proteins encoded by carboxysome superlocus genes are McdA (also previously annotated as ParF or ParA) and its interaction partner McdB (previously annotated as a hypothetical protein), which mediate β-carboxysome positioning in the cell and partitioning among the two daughter cells (Savage et al. 2010; Jain et al. 2012; MacCready et al. 2018). Their mechanism of action is discussed in greater detail in the section Carboxysome Positioning and Segregation.
CsoS2
The CsoS2 protein is encoded in the canonical cso operon that contains the majority of α-carboxysome structural genes. The protein is the largest, highly abundant, and most unique α-carboxysome shell component whose function had long remained unknown. Unlike all other known α-carboxysome proteins, CsoS2 has a very high isoelectric point in most bacteria. The protein exists in two isoforms that are present in approximately equimolar amounts in carboxysomes of some but not all bacteria (Baker et al. 1999; Roberts et al. 2012; Cannon and Shively 1983). The shorter isoform (CsoS2A) represents a C-terminally truncated version of the full-length polypeptide (CsoS2B) (Cai et al. 2015a) and is the product of programmed ribosomal frameshifting to the −1 reading frame at an intrinsic slippery sequence (Chaijarasphong et al. 2016). The presence of such a motif in the csoS2 coding sequence is predicted to correlate with the occurrence of two CsoS2 isoforms.
Three discrete regions can be distinguished in the CsoS2 primary sequence (Cai et al. 2015a): an N-terminal and a middle sequence, each with characteristic amino acid repeat motifs (Cannon et al. 2003), and a C-terminal region. Based on experimental and modelling evidence, CsoS2 likely has an elongated shape and is intrinsically disordered. In vitro binding studies revealed specific interactions of the protein with RubisCO and CsoS1 shell proteins and identified the location of several binding sites in the CsoS2 primary sequence (Cai et al. 2015a).
The essential role of the CsoS2 protein for α-carboxysome assembly is demonstrated by the phenotype of an H. neapolitanus csoS2 knockout mutant that does not produce CsoS2 protein. The mutant does not grow in air and does not produce carboxysomes (Cai et al. 2015a). Based on the collective data, a model was put forth in which CsoS2 is predicted to recruit shell proteins and RubisCO to the α-carboxysome biogenesis site. Insertion of the CsoS2 C-terminus into the developing shell facets and organization of RubisCO in the emerging lumen space occur simultaneously (Cai et al. 2015a). The model is consistent with the carboxysome biosynthesis intermediates that were observed in H. neapolitanus cells by cryo electron tomography (Iancu et al. 2010). Recent expression studies of wild-type and mutant H. neapolitanus cso operons in E. coli identified the small subunit (CbbS) of the carboxysomal RubisCO as the interaction partner of CsoS2 and the N-terminal domain of the large subunit (CbbL) as the portion of the enzyme that contacts all three BMC-H paralogs (Liu et al. 2018). Those results are consistent with the well-documented single layer of RubisCO holoenzyme molecules that line the inside of the carboxysome shell (Iancu et al. 2010; Schmid et al. 2006; Iancu et al. 2007) and remain shell-associated when carboxysomes are broken (Holthuijzen et al. 1986). The observed requirement for full-length CsoS2B to yield recombinant carboxysomes in E. coli (Chaijarasphong et al. 2016) confirms the crucial role of the protein’s C-terminal region in α-carboxysome assembly.
CcmM
The CcmM protein is encoded within the main β-carboxysome gene cluster alongside the genes for several shell proteins and CcmN (Sommer et al. 2017). The protein was shown to be crucial for β-carboxysome assembly and/or function; mutants that do not express functional CcmM protein are devoid of carboxysomes and have a strict high CO2-requiring (hcr) phenotype (Ludwig et al. 2000; Woodger et al. 2005; Berry et al. 2005; Emlyn-Jones et al. 2006a). The CcmM protein features an N-terminal domain that is homologous to γ-CA, and a C-terminal portion that contains three to five repeats with weak, albeit detectable similarity to the RubisCO small subunit (RbcS) sequence (Ludwig et al. 2000). The crystal structure of CcmM from Thermosynechococcus elongatus BP-1 (Peña et al. 2010) revealed that the N-terminal CA domain assembles into a trimer. Notably, the protein only acquires enzymatic activity after a key disulphide bond in this domain is formed (Peña et al. 2010), presumably in the oxidizing environment of the β-carboxysome interior (Chen et al. 2013; Peña et al. 2010).
In addition to full-length CcmM (CcmM58), β-carboxysomes also contain a truncated form (CcmM35), which consists of only the C-terminal portion of the protein (Long et al. 2007, 2010) and is generated by translation from an internal initiation site upstream of the C-terminal repeat region. Cameron and colleagues (2013) showed that the CcmM protein is needed during β-carboxysome biogenesis to condense RubisCO molecules into the core to which shell components are subsequently recruited. The reduced form of S. elongatus PCC 7942 CcmM35 rapidly condenses purified RubisCO holoenzyme into a liquid droplet network; when oxidized, its affinity for RubisCO is reduced. Mutants in which CcmM lacks one or more of the cysteines that form disulphide bonds in its C-terminal domain have an hcr phenotype and contain fewer and abnormally shaped (elongated) carboxysomes (Wang et al. 2019), emphasizing the crucial role of the CcmM protein and its oxidized form in β-carboxysome biogenesis that had been deduced earlier from mutant and carboxysome assembly studies.
