Abstract
There is an increasingly pressing need, by several applications in diverse domains, for developing techniques able to index and mine very large collections of sequences, or data series. It is not unusual for these applications to involve numbers of data series in the order of billions, which are often times not analyzed in their full detail due to their sheer size. In this work, we describe techniques for indexing and efficient similarity search in truly massive collections of data series, focusing on the iSAX family of data series indexes. We present their design characteristics, and describe their evolution to address different needs: bulk loading, adaptive indexing, parallelism and distribution, variable-length query answering, and bottom-up indexing. Based on this discussion, we conclude by presenting promising research directions.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Data series have gathered the attention of the data management community for almost three decades [54], and still represent an active and challenging research direction [7, 56, 83]. Data series are one of the most common data types, present in virtually every scientific and social domain [56]: they appear as audio sequences [34], shape and image data [76], financial [67], environmental monitoring [64], scientific data [30], and others. It is nowadays not unusual for applications to involve numbers of sequences in the order of billions [1, 2].
A data series, or data sequence, is an ordered sequence of data pointsFootnote 1. Formally, a data series \(T = (p_1,\,...\,p_n)\) is defined as a sequence of points \(p_i=(v_i, t_i)\), where each point is associated with a value \(v_i\) and a time \(t_i\) in which this recording was made, and n is the size (or length) of the series. If the dimension that imposes the ordering of the sequence is time then we talk about time series, though, a series can also be defined over other measures (e.g., angle in radial profiles in astronomy, mass in mass spectroscopy, position in biology, etc.).
A key observation is that analysts need to process and analyze a sequence (or subsequence) of values as a single object, rather than the individual points independently, which is what makes the management and analysis of data sequences a hard problem. In this context, Nearest Neighbor (NN) queries are of paramount importance, since they form the basis of virtually every data mining, or other complex analysis task involving data series [56]. However, NN queries on a large collection of data series are challenging, because data series collections grow very large in practice [13, 63]. Thus, methods for answering NN queries rely on two main techniques: data summarization and indexing. Data series summarization is used to reduce the dimensionality of the data series [3, 16, 35, 36, 43, 44, 62], and indexes are built on top of these summarizations [5, 62, 66, 70, 72].
In this study, we review the iSAX family of data series indexes, which all use the iSAX summarization technique to reduce the dimensionality of the original sequences. These indexes have attracted lots of attention, and represent the current state-of-the-art for several variations of the general problem. In particular, we present the iSAX summarization and discuss how it can be used to build the basic iSAX index [68, 69]. We describe iSAX2.0 [12] and iSAX2+ [13], the first data series indexes that inherently support bulk loading, allowing us to index datasets with 1 billion data series. We present the ADS and ADS+ indexes [78,79,80], which are the first adaptive data series indexes than can start answering queries correctly before the entire index has been built, as well as ADS-Full [80], which based on the same principles leads to an efficient 2-pass index creation strategy. We discuss ParIS [58] and ParIS+ [60], the first parallel data series indexes designed for modern hardware, and MESSI [59], a variation optimized for operation on memory-resident datasets. DPiSAX [42, 74, 75] is a distributed index that operates on top of Spark. We present ULISSE [45, 46], which is the first index that can inherently support queries of varying length. Finally, we describe Coconut [38,39,40], the first balanced index, which is built in a bottom-up fashion using a sortable iSAX-based summarization.
It is interesting to note that these indexes can be used not only for similarity search of data series, but also of general high-dimensional vectors [23, 24], leading to better performance than other high-dimensional techniques (including the popular LSH-based methods) [24].
By presenting all these indexes togetherFootnote 2, we contribute to the better understanding of the particular problems that each one solves, the way that their features could be combined, and the opportunities for future work.
2 Background and Preliminaries
[Data Series Queries]. Analysts need to perform (a) simple Selection-Projection-Transformation (SPT) queries, and (b) more complex Data-Mining (DM) queries. Simple SPT queries are those that select sequences and project points based on thresholds, point positions, or specific sequence properties (e.g., “above”, “first 10 points”, “peaks”), or queries that transform sequences using mathematical formulas (e.g., average). An example SPT query could be one that returns the first x points of all the sequences that have at least y points above a threshold. The majority of these queries could be handled (albeit not optimally) by current data management systems, which nevertheless, lack a domain specific query language that would support and facilitate such processing. DM queries on the other hand are more complex: they have to take into consideration the entire sequence, and treat it as a single object. Examples are: queries by content (range and similarity queries), clustering, classification, outlier, frequent sub-sequences, etc. These queries cannot be efficiently supported by current data management systems, since they require specialized data structures, algorithms, and storage methods.
