Abstract
Computational development of novel triazole-based aromatase inhibitors (AIs) was carried out followed by investigation of the possible interaction modes of these compounds with the enzyme and prediction of the binding affinity by tools of molecular modeling. In doing so, in silico design of potential AIs candidates fully satisfying the Lipinski’s “rule of five” was performed using the concept of click chemistry. Complexes of these drug-like molecules with the enzyme were then simulated by molecular docking and optimized by semiempirical quantum chemical method PM7. To identify the most promising compounds, stability of the PM7-based ligand/aromatase structures was estimated in terms of the values of binding free energies and dissociation constants. At the final stage, structures of the top ranking compounds bound to aromatase were analyzed by molecular dynamic simulations and binding free energy calculations. As a result, eight hits that specifically interact with the aromatase catalytic site and exhibit the high-affinity ligand binding were selected for the final analysis. The selected AIs candidates show strong attachment to the enzyme active site, suggesting that these small drug-like molecules may present good scaffolds for the development of novel potent drugs against breast cancer.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
In women organism during the fertile phase, estrogen synthesis occurs mainly in the ovaries. However, the intensity of estrogen synthesis in the ovaries decreases in post-menopause associated with about a third of cases of breast cancer [1,2,3]. At this phase, estrogens synthesized in the peripheral tissues using the cytochrome P450 complex, called aromatase. This complex consists of the heme-containing cytochrome P450 (CYP19A1) protein and flavoprotein NADPH-cytochrome P450 reductase [1,2,3]. Aromatase encoded by a single large gene, CYP19A1, catalyzes conversion of androgens to estrogens and exhibits biological activity in both peripheral target tissues and the mammary tumor tissues, providing a high level of estrogen concentration [1,2,3]. In estrogen-dependent malignant neoplasms, estrogens act as growth factors for tumor development. Therefore, inhibition of aromatase results in a decrease of estrogens level in the organism and prevents to growth and spread of cancer cells [1,2,3].
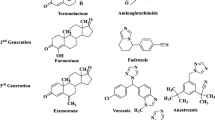
There are three generations of AIs among the drugs for treating hormone-dependent breast cancer. The disadvantage of the drugs of the first two generations (aminoglutethimide, fadrozole, formestane) is the lack of selectivity of action: besides aromatase, these drugs inhibit a number of other enzymes. The third-generation AIs vorozole, letrozole, anastrozole, and exemestane approved for clinical use by the USA Food and Drug Administration show greater specificity and efficacy. These inhibitors include drugs of two categories, namely i) irreversible steroidal inhibitor exemestane that is an androstenedione derivative and ii) reversible non-steroidal inhibitors vorozole, anastrozole and letrozole. Steroidal AIs and, in particular, exemestane are transformed by aromatase into compounds that irreversibly bind to the enzyme active site, completely disrupting its activity as a biocatalyst. After the termination of the action of these inhibitors, aromatase needs considerable time to be synthesized in the tissues again. Reversible nonsteroidal AIs vorozole, letrozole and anastrozole are triazole compounds that bind to the catalytic site of the enzyme by coordinating the heme iron of the CYP19A1 through a heterocyclic nitrogen lone pair. The third generation AIs are now the front-line drugs for treating the early and advanced stages of breast cancer in postmenopausal women.
Despite significant progress in the treatment of hormone-dependent breast cancer, this problem has not been completely solved. Unfortunately, the third-generation of aromatase inhibitors (AIs) cause a number of serious side effects, such as inhibition of muscle growth, arthralgia, decreased bone strength, impaired blood lipid profile, drop in libido, as well as deterioration of the general condition. In addition, resistance acquired after long-term therapy with these drugs also occurs. In this context, development of novel, more effective and less toxic AIs is of great value.