Because of sequence and structural homology between CcmM35 and RbcS, as well as the reported discrepancy in cellular abundance between RubisCO large and small subunits (Long et al. 2011), replacement of RbcS by CcmM35 in the RubisCO holoenzyme had been proposed as a possible means of modulating the activity of the CO2 fixing enzyme (Long et al. 2011). However, the RbcS-like domain of CcmM lacks the essential RbcL binding elements of RbcS (Ferlez et al. 2019). Furthermore, co-crystals of T. elongatus RubisCO holoenzyme and one RbcS-like CcmM35 domain (Ryan et al. 2019) revealed that CcmM35 binds at a separate site and does not induce release of endogenous RbcS, even if the holoenzyme contains severely truncated RbcS subunits. The authors suggested that CcmM35 binds peripherally to a cleft between two RbcL subunits in the holoenzyme complex. This binding mode was subsequently confirmed by cryo electron tomography analysis of S. elongatus PCC 7942 CcmM-RubisCO complexes (Wang et al. 2019).
4 Organization of the β-Carboxysome Interior
Considering the abundance of individual proteins in fractions highly enriched in β-carboxysomes, as well as resulting from in vivo and in vitro interaction studies (Long et al. 2007, 2010, 2011; Cot et al. 2008; Kinney et al. 2012; McGurn et al. 2016), a model for the molecular organization of the β-carboxysome interior has emerged in which the CcmN protein, which is essential for β-carboxysome assembly, tethers a peripheral layer of full-length CcmM58 trimers to the shell. The C-terminal domain of CcmN contacts CcmK2 and probably other BMC-H shell components through a conserved peptide motif. The protein interacts with the CcmM trimer through its N-terminal domain. In those cyanobacteria that also contain the β-CA CcaA, the N-terminal catalytic domains of the CcmM trimer form a complex with a hexamer of three CcaA dimers (McGurn et al. 2016). The C-terminal RbcS-like domains of CcmM bind a layer of RubisCO that abuts the shell. The truncated CcmM35 isoform, which is devoid of the N-terminal domain that mediates interactions with CcaA and CcmN, is thought to organize RubisCO into a paracrystalline array in the interior (Long et al. 2007, 2010, 2011; Cot et al. 2008; Peña et al. 2010). This model is attractive, because it considers the concentration gradient of bicarbonate from the shell towards the centre of the organelle and proposes efficient generation and trapping of the RubisCO substrate CO2 at the periphery immediately upon bicarbonate entry into the organelle lumen. However, high-resolution imaging studies (Niederhuber et al. 2017) have prompted a revision of the prevailing model. Using fluorescently tagged CcmM35 and CcmM58, the authors observed simultaneous assembly of both isoforms during carboxysome biogenesis. Furthermore, both isoforms appear to be distributed throughout the carboxysome lumen, suggesting that both C-terminal portion and full-length CcmM play a role in condensing the RubisCO core.
5 Positioning and Segregation of Carboxysomes
Early electron microscopic observations (reviewed in (Heinhorst et al. 2014)) frequently noted the close association of carboxysomes with the DNA fibrils of the bacterial nucleoid, a finding that was corroborated by Jain et al. (Jain et al. 2012). Using differential fluorescent staining, the authors showed that the multiple chromosome copies of S. elongatus PCC 7942 are regularly interspersed with carboxysomes along the longitudinal cell axis, suggesting that positioning and partitioning of DNA and carboxysomes are connected through shared molecular components.
Since movement of cell constituents is frequently brought about by components of the cytoskeleton and native, as well as recombinant BMCs were reported to be associated with filamentous molecular structures (Savage et al. 2010; Parsons et al. 2010), filaments of cytoskeleton proteins were implicated in the segregation and intracellular distribution of BMCs. A homolog of the DNA segregation protein ParA was shown to play an important role in β-carboxysome spacing and segregation during cell division in S. elongatus PCC 7942 (Savage et al. 2010; Jain et al. 2012). Savage and colleagues (2010) observed intracellular oscillations of fluorescently labelled ParA, which appears to be interspersed with carboxysomes, and found evidence for ParA filaments. Deletion of the protein leads to aberrantly distributed carboxysomes without, however, affecting the arrangement of the multiple chromosome copies in the cell (Jain et al. 2012).
MacCready et al. (2018) corroborated the S. elongatus PCC 7942 ParA oscillations observed previously (Savage et al. 2010) but found no evidence of filament formation. The authors showed that, like ParA in other bacteria, the cyanobacterial protein, which they re-named McdA (Maintenance of carboxysome distribution A), is a non-sequence-specific DNA binding protein when complexed with ATP. However, the genome of carboxysome-containing bacteria does not encode a recognizable ortholog of the known ParA partner protein, ParB. In a screen of deletion mutants of several nearby candidate genes, the researchers identified McdB, which has no homology to any known protein, as an McdA interaction partner that stimulates the ATPase activity of the latter. Schumacher et al. (2019) subsequently solved the crystal structures of McdA and McdB and showed interaction of purified McdA and McdB proteins from Cyanothece PCC 7424. Fluorescently tagged McdB co-localizes with carboxysomes in the cell, and two-hybrid screens showed that McdB, but not McdA, can directly interact with several carboxysome shell proteins (CcmK2, CcmK3, CcmK4 and CcmO). In addition, CcmK3 and CcmK4 deletion mutants of S. elongatus PCC 7942 contain aberrantly positioned carboxysomes (Rae et al. 2012). In a mutant that does not form carboxysomes, McdB exhibits a diffuse localization pattern, and oscillations of McdA were not detected. Collectively, these results suggest that McdB, when recruited to carboxysomes, affects the dynamics of McdA organization in the cell.
Since the findings of MacCready et al. (2018) rule out a cytoskeletal filament-based carboxysome movement and positioning in the cell, the authors proposed an alternative, Brownian ratchet mechanism. Supported by the observed movement of fluorescently tagged carboxysomes towards nucleoid areas, which are rich in McdA, in time lapse images, the ratchet model posits that DNA-bound McdA, upon stimulation of its ATPase activity by carboxysome-bound McdB, is released from the nucleoid, thus creating an McdA depletion zone in the vicinity of carboxysomes. McdB moves its carboxysome cargo along the McdA concentration gradient towards higher concentrations of nucleoid-bound McdA•ATP. Once a certain distance is reached between two carboxysomes, the nucleoid area between them repopulates with McdA•ATP. The authors further showed that the elongated shape of the nucleoid in S. elongatus PCC 7942 is responsible for the alignment of carboxysomes along the longitudinal cell axis in this cyanobacterium.