Note that the data series datasets and queries may refer to either static, or streaming data. In the case of streaming data series, we are interested in the sub-sequences defined by a sliding window. The same is also true for static data series of very large size (e.g., an electroencephalogram, or a genome sequence), which we divide into sub-sequences using a sliding (or shifting) window. The length of these sub-sequences is chosen so that they contain the patterns of interest.
One of the most basic data mining tasks is that of finding similar data series, or NN in a database [3]. Similarity search is an integral part of most data mining procedures, such as clustering [73], classification and deviation detection [11, 17]. Even though several distance measures have been proposed in the literature [6, 10, 18, 21, 51, 71], the Euclidean distance is the most widely used and one of the most effective for large data series collections [22]. We note that an additional advantage of Euclidean distance is that in the case of Z-normalized series (mean = 0, stddev = 1), which are very often used in practice [81, 82], it can be exploited to compute Pearson correlation [61].
[Data Series Summarizations]. A common approach for answering such queries is to perform a dimensionality reduction, or summarization technique. Several such summarizations have been proposed, such as the Discrete Fourier Transform (DFT) [3], the Discrete Wavelet Transform (DWT) [16], the Piecewise Aggregate Approximation (PAA) [36, 77], the Adaptive Piecewise Constant Approximation (APCA) [15], or the Symbolic Aggregate approXimation (SAX) [44]. Note that that on average, there is little difference among these summarizations in terms of fidelity of approximation [22, 57] (even though it is the case that certain representations favor particular data types, e.g., DFT for star-light-curves, APCA for bursty data, etc.).
These summarizations are usually accompanied by distance bounding functions that relate distances in the summarized space to distances in the original space through either lower or upper-bounding. With such bounding functions, we can index data series directly in the summarized space [5, 62, 66, 70, 72], and use these indexes to efficiently answer NN queries on large data series collections.
[Data Series Indexing]. Even though recent studies have shown that in certain cases sequential scans can be performed very efficiently [63], such techniques are only applicable when the database consists of a single, long data series, and queries are looking for potential matches in small subsequences of this long data series. Such approaches, however, do not bring benefit to the general case of querying a mixed database of several data series. Therefore, indexing is required in order to efficiently support data exploration tasks, which involve ad-hoc queries, i.e., the query workload is not known in advance.
A large set of indexing methods have been proposed for the different data series summarization methods, including traditional multidimensional [9, 29, 37, 62] and specialized [5, 66, 70, 72] indexes. Moreover, various distance measures have been presented that work on top of such indexes, e.g., Discrete Time Warping (DTW) and Euclidean Distance (ED).
Indexing can significantly reduce the time to answer DM queries. Nevertheless, recent studies have observed that the mere process of building the index can be prohibitively expensive in terms of time cost [12, 13, 78]: e.g., the process of creating the index for 1 billion data series takes several days to complete. This problem can be mitigated by the bulk loading technique. Bulk-loading has been studied in the context of traditional database indexes, such as B-trees and R-trees, and other multi-dimensional index structures [4, 25, 32, 33, 41, 65].
3 The iSAX Family of Indexes
In this section, we describe the iSAX family of indexes, that is, all the indexes that are designed based on the iSAX summarization, and discuss their evolution over time. Figure 1 depicts the lineage of these indexes, along with the corresponding timeline. We note that all these indexes support both Z-normalized and non Z-normalized series, and the same index can answer queries using both the Euclidean and Dynamic Time Warping (DTW) distances (in the way mentioned in [59]), for k-NN and \(\epsilon \)-range queries [23]. Finally, recent extensions of some of these indexes demonstrate that they can efficiently support approximate similarity search with quality guarantees (deterministic and probabilistic) [24], and that they dominate the state-of-the-art in the case of general high-dimensional vectors, as well [23, 24].
