Abstract
Semantic segmentation is an important task in medical image analysis. In general, training models with high performance needs a large amount of labeled data. However, collecting labeled data is typically difficult, especially for medical images. Several semi-supervised methods have been proposed to use unlabeled data to facilitate learning. Most of these methods use a self-training framework, in which the model cannot be well trained if the pseudo masks predicted by the model itself are of low quality. Co-training is another widely used semi-supervised method in medical image segmentation. It uses two models and makes them learn from each other. All these methods are not end-to-end. In this paper, we propose a novel end-to-end approach, called difference minimization network (DMNet), for semi-supervised semantic segmentation. To use unlabeled data, DMNet adopts two decoder branches and minimizes the difference between soft masks generated by the two decoders. In this manner, each decoder can learn under the supervision of the other decoder, thus they can be improved at the same time. Also, to make the model generalize better, we force the model to generate low-entropy masks on unlabeled data so the decision boundary of model lies in low-density regions. Meanwhile, adversarial training strategy is adopted to learn a discriminator which can encourage the model to generate more accurate masks. Experiments on a kidney tumor dataset and a brain tumor dataset show that our method can outperform the baselines, including both supervised and semi-supervised ones, to achieve the best performance.
This work is supported by the NSFC-NRF Joint Research Project (No. 61861146001).
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Semantic segmentation is of great importance in medical image analysis, because it can help detect the location and size of anatomical structures and aid in making therapeutic schedule. With the development of deep learning, deep neural networks especially fully convolutional networks (FCN) [12] have shown promising performance in segmenting both natural images and medial images. The models in these methods have millions of parameters to be optimized, thus a large amount of labeled data with pixel-level annotations is typically needed for training such models to achieve promising performance. However, it is generally difficult to collect a large amount of labeled data in medical image analysis. One main reason is that annotating medical images needs expertise knowledge but few experts have time for annotation. Another reason is that it is time-consuming to annotate medical images.
Semi-supervised learning can utilize a large amount of unlabeled data to improve model performance. semiFCN [2] proposes a semi-supervised network-based approach for medical image segmentation. In semiFCN, a network is trained to predict pseudo masks. The predicted pseudo masks are then used to update the network in turn. ASDNet [14] trains a confidence network to select regions with high confidence in soft masks for updating the segmentation network. Zhou et al. [18] propose to jointly improve the performance of disease grading and lesion segmentation by semi-supervised learning with an attention mechanism. Souly et al. [17] use weakly labeled data and unlabeled data to train a generative adversarial network (GAN) [8], which can force real data to be close in feature space and thus cluster together. These methods all use a self-training framework, in which the model is updated using pseudo masks predicted by the model itself. If the pseudo masks predicted by the model itself have low quality, the model will be updated using data with noise. On the other hand, co-training [4] uses two models and each model is updated using unlabeled data with pseudo masks predicted by the other model and labeled data with ground truth. In this manner, each model in co-training is supervised by the other model. So the two models can be improved in turn. Several methods [9, 15] explore co-training in deep learning. But they are not end-to-end methods.
In this paper, we propose a novel end-to-end approach, called difference minimization network (DMNet), for semi-supervised semantic segmentation in medical images. The contributions of our method can be listed as follows:
-
DMNet is a semi-supervised segmentation model, which can be trained with a limited amount of labeled data and a large amount of unlabeled data.
-
DMNet adopts the widely used encoder-decoder structure [1, 7, 16], but it has two decoder branches with a shared encoder. DMNet minimizes the difference between the soft masks predicted by the two decoders to utilize unlabeled data. Unlike co-training which is often not end-to-end, the two decoders in DMNet can be updated at the same time in an end-to-end way.
-
DMNet uses the sharpen [3] operation to force the model to generate predictions with low entropy on unlabeled data, which can improve the model performance.
-
DMNet adopts adversarial learning derived from GAN for further improvement.
-
Experiments on a kidney tumor dataset and a brain tumor dataset show that our method can outperform other baselines to achieve the best performance.