In several bacteria, particularly under physiological conditions that promote an increase in the number of carboxysomes, clustering of the organelles into groups has been observed (Sun et al. 2016) and reviewed in (Heinhorst et al. 2014; Sun et al. 2016). Whether this phenomenon reflects a physiological advantage, such as trapping of CO2 that has escaped from the lumen of a carboxysome by a neighbouring organelle (Ting et al. 2007), or is simply the result of molecular crowding (Iancu et al. 2010) was not resolved. MacCready et al. (2018) present evidence that carboxysome clustering or hexagonal packaging correlates with the number of carboxysomes per nucleoid surface. In addition, modelling of carboxysome distribution further predicts clustering above a certain threshold number and supports the Brownian ratchet mechanism as the driver of β-carboxysome positioning in the cell. Interestingly, mcdA and mcdB knockout and overexpression mutants, in addition to showing defects in carboxysome distribution, also display changes in organelle size and shape that suggest a possible role of McdA and McdB in organelle biogenesis.
6 Concluding Remarks
The BMC field has enjoyed a tremendous expansion in the past 10 years, thanks to a flurry of interest following the realization that BMCs constitute a strategy to compartmentalize key metabolic reactions that is widespread among bacteria. Furthermore, because of their small size and relative simplicity, their tremendous synthetic biology potential is increasingly being realized. Informed by the known structure and identity of individual BMC protein components and a growing understanding of their contributions to BMC biogenesis and function, impressive results have been achieved in designing BMCs with novel properties.
Paving the way for BMC engineering has been the development of heterologous expression systems for entire BMC operons (Bonacci et al. 2012; Liu et al. 2018; Parsons et al. 2008; Graf et al. 2018; Baumgart et al. 2017). The culmination of those efforts to date is the successful transfer of α- and β-carboxysome genes into higher plants and the targeting of their protein products to the chloroplast (Lin et al. 2014; Long et al. 2018), where the bacterial RubisCO is active in CO2 fixation (Long et al. 2018). Furthermore, empty BMC shells have been generated with specifically engineered architectures, sizes and properties that emphasize the structural and compositional flexibility of BMC shell protein-derived assemblies (reviewed in (Plegaria and Kerfeld 2018, Kirst and Kerfeld 2019).
Capitalizing on known mechanisms of protein encapsulation through short helical N- or C-terminal peptide sequences or utilizing endogenous BMC lumen proteins as vehicles (e.g. (Kinney et al. 2012; Fan et al. 2010; Fan and Bobik 2011; Menon et al. 2008, 2009, 2010); reviewed in (Plegaria and Kerfeld 2018)), orthologs of cargo proteins as well as entirely unrelated proteins have been targeted to the BMC lumen. In several cases, the resulting engineered BMCs are endowed with novel metabolic properties (Liang et al. 2017; Lawrence et al. 2014; Wagner et al. 2017) that demonstrate the ability of BMCs to accommodate substantial variations in their protein cargos.
Despite the scientific advances that have answered many questions related to BMC assembly, composition, structure and function, and the impressive developments in designing synthetic BMCs for specialty purposes, several basic questions remain that need to be addressed before a comprehensive understanding of BMC biology can be reached and the full potential of BMC engineering can be realized.
The most pressing questions in this regard relate to the permeability properties of the shell. At least one of the enzymes encapsulated in most, if not all BMCs is sensitive to oxygen, such as the enzymes of catabolic BMCs that rely on an oxygen-sensitive cofactor or operate through an oxygen-sensitive reaction mechanism. Likewise, for efficient CO2 fixation RubisCO must avoid O2, its competing, non-productive substrate. Although the shell is widely assumed to exclude oxygen from the BMC interior, experimental evidence for such selectively is lacking.
Despite the experimental evidence that supports the role of the pores in the BMC protein shell as the conduits through which substrates, products, possibly cofactors and regulatory metabolites enter and exit BMCs, the molecular mechanisms of their transport across the shell are not known. Additional open questions are the range of BMC-H proteins that are capable of forming heterohexamers, the dynamics of their associations with the array of hexamers in the BMC shell, and their significance in mediating and/or regulating metabolite traffic. Likewise, the proposed gating of metabolite transport through BMC-T pores and the properties that are ascribed to their internal nanocompartments constitute attractive potential regulatory strategies that await experimental support. Finally, establishing a complete inventory of pore permeabilities for the oligomers of the various BMC types will be the prerequisite for a more in-depth understanding of metabolic crosstalk between organelle lumen and the cytosol and for engineering efforts to manipulate the shell for the development of BMC-based nanoreactors with specific properties for specialty applications.