3.1 The iSAX Summarization and Basic Index
The Piecewise Aggregate Approximation (PAA) [36, 77] is a summarization technique that segments the data series in equal parts and calculates the average value for each segment. An example of a PAA representation can be seen in Fig. 2; in this case the original data series is divided into 4 equal parts. Based on PAA, Lin et al. [44] introduced the Symbolic Aggregate approXimation (SAX) representation that partitions the value space in segments of sizes that follow the normal distribution. Each PAA value can then be represented by a character (i.e., a small number of bits) that corresponds to the segment that it falls into. This leads to a representation with a very small memory footprint, an important advantage when managing large sequence collections. A segmentation of size 3 can be seen in Fig. 2, where the series is represented by SAX word “\(10\ 10\ 11\)”.
Lineage of the iSAX family of indexes. Timeline is depicted on the top; implementation languages are marked on the right. Solid arrows denote inheritance of the index design; dashed arrows denote inheritance of some of the design features; the two new versions of iSAX2+ and ADS+ marked with an asterisk support approximate similarity search with deterministic and probabilistic quality guarantees. Source code available by following the links in the corresponding papers.
The SAX representation was later extended to indexable SAX (iSAX) [70], which allows variable cardinality for each character of a SAX representation. An iSAX representation is composed of a set of characters that form a word, and each word represents a data series. In the case of a binary alphabet, with a word size of 3 characters and a maximum cardinality of 2 bits, we could have a set of data series (two in the following example) represented with the following words: \(00_2 10_2 01_2\), \(00_2 11_2 01_2\), where each character has a full cardinality of 2 bits and each word corresponds to one data series. Reducing the cardinality of the second character in each word, we get for both words the same iSAX representation: \(00_2 1_1 01_2\) (\(1_1\) corresponds to both 10 and 11, since the last bit is trailed when the cardinality is reduced). Starting with a cardinality of 1 for each character in the root node and gradually splitting by increasing the cardinality one character at a time, we can build in a top-down fashion the (non-balanced) iSAX tree index [69, 70]. These algorithms can be efficiently implemented with bit-wise operations.
An example of iSAX and SAX representations [78].
The iSAX index supports both approximate and exact similarity search [23]: approximate does not guarantee that it will always find the correct answers (though, in most cases it returns high-quality results [24, 70]); exact guarantees that it will always return the correct results. In approximate search, the algorithm uses the iSAX summaries to traverse a single path of the index tree from the root to the most promising leaf, then computes the raw distances between the query and each series in the leaf, and return the series with the smallest distance, i.e., the Best-So-Far distance (BSF). Exact search starts with an approximate search that returns a BSF, which is then used to prune the rest of the index leaves; the leaves that cannot be pruned are visited, the raw distances of the series to the query are computed, and the BSF is updated (if needed). At the end of this process, we get the exact answer.
3.2 Bulk-Loading: iSAX 2.0 and iSAX2+
Inserting a large collection of data series into the index iteratively is an expensive operation, involving a high number of disk I/O operations [12, 13]. This is because for each time series, we have to store the raw data series on disk, and insert into the index the corresponding iSAX representation. In order to speedup the process of building the index, iSAX 2.0 [12] and iSAX2+ [13] were the first data series indexes (based on the iSAX index) with a bulk loading strategy.
The key idea is to effectively group the data series that will end up in a particular subtree of the index, and process them all together. In order to achieve this goal, we use two main memory buffer layers, namely, the First Buffer Layer (FBL), and the Leaf Buffer Layer (LBL) [13]. The FBL corresponds to the children of the root of the index, while the LBL corresponds to the leaf nodes. The role of the buffers in FBL is to cluster together data series that will end up in the same subtree of the index, rooted in one of the direct children of the root. In contrast, the buffers in LBL are used to gather all the data series of leaf nodes, and flush them to disk.
The algorithm operates in two phases, which alternate until the entire dataset is processed, as follows (for more details, refer to [13]). During Phase 1, the algorithm reads data series and inserts them in the corresponding buffer in the FBL. This phase continues until the main memory is full. Then Phase 2 starts, where the algorithm proceeds by moving the data series contained in each FBL buffer to the appropriate LBL buffers. During this phase, the algorithm processes the buffers in FBL sequentially. For each FBL buffer, the algorithm creates all the necessary internal and leaf nodes, in order to index these data series. When all data series of a specific FBL buffer have been moved down to the corresponding LBL buffers, the algorithm flushes these LBL buffers to disk.