2 Notation
We use \(\textit{\textbf{X}} \in \mathcal {R}^{H \times W}\) to denote an image in the labeled training set, and \(\textit{\textbf{Y}} \in \{0, 1\}^{H \times W \times K}\) to denote the corresponding ground-truth label which is encoded into a one-hot format. Here, K is the number of classes, H and W are the height and width of the image respectively. DMNet has two segmentation branches, and we denote the class probability maps generated by the two segmentation branches as \(\hat{\textit{\textbf{Y}}}^{(1)}, \hat{\textit{\textbf{Y}}}^{(2)} \in \mathcal {R}^{H \times W \times K}\). Furthermore, we denote an unlabeled image as \(\textit{\textbf{U}} \in \mathcal {R}^{H \times W}\). We use [1 : N] to denote \([1,2,\cdots ,N]\).
3 Method
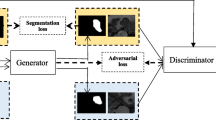
The framework of DMNet is shown in Fig. 1, which is composed of a segmentation network with two decoder branches, a sharpen operation for unlabeled data and a discriminator for both labeled and unlabeled data. Each component will be described detailedly in the following subsections.
3.1 Segmentation Network
As shown in Fig. 1, the segmentation network in DMNet adopts the widely used encoder-decoder architecture, which is composed of a shared encoder and two different decoders. By sharing an encoder, our segmentation network has some advantages. First, it can save GPU memory compared to the architecture in which two decoders use separate encoders. Second, since the encoder is shared by two decoders, it can be updated by the information from two decoders. Therefore it can learn better features from the difference between soft masks generated by two decoders, which can lead to better performance. This will be verified by our experimental results in Sect. 4. The two decoders in DMNet use different architectures to introduce diversity. By adopting different architectures, the two decoders will not typically output exactly the same segmentation masks and they can learn from each other. By using labeled and unlabeled data in turn, DMNet can utilize unlabeled data adequately to improve segmentation performance. DMNet is a general framework, and any segmentation network with an encoder-decoder architecture, such as UNet [16], VNet [13], SegNet [1] and DeepLab v3+ [7], can be used in DMNet. In this paper, we adopt UNet [16] and DeepLab v3+ [7] for illustration. The shared encoder can extract latent representation with high-level semantic information of the input image. Then we use the ground truth to supervise the learning of segmentation network for labeled data while minimizing the difference between the masks generated by the two decoders to let them learn from each other for unlabeled data.
We use Dice loss [13] to train our segmentation network on labeled data, which is defined as follows:
where \(\textit{\textbf{Y}}_{h,w,k} = 1\) when the pixel at position (h, w) belongs to class k, and other values in \(\textit{\textbf{Y}}_{h, w, k}\) is set to be 0. \(\hat{\textit{\textbf{Y}}}^{(i)}_{h,w,k}\) is the probability that the pixel at position (h, w) belongs to class k predicted by the segmentation branch i. \(\theta _s\) is the parameter of the segmentation network.
The loss function used for unlabeled data is described in Sect. 3.3.
3.2 Sharpen Operation
Given an unlabeled data \(\textit{\textbf{U}}\), our segmentation network can generate soft masks \(\hat{\textit{\textbf{Y}}}^{(1)}\) and \(\hat{\textit{\textbf{Y}}}^{(2)}\). To make the predictions of the segmentation networks have low entropy or high confidence, we adopt the sharpen operation [3] to reduce the entropy of predictions on unlabeled data, which is defined as follows:
where \(\hat{\textit{\textbf{Y}}}^{(i)}\) is the soft mask predicted by decoder branch i and temperature T is a hyperparameter.
3.3 Difference Minimization for Semi-supervised Segmentation
As described in Sect. 3.1, two decoders can generate two masks on unlabeled data. If the two masks vary from each other, it means the model is unsure about the predictions and thus the model cannot generalize well. Therefore, we minimize the difference between the two masks to make the two decoders generate consistent masks on the same unlabeled data. In other words, the two decoders can learn under the supervision of each other.
More specifically, given an unlabeled data \(\textit{\textbf{U}}\), the two decoder branches can generate two probability masks \(\hat{\textit{\textbf{Y}}}^{(1)}\) and \(\hat{\textit{\textbf{Y}}}^{(2)}\) which are processed by the sharpen operation. Since dice loss can measure the similarity of two segmentation masks and the loss can be backpropogated through two terms, we extend dice loss to the unlabeled setting and get the corresponding loss \(L_{semi}\) as follows:
From the definition of \(L_{semi}\), we can see that the two decoders can be updated by minimizing the difference between the masks they generate.