References
Axen SD, Erbilgin O, Kerfeld CA (2014) A taxonomy of bacterial microcompartment loci constructed by a novel scoring method. PLoS Comput Biol 10:e1003898
Baker SH, Lorbach SC, Rodriguez-BUEY M, Williams DS, Aldrich HC, Shively JM (1999) The correlation of the gene csoS2 of the carboxysome operon with two polypeptides of the carboxysome in Thiobacillus neapolitanus. Arch Microbiol 172:233–239
Baumgart M, Huber I, Abdollahzadeh I, Gensch T, Frunzke J (2017) Heterologous expression of the Halothiobacillus neapolitanus carboxysomal gene cluster in Corynebacterium glutamicum. J Biotechnol 258:126–135
Berry S, Fischer JH, Kruip J, Hauser M, Wildner GF (2005) Monitoring cytosolic pH of carboxysome-deficient cells of Synechocystis sp. PCC 6803 using fluorescence analysis. Plant Biol (Stuttg) 7:342–347
Bobik TA, Xu Y, Jeter RM, Otto KE, Roth JR (1997) Propanediol utilization genes (pdu) of Salmonella typhimurium: three genes for the propanediol dehydratase. J Bacteriol 179:6633–6639
Bobik TA, Havemann GD, Busch RJ, Williams DS, Aldrich HC (1999) The propanediol utilization (pdu) operon of Salmonella enterica serovar typhimurium LT2 includes genes necessary for formation of polyhedral organelles involved in coenzyme B(12)-dependent 1, 2-propanediol degradation. J Bacteriol 181:5967–5975
Bonacci W, Teng PK, Afonso B, Niederholtmeyer H, Grob P, Silver PA, Savage DF (2012) Modularity of a carbon-fixing protein organelle. Proc Natl Acad Sci U S A 109:478–483
Cai F, Menon BB, Cannon GC, Curry KJ, Shively JM, Heinhorst S (2009) The pentameric vertex proteins are necessary for the icosahedral carboxysome shell to function as a CO2 leakage barrier. PLoS One 4:e7521
Cai F, Sutter M, Cameron JC, Stanley DN, Kinney JN, Kerfeld CA (2013) The structure of CcmP, a tandem bacterial microcompartment domain protein from the beta-carboxysome, forms a subcompartment within a microcompartment. J Biol Chem 288:16055–16063
Cai F, Dou Z, Bernstein SL, Leverenz R, Williams EB, Heinhorst S, Shively J, Cannon GC, Kerfeld CA (2015a) Advances in understanding carboxysome assembly in Prochlorococcus and Synechococcus implicate CsoS2 as a critical component. Life (Basel) 5:1141–1171
Cai F, Sutter M, Bernstein SL, Kinney JN, Kerfeld CA (2015b) Engineering bacterial microcompartment shells: chimeric shell proteins and chimeric carboxysome shells. ACS Synth Biol 4:444–453
Cameron JC, Wilson SC, Bernstein SL, Kerfeld CA (2013) Biogenesis of a bacterial organelle: the carboxysome assembly pathway. Cell 155:1131–1140
Cannon GC, Shively JM (1983) Characterization of a homogenous preparation of carboxysomes from Thiobacillus neapolitanus. Arch Microbiol 134:52–59
Cannon GC, Baker SH, Soyer F, Johnson DR, Bradburne CE, Mehlman JL, Davies PS, Jiang QL, Heinhorst S, Shively JM (2003) Organization of carboxysome genes in the thiobacilli. Curr Microbiol 46:115–119
Chaijarasphong T, Nichols RJ, Kortright KE, Nixon CF, Teng PK, Oltrogge LM, Savage DF (2016) Programmed ribosomal frameshifting mediates expression of the alpha-carboxysome. J Mol Biol 428:153–164
Chen AH, Robinson-Mosher A, Savage DF, Silver PA, Polka JK (2013) The bacterial carbon-fixing organelle is formed by shell envelopment of preassembled cargo. PLoS One 8:e76127
Chowdhury C, Chun S, Pang A, Sawaya MR, SINHA S, Yeates TO, Bobik TA (2015) Selective molecular transport through the protein shell of a bacterial microcompartment organelle. Proc Natl Acad Sci U S A 112:2990–2995
Cot SS-W, So AK-C, Espie GS (2008) A multiprotein bicarbonate dehydration complex essential to carboxysome function in cyanobacteria. J Bacteriol 190:936–945
Craciun S, Balskus EP (2012) Microbial conversion of choline to trimethylamine requires a glycyl radical enzyme. Proc Natl Acad Sci U S A 109:21307–21312
Craciun S, Marks JA, Balskus EP (2014) Characterization of choline trimethylamine-lyase expands the chemistry of glycyl radical enzymes. ACS Chem Biol 9:1408–1413
Craciun S, Marks JA, Balskus EP (2016) Correction to characterization of choline trimethylamine-Lyase expands the chemistry of Glycyl radical enzymes. ACS Chem Biol 11:2068
Crowley CS, Sawaya MR, Bobik TA, Yeates TO (2008) Structure of the PduU shell protein from the Pdu microcompartment of Salmonella. Structure 16:1324–1332
Crowley CS, Cascio D, Sawaya MR, Kopstein JS, Bobik TA, Yeates TO (2010) Structural insight into the mechanisms of transport across the Salmonella enterica Pdu microcompartment shell. J Biol Chem 285:37838–37846
Dou Z, Heinhorst S, Williams EB, Murin CD, Shively JM, Cannon GC (2008) CO2 fixation kinetics of Halothiobacillus neapolitanus mutant carboxysomes lacking carbonic anhydrase suggest the shell acts as a diffusional barrier for CO2. J Biol Chem 283:10377–10384
Emlyn-Jones D, Woodger FJ, Andrews TJ, Price GD, Whitney SM (2006a) A Synechococcus PCC 7942 ΔCcmM (cyanophyceae) mutant pseudoreverts to air growth without regaining carboxysomes. J Phycol 42:769–777
Emlyn-Jones D, Woodger FJ, Price GD, Whitney SM (2006b) RbcX can function as a rubisco chaperonin, but is non-essential in Synechococcus PCC7942. Plant Cell Physiol 47:1630–1640