The difference between iSAX 2.0 [12] and iSAX2+ [13] is that the former treats the data series raw values (i.e., the detailed sequence of all the values of the data series) and their summarizations (i.e., the iSAX representations) together, while the latter uses just the summarizations in order to build the index, and only processes the raw values in order to insert them to the correct leaf node. In both cases, the goal is to minimize the random disk I/O, by making sure that the data series that end up in the same leaf node of the index are (temporarily) stored in the same (or contiguous) disk pages. The experiments demonstrate that iSAX 2.0 and iSAX2+ significantly outperform previous approaches, reducing the time required to index 1 billion data series by 72% and 82%, respectively. A recent extension of iSAX2+ supports approximate answers with quality guarantees [24].
3.3 Adaptive Indexing: ADS, ADS+, ADS-Full
Even though iSAX 2.0 and iSAX2+ can effectively cope with very large data series collections, users still have to wait for extended periods of time before the entire index is built and being able to start answering queries.
The Adaptive Data Series (ADS) and ADS+ indexes [78, 79] are based on the iSAX 2.0 index, and address the above problem. They perform only a few basic steps, mainly creating the basic skeleton of the index tree, which contains condensed information on the input data series, and are then ready to start answering queries. As queries arrive, ADS fetches data series from the raw data and moves only those data series needed to correctly answer the queries inside the index. Future queries may be completely covered by the contents of the index, or alternatively ADS adaptively and incrementally fetches any missing data series directly from the raw data set. When the workload stabilizes, ADS can quickly serve fully contained queries while as the workload shifts, ADS may temporarily need to perform some extra work to adapt before stabilizing again.
The additional feature of ADS+ (when compared to ADS) is that it does not require a fixed leaf size: it dynamically and adaptively adjusts the leaf size in hot areas of the index. ADS+ uses two different leaf sizes: a big build-time leaf size for optimal index construction, and a small query-time leaf size for optimal access costs. Initially, the index tree is built as in plain ADS, with a constant leaf size, equal to build-time leaf size. In traditional indexes, this leaf size remains the same across the life-time of the index. In our case, when a query that needs to search a partial leaf arrives, ADS+ refines its index structure on-the-fly by recursively splitting the target leaf, until the target sub-leaf becomes smaller or equal to the query-time leaf size.
ADS and ADS+ support the same query answering mechanisms as iSAX2.0 and iSAX2+, but they also introduced the Scan of In-Memory Summarizations (SIMS) algorithm for exact query answering. SIMS starts by an approximate search to compute the BSF, which is then used to compare to the in-memory iSAX summaries of all the series in the collection, and finally, performs a skip-sequential scan of the raw series that were not pruned in the previous step.
Experiments with up to 1 billion data series and \(10^5\) random approximate queries show that ADS+ answers all queries in less than 5 h, while iSAX 2.0 needs more than 35 h. In turn, ADS+ and iSAX 2.0 are orders of magnitude faster in index creation than KD-Tree [8], R-Tree [29], and X-Tree [9].
In settings where a complete index is required, i.e., when there is a completely random and very large work-load, a full index can also be efficiently constructed using ADS-Full [80]. In the first step, the ADS structure is built by performing a full pass over the raw data file, storing only the iSAX representations at each leaf. In the second step, one more sequential pass over the raw data file is performed, and data series are moved in the correct pages on disk. The benefit of this process is that it completely skips costly split operations on raw data series, leading to a 2x–3x faster creation of the full index, when compared to iSAX 2.0. A recent extension of ADS+ supports approximate answers with quality guarantees [24].
3.4 Parallel and Distributed: ParIS, ParIS+, MESSI, DPiSAX
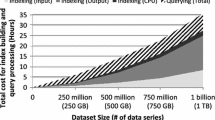
The continued increase in the rate and volume of data series production with collections that grow to several terabytes in size [53] renders single-core data series indexing technologies inadequate. For example, ADS+ [80], requires >4 min to answer a single exact query on a moderately sized 250 GB sequence collection.
The Parallel Index for Sequences (ParIS) [58], based on ADS+, is the first data series index that takes advantage of modern hardware parallelization, and incorporate the state-of-the-art techniques in sequence indexing, in order to accelerate processing times. ParIS, which is a disk-based index, can effectively operate on multi-core and multi-socket architectures, in order to distribute and execute in parallel the computations needed for both index construction and query answering. Moreover, ParIS exploits the Single Instruction Multiple Data (SIMD) capabilities of modern CPUs, to further parallelize the execution of individual instructions inside each core. Overall, ParIS achieves very good overlap of the CPU computation with the required disk I/O. ParIS+ [60], an alternative of ParIS, completely removes the CPU cost during index creation, resulting in index creation that is purely I/O bounded, and 2.6x faster than ADS+. ParIS+ achieves this by reorganizing the way that the workload is distributed among the worker threads. ParIS and ParIS+ employ the same algorithmic techniques for query answering. The experiments also demonstrate their effectiveness in exact query answering: they are up to 1 order of magnitude faster than ADS+, and up to 3 orders of magnitude faster than the state-of-the-art optimized serial scan method, UCR Suite [63]. We also note that ParIS and ParIS+ have the potential to deliver more benefit as we move to faster storage media.