2 Methods
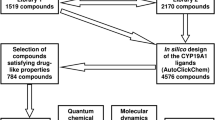
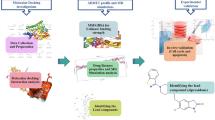
In this study, computational development of novel triazole-based AIs was carried out followed by evaluation of their antitumor activity by tools of molecular modeling. In doing so, the following studies were performed: i) in silico design of potential aromatase inhibitor candidates by the AutoClickChem techniques [4]; ii) identification of compounds satisfying the Lipinski’s “rule of five” [5]; iii) molecular docking of these drug-like compounds with the enzyme active site using the QuickVina 2 program [6]; iv) refinement of the ligand-binding poses by the PM7 semiempirical quantum chemical method [7]; v) prediction of the interaction modes dominating the binding; vi) calculation of the values of binding free energy and dissociation constant (Kd) for the ligand/CYP19A1 complexes; vii) prediction of the binding affinity between the identified compounds and aromatase by molecular dynamics simulations and binding free energy calculations [8]; and viii) selection of molecules most promising for synthesis and biochemical trials.
3 Results
Based on the analysis of the data obtained, eight top-ranked compounds that exhibited the low values of binding free energy (<−7 kcal/mol) in the PM7-based ligand/aromatase complexes were selected for the final analysis. Depending on the mechanism of binding to the active site of CYP19A1, these compounds were divided into two structural groups designated as groups 1 and 2. The data of molecular modeling indicate that each of the identified compounds of group 1 (Fig. 1) shows peculiar interactions with the enzyme binding pocket, the interaction being realized between the triazole ring and the heme iron, van der Waals interactions with the hydrophobic pocket lined by Arg-115, Ile-133, Phe-134, Trp-224, Thr-310, Val-370, Met-374, Leu-477, Ser-478, and, except compound II, the hydrogen bond with Met-374, which is also involved in hydrogen bonding with the natural substrate androstenedione. In addition, some identified compounds form van der Waals contacts with the heme of CYP19A1, and π-conjugated systems of individual molecules participate in specific π-π interactions with the pyrrole rings of the heme group. Finally, the selected AIs candidates expose strong attachment to the enzyme active site, in line with the low values of binding free energy and Kd (Table 1). In summary, the conclusions that can be made by the new identified AIs from group 1 are that, in addition to the interaction between the triazole rings and the heme iron, hydrophobic contacts play a pivotal role in the ligand binding, and hydrogen bond involving Met-374 is essential for the ligand recognition (Fig. 1).
Unlike the molecules of group 1, the ligands of group 2 demonstrate a mechanism of binding to aromatase uncharacteristic for triazole-based compounds generally coordinating the iron atom of the heme via a heterocyclic nitrogen lone pair. According to the calculated data (Fig. 2), the ligands of interest coordinate the iron atom of the CYP19A1 heme group through the lone pairs of their oxygen atoms. Similarly to the molecules of group 1, all these compounds target the well-conserved hotspots of the aromatase catalytic site using multiple van der Waals interactions with the critically important residues of this hydrophobic pocket.
Molecular dynamics insights into the ligand/aromatase complexes validate the main findings derived from the analysis of their static structures. These complexes are relatively stable during the MD simulations, which is supported by the averages of binding free energies, their enthalpic components, and corresponding standard deviations (Table 3). The averages of binding free energy predicted for the designed compounds in the complexes with aromatase are comparable with the value calculated for the letrozole/aromatase complex by the identical computational protocol (Table 3). Furthermore, these averages are also comparable with the values estimated for the static models of the ligand/aromatase complexes (Tables 1, 2) as well as to that of −12.06 kcal/mol obtained with the formula ∆G = R × T × ln(Kd) [9] at temperature T = 310 K using the experimental value of Kd for letrozole bound to aromatase [10]
Besides, compound II participates in specific π-π interactions with the pyrrole rings of the CYP19A1 heme group and form hydrogen bonds with residues Met-374 and Thr-310, and compound I forms a salt bridge with the enzyme heme.
Finally, analysis of the data of Table 2 indicates that, like the molecules from group 1, the compounds of group 2 exhibit a high binding affinity in the complexes with aromatase, as evidence with the low values of dissociation constant and binding free energy.
Unlike the molecules of group 1, the ligands of group 2 demonstrate a mechanism of binding to aromatase uncharacteristic for triazole-based compounds generally coordinating the iron atom of the heme via a heterocyclic nitrogen lone pair. According to the calculated data, the ligands of interest coordinate the iron atom of the CYP19A1 heme group through the lone pairs of their oxygen atoms. Similarly to the molecules of group 1, all these compounds target the well-conserved hotspots of the aromatase catalytic site using multiple van der Waals interactions with the critically important residues of this hydrophobic pocket. Besides, compound II participates in specific π-π interactions with the pyrrole rings of the CYP19A1 heme group and form hydrogen bonds with residues Met-374 and Thr-310, and compound I forms a salt bridge with the enzyme heme.