Erbilgin O, Mcdonald KL, Kerfeld CA (2014) Characterization of a planctomycetal organelle: a novel bacterial microcompartment for the aerobic degradation of plant saccharides. Appl Environ Microbiol 80:2193–2205
Fan C, Bobik TA (2011) The N-terminal region of the medium subunit (PduD) packages adenosylcobalamin-dependent diol dehydratase (PduCDE) into the Pdu microcompartment. J Bacteriol 193:5623–5628
Fan C, Cheng S, Liu Y, Escobar CM, Crowley CS, Jefferson RE, Yeates TO, Bobik TA (2010) Short N-terminal sequences package proteins into bacterial microcompartments. Proc Natl Acad Sci U S A 107:7509–7514
Faulkner M, Rodriguez-Ramos J, Dykes GF, Owen SV, Casella S, Simpson DM, Beynon RJ, Liu LN (2017) Direct characterization of the native structure and mechanics of cyanobacterial carboxysomes. Nanoscale 9:10662–10673
Ferlez B, Sutter M, Kerfeld CA (2019) Glycyl radical enzyme-associated microcompartments: redox-replete bacterial organelles. mBio 10:e02327–e02318
Garcia-Alles LF, Root K, Maveyraud L, Aubry N, Lesniewska E, Mourey L, Zenobi R, Truan G (2019) Occurrence and stability of hetero-hexamer associations formed by beta-carboxysome CcmK shell components. PLoS One 14:e0223877
Graf L, Wu K, Wilson JW (2018) Transfer and analysis of Salmonella pdu genes in a range of gram-negative bacteria demonstrate exogenous microcompartment expression across a variety of species. Microb Biotechnol 11:199–210
Havemann GD, Bobik TA (2003) Protein content of polyhedral organelles involved in coenzyme B12-dependent degradation of 1,2-propanediol in Salmonella enterica serovar typhimurium LT2. J Bacteriol 185:5086–5095
Heinhorst S, Cannon GC, Shively JM (2014) Carboxysomes and their structural organization in prokaryotes. In: BARTON LL, Bazylinski DA, Xu, H. (eds) Nanomicrobiology. Springer, New York, pp 75–101
Heldt D, Frank S, Seyedarabi A, Ladikis D, Parsons JB, Warren MJ, Pickersgill RW (2009) Structure of a trimeric bacterial microcompartment shell protein, EtuB, associated with ethanol utilization in Clostridium kluyveri. Biochem J 423:199–207
holthuijzen YA, Breemen JFL, Konings WN, Bruggen EFJ (1986) Electron microscopic studies of carboxysomes of Thiobacillus neapolitanus. Arch Microbiol 144:258–262
Huang F, Vasieva O, Sun Y, Faulkner M, Dykes GF, Zhao Z, Liu L-N (2019) Roles of RbcX in carboxysome biosynthesis in the cyanobacterium Synechococcus elongatus PCC7942. Plant Physiol 179:184–194
Iancu CV, Ding HJ, Morris DM, Dias DP, Gonzales AD, Martino A, Jensen GJ (2007) The structure of isolated Synechococcus strain WH8102 carboxysomes as revealed by electron cryotomography. J Mol Biol 372:764–773
Iancu CV, Morris DM, Dou Z, Heinhorst S, Cannon GC, Jensen GJ (2010) Organization, structure, and assembly of α-carboxysomes determined by electron cryotomography of intact cells. J Mol Biol 396:105–117
Jain IH, Vijayan V, O’Shea EK (2012) Spatial ordering of chromosomes enhances the fidelity of chromosome partitioning in cyanobacteria. Proc Natl Acad Sci U S A 109:13638–13643
Keeling TJ, Samborska B, Demers RW, Kimber MS (2014) Interactions and structural variability of beta-carboxysomal shell protein CcmL. Photosynth Res 121:125–133
Kerfeld CA, Sawaya MR, Tanaka S, Nguyen CV, Phillips M, Beeby M, Yeates TO (2005) Protein structures forming the shell of primitive bacterial organelles. Science 309:936–938
Kerfeld CA, Heinhorst S, Cannon GC (2010) Bacterial microcompartments. Ann Rev Microbiol 64:391–408
Kerfeld CA, Aussignargues C, Zarzycki J, Cai F, SUTTER M (2018) Bacterial microcompartments. Nat Rev Microbiol 16:277–290
Kinney JN, Salmeen A, Cai F, Kerfeld CA (2012) Elucidating essential role of conserved carboxysomal protein CcmN reveals common feature of bacterial microcompartment assembly. J Biol Chem 287:17729–17736
Kirst H, Kerfeld CA (2019) Bacterial microcompartments: catalysis-enhancing metabolic modules for next generation metabolic and biomedical engineering. BMC Biol 17:79
Klein MG, Zwart P, Bagby SC, Cai F, Chisholm SW, Heinhorst S, Cannon GC, Kerfeld CA (2009) Identification and structural analysis of a novel carboxysome shell protein with implications for metabolite transport. J Mol Biol 392:319–333
Kofoid E, Rappleye C, Stojiljkovic I, Roth J (1999) The 17-gene ethanolamine (eut) operon of Salmonella typhimurium encodes five homologues of carboxysome shell proteins. J Bacteriol 181:5317–5329
Larsson AM, Hasse D, Valegard K, Andersson I (2017) Crystal structures of beta-carboxysome shell protein CcmP: ligand binding correlates with the closed or open central pore. J Exp Bot 68:3857–3867
Lawrence AD, Frank S, Newnham S, Lee MJ, Brown IR, Xue WF, Rowe ML, Mulvihill DP, Prentice MB, Howard MJ, Warren MJ (2014) Solution structure of a bacterial microcompartment targeting peptide and its application in the construction of an ethanol bioreactor. ACS Synth Biol 3:454–465
Li LA, Tabita FR (1997) Maximum activity of recombinant ribulose 1,5-bisphosphate carboxylase/oxygenase of Anabaena sp. strain CA requires the product of the rbcX gene. J Bacteriol 179:3793–3796