Still, ParIS+ is designed for disk-resident data and therefore its performance is dominated by the I/O costs it encounters. For instance, ParIS+ answers a 1-NN exact query on a 100 GB dataset in 15 s, which is above the limit for keeping the user’s attention (i.e., 10 s), let alone for supporting interactivity in the analysis process (i.e., 100 ms) [26]. The in-MEmory data SerieS Index (MESSI) [59] is based on ParIS+, and is the first parallel index designed for memory-resident datasets. MESSI effectively uses multi-core and multi-socket architectures in order to concurrently execute the computations needed for both index construction and query answering, and it exploits SIMD. Since MESSI copes with in-memory data series, no CPU cost can be hidden under I/O, and required more careful design choices and coordination of the parallel workers when accessing the required data structures, in order to improve its performance. This led to the development of a more subtle design for the construction of the index and on the development of new algorithms for answering similarity search queries on this index. The results show a further \(\sim \)4x speedup in index creation time, in comparison to an in-memory version of ParIS+. Furthermore, MESSI answers exact 1-NN queries on 100 GB datasets 6-11x faster than ParIS+, achieving for the first time interactive exact query answering times, at \(\sim \)50 ms.
In order to exploit parallelism across compute nodes, the Distributed Partitioned iSAX (DPiSAX) [42, 74, 75] index was developed. DPiSAX is based on iSAX2+, and was designed to operate on top of Spark. DPiSAX uses a sampling phase that allows to balance the partitions of data series across the compute nodes (according to their iSAX representations), which is necessary for efficient query processing. DPiSAX gracefully scales to billions of time series, and a parallel query processing strategy that, given a batch of queries, efficiently exploits the index. The experiments show that DPiSAX can build its index on 4 billion data series in less than 5 h (and one order of magnitude faster than iSAX2+). Also, DPiSAX processes 10 millions 10-NN approximate queries on a 1 billion data series collection in 140 s.
The DPiSAX solution is complementary to the ParIS+ and MESSI solutions, and they could be combined in order to exploit both parallelism and distribution.
3.5 Variable-Length: ULISSE
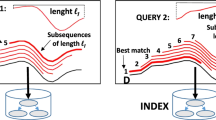
Despite the fact that data series indexes enable fast similarity search, all existing indexes can only answer queries of a single length (fixed at index construction time), which is a severe limitation. The ULtra compact Index for variable-length Similarity SEarch (ULISSE) [45, 46] is the first, single data series index structure designed for answering similarity search queries of variable length. ULISSE introduces a novel envelope representation that effectively and succinctly summarizes multiple sequences of different lengths. These envelopes are then used to build a tree index that resembles to iSAX2+. ULISSE supports both approximate and exact similarity search, combining disk based index visits with in-memory sequential scans, inspired by ADS+. ULISSE supports non Z-normalized and Z-normalized sequences, and can be used with no changes with both Euclidean Distance and Dynamic Time Warping, for answering k-NN and \(\epsilon \)-range queries [47].
The experimental results show that ULISSE is several times, and up to orders of magnitude more efficient in terms of both space and time cost, when compared to competing approaches (i.e., UCR Suite, MASS, and CMRI) [45, 47].
3.6 Sortable Summarizations: Coconut-Trie/Tree/LSM
We observe that a shortcoming of the indexes presented earlier is that their design is based on summarizations [14, 44] (used as keys by the index) that are unsortable. Thus, sorting based on these summarizations would place together data series that are similar in terms of their beginning, i.e., the first segment, yet arbitrarily far in terms of the rest of the segments. Hence, existing summarizations cannot be sorted while keeping similar data series next to each other in the sorted order. This leads to top-down index building (resulting in many small random disk I/Os and non-contiguous nodes), and prefix-based node-splitting (resulting in low fill-factors for leaf nodes), which negatively affect time performance and disk space occupancy.