3.4 Discriminator
In DMNet, we also adopt adversarial learning to learn a discriminator. Unlike the original discriminator in GAN which discriminates whether an image is generated or is real, our discriminator adopts a fully convolutional network (FCN). The FCN discriminator is composed of three convolutional layers whose stride is 2 for downsampling and three corresponding upsampling layers. Each convolutional layer is followed by a ReLU layer. It can discriminate whether a region or some pixels are predicted or from ground truth.
Adversarial Loss for Discriminator. The objective function of discriminator can be written as follows:
where \(\theta _d\) is the parameter of the discriminator \(D(\cdot )\). \(\textit{\textbf{1}}\) and \(\textit{\textbf{0}}\) are tensors filled with 1 or 0 respectively, with the same size as that of the outputs of \(D(\cdot )\). The term \(L_{bce}(D(\textit{\textbf{Y}}), \textit{\textbf{1}})\) in \(L_{dis}(\hat{\textit{\textbf{Y}}}^{(1)}, \hat{\textit{\textbf{Y}}}^{(2)}, \textit{\textbf{Y}};\theta _d)\) is used only when the input data is labeled and is ignored when the input data is unlabeled data. \(L_{bce}\) is defined as follows:
where \(\theta \) is the parameter of \(\textit{\textbf{A}}\).
Adversarial Loss for Segmentation Network. In the adversarial learning scheme, the segmentation network tries to fool the discriminator. Hence, there is an adversarial loss \(L_{adv}\) for segmentation network to learn consistent features:
where \(\textit{\textbf{O}}\) denotes either a labeled image or an unlabeled image, \(\hat{\textit{\textbf{Y}}}^{(1)}\) and \(\hat{\textit{\textbf{Y}}}^{(2)}\) are the corresponding masks predicted by the two decoder branches in the segmentation network.
3.5 Total Loss
Based on the above results, the loss function for the segmentation network can be written as follows:
where \(\lambda _1\) and \(\lambda _2\) are two balance parameters. By integrating the discriminator, the objective of DMNet can be written as follows:
4 Experiments
We adopt two real datasets to evaluate DMNet and other baselines, including supervised baselines and semi-supervised baselines.
4.1 Dataset and Evaluation Metric
We conduct our experiments on the KiTS19Footnote 1 dataset and BraTS18Footnote 2 dataset. KiTS19 dataset is a kidney tumor dataset. It contains 210 labeled 3D computed tomography (CT) images for training and validation, and 90 CT images whose annotation is not published for testing. In our experiments, we use the 210 CT images with annotation to verify the effectiveness of our DMNet.
BraTS18 dataset is a brain tumor dataset. It contains 385 labeled 3D MRI scans and each MRI scan has four modalities (T1, T1 contrast-enhanced, T2 and FLAIR). We use T1, T1 contrast-enhanced and T2 modality to form a three-channel input. This dataset divides the brain tumor into four categories: whole tumor, tumor core, enhancing tumor structures and cystic/necrotic components. In our experiments, we combine these four categories so there are two classes in our experiment: tumor and background.
For each patient in KiTS19 and BraTS18, we choose one slice with its ground-truth label as a labeled image, and choose two slices as unlabeled images by discarding their labels. We split all labeled data into three subsets for training, validation and testing according to the proportion of 7:1:2. The unlabeled data is used for training only. Training data, validation data and testing data have no patient-level overlap to make sure that our model has never seen slices from validation patient or testing patient during training.
Mean Intersection over Union (mIoU) [11] can measure the similarity of any two shapes and is widely used in semantic segmentation. We also adopt mIoU as the evaluation metric.
4.2 Implementation Detail
We use PytorchFootnote 3 to implement DMNet on a workstation with an Intel (R) CPU E5-2620V4@2.1G of 8 cores, 128G RAM and an NVIDIA (R) GPU TITAN Xp. Our encoder network is ResNet101 [10] and we use it for all experiments. In the training phase, we resize the input image to 224 \(\times \) 224 for KiTS19 and 240 \(\times \) 240 for BraTS18, and randomly flip it horizontally with a probability of 0.5. In the inference phase, we use the average result of two segmentation branches as the final result. We train our model from scratch using Adam algorithm. The initial learning rate for segmentation network and discriminator is set to be 1e-4 and 1e-5, respectively. The weight decay is set to be 5e-5. We train our model for 150 epochs and decrease the learning rate according to a poly scheme [6]. In our experiment, \(\beta \) in poly is set to be 0.9. Without explicit statement, we set \(\lambda _1\) and \(\lambda _2\) to be 0.01 and 0.1 respectively and set temperature T to be 0.5.