Liang M, Frank S, Lünsdorf H, Warren MJ, Prentice MB (2017) Bacterial microcompartment-directed polyphosphate kinase promotes stable polyphosphate accumulation in E coli. Biotechnol J 68:1600415
Lin MT, Occhialini A, Andralojc PJ, Devonshire J, Hines KM, Parry MA, Hanson MR (2014) Beta-Carboxysomal proteins assemble into highly organized structures in Nicotiana chloroplasts. Plant J 79:1–12
Liu Y, He X, Lim W, Mueller J, Lawrie J, Kramer L, Guo J, Niu W (2018) Deciphering molecular details in the assembly of alpha-type carboxysome. Sci Rep 8:15062
Long BM, Badger MR, Whitney SM, Price GD (2007) Analysis of carboxysomes from Synechococcus PCC7942 reveals multiple RubisCO complexes with carboxysomal proteins CcmM and CcaA. J Biol Chem 282:29323–29335
Long BM, Tucker L, Badger MR, Price GD (2010) Functional cyanobacterial β-carboxysomes have an absolute requirement for both long and short forms of the CcmM protein. Plant Physiol 153:285–293
Long BM, Rae BD, Badger MR, Price GD (2011) Over-expression of the beta-carboxysomal CcmM protein in Synechococcus PCC7942 reveals a tight co-regulation of carboxysomal carbonic anhydrase (CcaA) and M58 content. Photosynth Res 109:33–45
Long BM, Hee WY, Sharwood RE, Rae BD, Kaines S, Lim YL, Nguyen ND, Massey B, Bala S, von Caemmerer S, Badger MR, Price GD (2018) Carboxysome encapsulation of the CO2-fixing enzyme rubisco in tobacco chloroplasts. Nat Commun 9:3570
Ludwig M, Sültemeyer D, Price GD (2000) Isolation of ccmKLMN genes from the marine cyanobacterium Synechococcus sp. PCC7002 (cyanophyceae), and evidence that CcmM is essential for carboxysome assembly. J Phycol 36:1109–1119
MacCready JS, Hakim P, Young EJ, Hu L, Liu J, Osteryoung KW, Vecchiarelli AG, Ducat DC (2018) Protein gradients on the nucleoid position the carbon-fixing organelles of cyanobacteria. elife 7:e39723
Mahinthichaichan P, Morris DM, Wang Y, Jensen GJ, Tajkhorshid E (2018) Selective permeability of carboxysome shell pores to anionic molecules. J Phys Chem B 122:9110–9118
Mallette E, Kimber MS (2017) A complete structural inventory of the mycobacterial microcompartment shell proteins constrains models of global architecture and transport. J Biol Chem 292:1197–1210
McGurn LD, Moazami-Goudarzi M, White SA, Suwal T, Brar B, Tang JQ, Espie GS, Kimber MS (2016) The structure, kinetics and interactions of the beta-carboxysomal beta-carbonic anhydrase, CcaA. Biochem J 473:4559–4572
Menon BB, Dou Z, Heinhorst S, Shively JM, Cannon GC (2008) Halothiobacillus neapolitanus carboxysomes sequester heterologous and chimeric RubisCO species. PLoS One 3:e3570
Menon BB, Dou Z, Milam J, Shively JM, Heinhorst S, Cannon GC (2009) Phenotypic analysis of a Halothiobacillus neapolitanus mutant harboring beta-cyanobacterial form IB RubisCO. Amercian Society for Microbiology 109th General Meeting, Philadelphia, PA
Menon BB, Heinhorst S, Shively JM, Cannon GC (2010) The carboxysome shell is permeable to protons. J Bacteriol 192:5881–5886
Mueller-Cajar O, Stotz M, Wendler P, Hartl FU, Bracher A, Hayer-Hartl M (2011) Structure and function of the AAA+ protein CbbX, a red-type rubisco activase. Nature 479:194–199
Niederhuber MJ, Lambert TJ, Yapp C, Silver PA, Polka JK (2017) Superresolution microscopy of the beta-carboxysome reveals a homogeneous matrix. Mol Biol Cell 28:2734–2745
Onizuka T, Endo S, Akiyama H, Kanai S, Hirano M, Yokota A, Tanaka S, Miyasaka H (2004) The rbcX gene product promotes the production and assembly of ribulose-1,5-bisphosphate carboxylase/oxygenase of Synechococcus sp. PCC7002 in Escherichia coli. Plant Cell Physiol 45:1390–1395
Pang A, Warren MJ, Pickersgill RW (2011) Structure of PduT, a trimeric bacterial microcompartment protein with a 4Fe-4S cluster-binding site. Acta Crystallogr D Biol Crystallogr 67:91–96
Pang A, Liang M, Prentice MB, Pickersgill RW (2012) Substrate channels revealed in the trimeric Lactobacillus reuteri bacterial microcompartment shell protein PduB. Acta Crystallogr D Biol Crystallogr 68:1642–1652
Park J, Chun S, Bobik TA, Houk KN, Yeates TO (2017) Molecular dynamics simulations of selective metabolite transport across the propanediol bacterial microcompartment shell. Journal Phys Chem B 121:8149–8154
Parsons JB, Dinesh SD, Deery E, Leech HK, Brindley AA, Heldt D, Frank S, Smales CM, Lunsdorf H, Rambach A, Gass MH, Bleloch A, Mcclean KJ, Munro AW, Rigby SEJ, Warren MJ, Prentice MB (2008) Biochemical and structural insights into bacterial organelle form and biogenesis. J Biol Chem 283:14366–14375
Parsons JB, Frank S, Bhella D, Liang M, Prentice MB, Mulvihill DP, warren MJ (2010) Synthesis of empty bacterial microcompartments, directed organelle protein incorporation, and evidence of filament-associated organelle movement. Mol Cell 38:305–315
Peña KL, Castel SE, de Araujo C, Espie GS, Kimber MS (2010) Structural basis of the oxidative activation of the carboxysomal γ-carbonic anhydrase, CcmM. Proc Natl Acad Sci U S A 107:2455–2460