Finally, analysis of the ligand/aromatase complexes indicates a high binding affinity between the identified compounds and the enzyme, in agreement with the low values of binding free energy calculated both for their static and dynamic models.
4 Conclusions
Thus, the data of molecular modeling indicate that each of the identified compounds of group 1 shows peculiar interactions with the enzyme binding pocket, the interaction being realized between the triazole ring and the heme iron, van der Waals interactions with the hydrophobic pocket lined by Arg-115, Ile-133, Phe-134, Trp-224, Thr-310, Val-370, Met-374, Leu-477, Ser-478, and, except compound II, the hydrogen bond with Met-374, which is also involved in hydrogen bonding with the natural substrate androstenedione. In addition, some identified compounds form van der Waals contacts with the heme of CYP19A1, and π-conjugated systems of individual molecules participate in specific π-π interactions with the pyrrole rings of the heme group. Finally, the selected AIs candidates expose strong attachment to the enzyme active site, in line with the low values of binding free energy and Kd. In summary, the conclusions that can be made by the new identified AIs are that, in addition to the interaction between the triazole rings and the heme iron, hydrophobic contacts play a pivotal role in the ligand binding, and hydrogen bond involving Met-374 is also essential for the ligand recognition.
References
Macedo, L.F., Sabnis, G., Brodie, A.: Aromatase inhibitors and breast cancer. Ann. N.Y. Acad. Sci. 1155, 162–173 (2009)
Ghosh, D., Griswold, J., Erman, M., Pangborn, W.: Structural basis for androgen specificity and oestrogen synthesis in human aromatase. Nature 457, 219–223 (2009)
Hong, Y., Chen, S.: Aromatase inhibitors: structural features and biochemical characterization. Ann. N.Y. Acad. Sci. 1089, 237–251 (2006)
Durrant, J.D., McCammon, J.A.: AutoClickChem: click chemistry in silico. PLoS Comput. Biol. 8, e1002397 (2012)
Lipinski, C.A., Lombardo, F., Dominy, B.W., Feeney, P.J.: Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 46, 3–26 (2001)
Handoko, S.D., Ouyang, X., Su, C.T.T., Kwoh, C.K., Ong, Y.S.: QuickVina: accelerating AutoDock vina using gradient-based heuristics for global optimization. IEEE/ACM Trans. Comput. Biol. Bioinform. 9, 1266–1272 (2012)
Stewart, J.J.P.: Optimization of parameters for semiempirical methods VI: more modifications to the NDDO approximations and re-optimization of parameters. J. Mol. Model. 19, 1–32 (2013)
Case, D.A., et al.: AMBER 2016. University of California, San Francisco (2016)
Sharma, G., First, E.A.: Thermodynamic analysis reveals a temperature-dependent change in the catalytic mechanism of bacillus stearothermophilus tyrosyl-tRNA synthetase. J. Biol. Chem. 284, 4179–4190 (2009)
Adamchik, S., et al.: Synthesis and properties of new triazole aromatase inhibitors. In: Proceedings of Belarusian State University, vol. 11, pp. 280–290 (2016). (in Russian)
Acknowledgments
This study was supported by a grant of the State Program of Scientific Research “Chemical Technologies and Materials” (subprogram 2.2 “Biologically active compounds”, project 2.15).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 Springer Nature Switzerland AG
About this paper
Cite this paper
Andrianov, A.M., Nikolaev, G.I., Kornoushenko, Y.V., Usanov, S.A. (2020). In Silico Design and Evaluation of Novel Triazole-Based Compounds as Promising Drug Candidates Against Breast Cancer. In: Cai, Z., Mandoiu, I., Narasimhan, G., Skums, P., Guo, X. (eds) Bioinformatics Research and Applications. ISBRA 2020. Lecture Notes in Computer Science(), vol 12304. Springer, Cham. https://doi.org/10.1007/978-3-030-57821-3_29
Download citation
DOI: https://doi.org/10.1007/978-3-030-57821-3_29
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-57820-6
Online ISBN: 978-3-030-57821-3
eBook Packages: Computer ScienceComputer Science (R0)