4.3 Baselines
Several semi-supervised methods are adopted as baselines for comparison. More specifically, we compare DMNet to semiFCN [2] and SDNet [5]. semiFCN is a relatively early method in semi-supervised segmentation used for medical image analysis. SDNet is a state-of-the-art method in medical image segmentation. We carefully reimplement semiFCN and SDNet. We adopt ResNet101 as backbone for both methods for fair comparison.
We also design several supervised counterparts of DMNet to demonstrate the usefulness of unlabeled data and design some semi-supervised counterparts to demonstrate the usefulness of each component of DMNet. Supervised DMNet without adv denotes a supervised variant which adopts only labeled data for training without adversarial learning. Supervised DMNet with adv denotes a supervised variant which adopts only labeled data for training but the adversarial learning is adopted. Both variants do not minimize the difference between two decoder branches. Separate DMNet denotes a semi-supervised variant which adopts two separate encoders. That’s to say, Separate DMNet is composed of two separate encoder-decoder networks. DMNet_wo_adv_wo_sharpen denotes a semi-supervised variant which does not adopt the adversarial training strategy and sharpen operation. DMNet_wo_sharpen denotes a semi-supervised variant which does not adopt the sharpen operation on unlabeled data but adopts adversarial learning.
4.4 Comparison with Baselines
We compare our DMNet to baselines, including semiFCN [2] and SDNet [5], on KiTS19 dataset and BraTS18 dataset. The results are shown in Table 1. From the results, we can see that our DMNet outperforms these methods and achieves the best results, when trained with different amount of labeled data. DMNet has obvious advantage over other methods when the amount of labeled data is limited. When we use only 10% of the labeled data and all unlabeled data, DMNet can achieve 88.4% and 78.7% mIoU on KiTS19 and BraTS18, which outperforms semiFCN by 12.3% and 15.1%, and outperforms SDNet by 5.2% and 3.6%, respectively.
4.5 Ablation Study
We also perform ablation study on BraTS18 to show the effectiveness of each component used in DMNet.
Table 2 shows the results of Supervised DMNet without adv trained with 100% of the labeled data using different loss functions. From the results of Table 2, we can see that Dice loss can surpass the performance of cross entropy loss.
Table 3 shows the results of DMNet and its variants introduced in Subsect. 4.3. From the results of Separate DMNet, we can see that our architecture design, in which the two decoders share an encoder, has better performance than the architecture in which two decoders use separate encoders. Therefore, it proves that the architecture of DMNet has advantages. More specifically, it can save GPU memory and achieve better performance. Comparing the results between DMNet_wo_adv_wo_sharpen and DMNet_wo_sharpen, and the results between Supervised DMNet without adv and Supervised DMNet with adv, we can see that adversarial learning strategy can improve the performance whether in supervised setting or semi-supervised setting. From the results of DMNet_wo_sharpen and DMNet, we can see that the sharpen operation can benefit the learning on unlabeled data. Comparing the results of DMNet to those of supervised variants, we can conclude that the proposed DMNet can utilize unlabeled data to improve the segmentation performance, especially when the amount of labeled data is limited. When only 10% of labeled data is available, DMNet can improve the mIoU from 67.0% to 78.7%. When all labeled data is available, in which case the amount of unlabeled data is almost the same as that of labeled data, DMNet can also improve the mIoU from 84.2% to 87.0%.
5 Conclusion
In this paper, we propose a novel semi-supervised method, called DMNet, for semantic segmentation in medical image analysis. DMNet can be trained with a limited amount of labeled data and a large amount of unlabeled data. Hence, DMNet can be used to solve the problem that it is typically difficult to collect a large amount of labeled data in medical image analysis. Experiments on a kidney tumor dataset and a brain tumor dataset show that DMNet can outperform other baselines, including both supervised ones and semi-supervised ones, to achieve the best performance.