Penrod JT, Roth JR (2006) Conserving a volatile metabolite: a role for carboxysome-like organelles in Salmonella enterica. J Bacteriol 188:2865–2874
Petit E, Latouf WG, Coppi MV, Warnick TA, Currie D, Romashko I, Deshpande S, Haas K, Alvelo-Maurosa JG, Wardman C, Schnell DJ, Leschine SB, Blanchard JL (2013) Involvement of a bacterial microcompartment in the metabolism of fucose and rhamnose by Clostridium phytofermentans. PLoS One 8:e54337
Pitts AC, Tuck LR, Faulds-Pain A, Lewis RJ, Marles-Wright J (2012) Structural insight into the Clostridium difficile ethanolamine utilisation microcompartment. PLoS One 7:e48360
Plegaria JS, Kerfeld CA (2018) Engineering nanoreactors using bacterial microcompartment architectures. Curr Opin Biotechnol 51:1–7
Price GD, Badger MR (1991) Evidence for the role of carboxysomes in the cyanobacterial CO2-concentrating mechanism. Can J Bot 69:963–973
Rae BD, Long BM, Badger MR, Price GD (2012) Structural determinants of the outer shell of β-carboxysomes in Synechococcus elongatus PCC 7942: Roles for CcmK2, K3-K4, CcmO, and CcmL. PLoS One 7:e43871
Ravcheev DA, Moussu L, Smajic S, Thiele I (2019) Comparative genomic analysis reveals novel microcompartment-associated metabolic pathways in the human gut microbiome. Front Genet 10:636
Roberts EW, Cai F, Kerfeld CA, Cannon GC, Heinhorst S (2012) Isolation and characterization of the Prochlorococcus carboxysome reveal the presence of the novel shell protein CsoS1D. J Bacteriol 194:787–795
Ryan P, Forrester TJB, Wroblewski C, Kenney TMG, Kitova EN, Klassen JS, Kimber MS (2019) The small RbcS-like domains of the beta-carboxysome structural protein CcmM bind RubisCO at a site distinct from that binding the RbcS subunit. J Biol Chem 294:2593–2603
Sagermann M, Ohtaki A, Nikolakakis K (2009) Crystal structure of the EutL shell protein of the ethanolamine ammonia lyase microcompartment. Proc Natl Acad Sci U S A 106:8883–8887
Samborska B, Kimber MS (2012) A dodecameric CcmK2 structure suggests beta-carboxysomal shell facets have a double-layered organization. Structure 20:1353–1362
Savage DF, Afonso B, Chen AH, Silver PA (2010) Spatially ordered dynamics of the bacterial carbon fixation machinery. Science 327:1258–1261
Schindel HS, Karty JA, Mckinlay JB, Bauer CE (2019) Characterization of a glycyl radical enzyme bacterial microcompartment pathway in Rhodobacter capsulatus. J Bacteriol 201:e00343–e00318
Schmid MF, Paredes AM, Khant HA, Soyer F, Aldrich HC, Chiu W, Shively JM (2006) Structure of Halothiobacillus neapolitanus carboxysomes by cryo-electron tomography. J Mol Biol 364:526–535
Schumacher MA, Henderson M, Zhang H (2019) Structures of maintenance of carboxysome distribution Walker-box McdA and McdB adaptor homologs. Nucleic Acids Res 47:5950–5962
Seedorf H, Fricke WF, Veith B, Bruggemann H, Liesegang H, Strittmatter A, Miethke M, Buckel W, hinderberger J, Li F, Hagemeier C, Thauer RK, Gottschalk G (2008) The genome of Clostridium kluyveri, a strict anaerobe with unique metabolic features. Proc Natl Acad Sci U S A 105:2128–2133
Shively JM, Ball F, Brown DH, Saunders RE (1973) Functional organelles in prokaryotes: polyhedral inclusions (carboxysomes) of Thiobacillus neapolitanus. Science 182:584–586
Shively JM, Bradburne CE, Aldrich HC, Bobik TA, Mehlman JL, Jin S, Baker SH (1998) Sequence homologs of the carboxysomal polypeptide CsoS1 of the thiobacilli are present in cyanobacteria and enteric bacteria that form carboxysomes-polyhedral bodies. Can J Bot 76:906–916
Slininger Lee MF, Jakobson CM, Tullman-Ercek D (2017) Evidence for improved encapsulated pathway behavior in a bacterial microcompartment through shell protein engineering. ACS Synth Biol 6:1880–1891
Sommer M, Cai F, Melnicki M, Kerfeld CA (2017) β-Carboxysome bioinformatics: identification and evolution of new bacterial microcompartment protein gene classes and core locus constraints. J Exp Bot 68:3841–3855
Sommer M, Sutter M, Gupta S, Kirst H, Turmo A, Lechno-Yossef S, Burton RL, Saechao C, Sloan NB, Cheng X, Chan LG, Petzold CJ, Fuentes-Cabrera M, Ralston CY, Kerfeld CA (2019) Heterohexamers formed by CcmK3 and CcmK4 increase the complexity of beta carboxysome shells. Plant Physiol 179:156–167
Stojiljkovic I, Baumler AJ, Heffron F (1995) Ethanolamine utilization in Salmonella typhimurium: nucleotide sequence, protein expression, and mutational analysis of the cchA cchB eutE eutJ eutG eutH gene cluster. J Bacteriol 177:1357–1366
Sun Y, Casella S, Fang Y, Huang F, Faulkner M, Barrett S, Liu LN (2016) Light modulates the biosynthesis and organization of cyanobacterial carbon fixation machinery through photosynthetic electron flow. Plant Physiol 171:530–541
Sutter M, Roberts EW, Gonzalez RC, Bates C, Dawoud S, Landry K, Cannon GC, Heinhorst S, Kerfeld CA (2015) Structural characterization of a newly identified component of alpha-carboxysomes: the AAA+ domain protein CsoCbbQ. Sci Rep 5:16243
Sutter M, Greber B, Aussignargues C, Kerfeld CA (2017) Assembly principles and structure of a 6.5-MDa bacterial microcompartment shell. Science 356:1293–1297
Tabita FR, Hanson TE, Li H, Satagopan S, Singh J, Chan S (2007) Function, structure, and evolution of the RubisCO-like proteins and their RubisCO homologs. Microbiol Mol Biol Rev 71:576–599
Tabita FR, Satagopan S, Hanson TE, Kreel NE, Scott SS (2008) Distinct form I, II, III, and IV rubisco proteins from the three kingdoms of life provide clues about rubisco evolution and structure/function relationships. J Exp Bot 59:1515–1524
Takenoya M, Nikolakakis K, Sagermann M (2010) Crystallographic insights into the pore structures and mechanisms of the EutL and EutM shell proteins of the ethanolamine-utilizing microcompartment of Escherichia coli. J Bacteriol 192:6056–6063
Tanaka S, Kerfeld CA, Sawaya MR, Cai F, Heinhorst S, Cannon GC, Yeates TO (2008) Atomic-level models of the bacterial carboxysome shell. Science 319:1083–1086
Tanaka S, Sawaya MR, Phillips M, Yeates TO (2009) Insights from multiple structures of the shell proteins from the beta-carboxysome. Protein Sci 18:108–120
Tanaka S, Sawaya MR, Yeates TO (2010) Structure and mechanisms of a protein-based organelle in Escherichia coli. Science 327:81–84
Thompson MC, Crowley CS, Kopstein J, Bobik TA, Yeates TO (2014) Structure of a bacterial microcompartment shell protein bound to a cobalamin cofactor. Acta Crystallogr F Struct Biol Commun 70:1584–1590
Ting CS, Hsieh C, Sundararaman S, Mannella C, Marko M (2007) Cryo-electron tomography reveals the comparative three-dimensional architecture of Prochlorococcus, a globally important marine cyanobacterium. J Bacteriol 189:4485–4493
Tsai Y, Sawaya MR, Cannon GC, Cai F, Williams EB, Heinhorst S, Kerfeld CA, Yeates TO (2007) Structural analysis of CsoS1A and the protein shell of the Halothiobacillus neapolitanus carboxysome. PLoS Biol 5:e144
Tsai Y, Sawaya MR, Yeates TO (2009) Analysis of lattice-translocation disorder in the layered hexagonal structure of carboxysome shell protein CsoS1C. Acta Crystallogr D Biol Crystallogr 65:980–988
Tsai YC, Lapina MC, Bhushan S, Mueller-Cajar O (2015) Identification and characterization of multiple rubisco activases in chemoautotrophic bacteria. Nat Commun 6:8883
Urano N, Kataoka M, Ishige T, Kita S, Sakamoto K, Shimizu S (2011) Genetic analysis around aminoalcohol dehydrogenase gene of Rhodococcus erythropolis MAK154: a putative GntR transcription factor in transcriptional regulation. Appl Microbiol Biotechnol 89:739–746
Wagner HJ, Capitain CC, Richter K, Nessling M, Mampel J (2017) Engineering bacterial microcompartments with heterologous enzyme cargos. Eng Life Sci 17:36–46
Wang H, Yan X, Aigner H, Bracher A, Nguyen ND, Hee WY, Long BM, Price GD, Hartl FU, Hayer-Hartl M (2019) Rubisco condensate formation by CcmM in β-carboxysome biogenesis. Nature 566:131–135
Wheatley NM, Gidaniyan SD, Liu Y, Cascio D, Yeates TO (2013) Bacterial microcompartment shells of diverse functional types possess pentameric vertex proteins. Protein Sci 22:660–665
Wheatley NM, Sundberg CD, Gidaniyan SD, Cascio D, Yeates TO (2014) Structure and identification of a pterin dehydratase-like protein as a ribulose-bisphosphate carboxylase/oxygenase (RuBisCO) assembly factor in the alpha-carboxysome. J Biol Chem 289:7973–7981
Woodger FJ, Badger MR, Price GD (2005) Sensing of inorganic carbon limitation in Synechococcus PCC 7942 is correlated with the size of the internal inorganic carbon pool and involves oxygen. Plant Physiol 139:1959–1969
Yeates TO, Jorda J, Bobik TA (2013) The shells of BMC-type microcompartment organelles in bacteria. J Mol Microbiol Biotechnol 23:290–299
Zarzycki J, Sutter M, Cortina NS, Erb TJ, Kerfeld CA (2017) In vitro characterization and concerted function of three core enzymes of a glycyl radical enzyme – associated bacterial microcompartment. Sci Rep 7:42757
Zhang S, Laborde SM, Frankel LK, Bricker TM (2004) Four novel genes required for optimal photoautotrophic growth of the cyanobacterium Synechocystis sp. strain PCC 6803 identified by in vitro transposon mutagenesis. J Bacteriol 186:875–879
Acknowledgements
First and foremost, we would like to acknowledge the generous financial support for our carboxysome research throughout the years from the National Science Foundation of the USA. Thanks go to our collaborators, Cristina Iancu and Grant J. Jensen, for providing us with the cryo electron tomogram image for Fig. 1, and to Markus Sutter and Cheryl Kerfeld for generating yet another beautiful BMC schematic (Fig. 2) for us. Their help is greatly appreciated.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 Springer Nature Switzerland AG
About this chapter
Cite this chapter
Heinhorst, S., Cannon, G.C. (2020). Bacterial Microcompartments. In: Jendrossek, D. (eds) Bacterial Organelles and Organelle-like Inclusions. Microbiology Monographs, vol 34. Springer, Cham. https://doi.org/10.1007/978-3-030-60173-7_6
Download citation
DOI: https://doi.org/10.1007/978-3-030-60173-7_6
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-60172-0
Online ISBN: 978-3-030-60173-7
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)