References
Badrinarayanan, V., Kendall, A., Cipolla, R.: SegNet: a deep convolutional encoder-decoder architecture for image segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 39(12), 2481–2495 (2017)
Bai, W., et al.: Semi-supervised learning for network-based cardiac MR image segmentation. In: Descoteaux, M., Maier-Hein, L., Franz, A., Jannin, P., Collins, D.L., Duchesne, S. (eds.) MICCAI 2017. LNCS, vol. 10434, pp. 253–260. Springer, Cham (2017). https://doi.org/10.1007/978-3-319-66185-8_29
Berthelot, D., Carlini, N., Goodfellow, I.J., Papernot, N., Oliver, A., Raffel, C.: MixMatch: a holistic approach to semi-supervised learning. CoRR (2019)
Blum, A., Mitchell, T.M.: Combining labeled and unlabeled data with co-training. In: Proceedings of Annual Conference on Computational Learning Theory (COLT) (1998)
Chartsias, A., et al.: Factorised spatial representation learning: application in semi-supervised myocardial segmentation. In: Frangi, A.F., Schnabel, J.A., Davatzikos, C., Alberola-López, C., Fichtinger, G. (eds.) MICCAI 2018. LNCS, vol. 11071, pp. 490–498. Springer, Cham (2018). https://doi.org/10.1007/978-3-030-00934-2_55
Chen, L., Papandreou, G., Kokkinos, I., Murphy, K., Yuille, A.L.: Semantic image segmentation with deep convolutional nets and fully connected CRFs. In: Proceedings of International Conference on Learning Representations (ICLR) (2015)
Chen, L., Zhu, Y., Papandreou, G., Schroff, F., Adam, H.: Encoder-decoder with atrous separable convolution for semantic image segmentation. In: Proceedings of European Conference on Computer Vision (ECCV) (2018)
Goodfellow, I.J., et al.: Generative adversarial nets. In: Proceedings of Neural Information Processing Systems (NIPS) (2014)
Han, B., Yao, Q., Yu, X., Niu, G., Xu, M., Hu, W., Tsang, I.W., Sugiyama, M.: Co-teaching: robust training of deep neural networks with extremely noisy labels. In: Proceedings of Neural Information Processing Systems (NIPS) (2018)
He, K., Zhang, X., Ren, S., Sun, J.: Deep residual learning for image recognition. In: Proceedings of Computer Vision and Pattern Recognition (CVPR) (2016)
Jaccard, P.: Étude comparative de la distribution florale dans une portion des alpes et des jura. Bull. Soc. Vaudoise Sci. Nat. 37, 547–579 (1901)
Long, J., Shelhamer, E., Darrell, T.: Fully convolutional networks for semantic segmentation. In: Proceeding of Computer Vision and Pattern Recognition (CVPR) (2015)
Milletari, F., Navab, N., Ahmadi, S.: V-net: fully convolutional neural networks for volumetric medical image segmentation. In: Proceeding of 3D Vision (3DV) (2016)
Nie, D., Gao, Y., Wang, L., Shen, D.: ASDNet: attention based semi-supervised deep networks for medical image segmentation. In: Frangi, A.F., Schnabel, J.A., Davatzikos, C., Alberola-López, C., Fichtinger, G. (eds.) MICCAI 2018. LNCS, vol. 11073, pp. 370–378. Springer, Cham (2018). https://doi.org/10.1007/978-3-030-00937-3_43
Qiao, S., Shen, W., Zhang, Z., Wang, B., Yuille, A.L.: Deep co-training for semi-supervised image recognition. In: Proceedings of European Conference on Computer Vision (ECCV) (2018)
Ronneberger, O., Fischer, P., Brox, T.: U-Net: convolutional networks for biomedical image segmentation. In: Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F. (eds.) MICCAI 2015. LNCS, vol. 9351, pp. 234–241. Springer, Cham (2015). https://doi.org/10.1007/978-3-319-24574-4_28
Souly, N., Spampinato, C., Shah, M.: Semi and weakly supervised semantic segmentation using generative adversarial network. CoRR (2017)
Zhou, Y., et al.: Collaborative learning of semi-supervised segmentation and classification for medical images. In: Proceeding of Computer Vision and Pattern Recognition (CVPR) (2019)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 Springer Nature Switzerland AG
About this paper
Cite this paper
Fang, K., Li, WJ. (2020). DMNet: Difference Minimization Network for Semi-supervised Segmentation in Medical Images. In: Martel, A.L., et al. Medical Image Computing and Computer Assisted Intervention – MICCAI 2020. MICCAI 2020. Lecture Notes in Computer Science(), vol 12261. Springer, Cham. https://doi.org/10.1007/978-3-030-59710-8_52
Download citation
DOI: https://doi.org/10.1007/978-3-030-59710-8_52
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-59709-2
Online ISBN: 978-3-030-59710-8
eBook Packages: Computer ScienceComputer Science (R0)