The Compact and Contiguous Sequence Infrastructure (Coconut) index [38, 39] was developed in order to address these problems, by transforming the iSAX summarization into a sortable summarization. The core idea is interweaving the bits that represent the different segments, such that the more significant bits across all segments precede all less significant bits. As a result, Coconut is the first technique for sorting data series based on their summarizations that can lead to bottom-up creation of balanced indexes: the series are positioned on a z-order curve [52], in a way that similar data series are close to each other. Indexing based on sortable summarizations has the same ability as existing summarizations to prune the search space. Coconut supports bulk-loading techniques and log-structured updates to enable maintaining a contiguous index. This eliminates random I/O during construction, updating and querying. Furthermore, Coconut is able to split data series across nodes by sorting them and using the median value as a splitting point, leading to data series being packed more densely into leaf nodes (i.e., at least half full). We studied Coconut-Trie and Coconut-Tree, which split data series across nodes based on common prefixes and median values, respectively. Coconut-Trie, which is similar to an ADS+ index in structure, dominates the state-of-the-art in terms of query speed because it creates contiguous leaves. Coconut-Tree, based on a B+-Tree index, dominates Coconut-Trie and the state-of-the-art in terms of index construction speed, query (using SIMS) speed and storage overheads because it creates a contiguous, balanced index that is also densely populated. Finally, Coconut-LSM [39, 40], that is based on an LSM tree index, supports efficient log-structured updates and variable-size window queries over different windows of the data based on recency.
Overall, across a wide range of workloads and datasets, Coconut-Tree improves both construction speed and storage overheads by one order of magnitude and query speed by two orders of magnitude relative to DSTree and ADS. Coconut-LSM supports updates without degrading query throughput, and is able to narrow the search scope temporally. This improves query throughput by a further 2–3 orders of magnitudes in our experiments for queries over recent data, thus, making Coconut-LSM an efficient solution for streaming data series.
4 Discussion and Open Research Directions
Despite the strong increasing interest in data series management systems [83], existing approaches (e.g., based on DBMSs, Column Stores, TSMSs, or Array Databases) do not provide a viable solution, since they have not been designed for managing and processing sequence data as first class citizens: they do not offer a suitable storage model, declarative query language, or optimization mechanism. Moreover, they lack auxiliary data structures (such as indexes), that can support a variety of sequence query workloads in an efficient manner. For example, they do not have native support for similarity search [31, 53], and therefore, cannot efficiently support complex analytics on very large data series collections.
Current solutions for processing data series collections, in various domains, are mostly ad hoc (and hardly scalable), requiring huge investments in time and effort, and duplication of effort across different teams. For this reason new data management technologies should be developed; albeit ones that will meet their requirements for processing and analyzing very large sequence collections.
An interesting and challenging research direction is to design and develop a general purpose Sequence Management System, able to cope with big data series (very large and continuously growing collections of data series with diverse characteristics, which may have uncertainty in their values), by transparently optimizing query execution, and taking advantage of new management and query answering techniques, as well as modern hardware [53, 55]. Just like databases abstracted the relational data management problem and offered a black box solution that is now omnipresent, the proposed system will enable users and analysts that are not experts in data series management to tap in the goldmine of the massive and ever-growing data series collections they (already) have.
Our preliminary results, including the first data series similarity search benchmark [81, 82], and indexing algorithms that can be efficiently bulk-loaded [12, 13, 38,39,40], adapt to the query workload [78,79,80], support similarity queries of varying length [45, 46, 48, 49], take into account uncertainty [19, 20], and exploit multi-cores [58,59,60] and distributed platforms (e.g., Apache Spark) [42, 74, 75], are promising first steps. Nevertheless, much progress is still needed along the directions mentioned above. This is especially true for query optimization, since earlier work has shown that different techniques and algorithms perform better for different query workloads and data and hardware characteristics [23]. Trying to further optimize query execution times, techniques that provide approximate answers, and in particular answers with (deterministic, or probabilistic) guarantees on the associated error bounds [23, 24], can be very useful. The same is true for techniques that provide progressive answers [28], which can also lead to significant speedup, while guaranteeing the desired levels of accuracy.
It would also be interesting to develop an index that combines all (or most of) the features mentioned earlier, namely, support for progressive exact and approximate queries of variable length, running on modern hardware in parallel and distributed environments. Given the way that these index solutions have been developed, i.e., by building on top of one another, combining the various features in a single solution seems feasible.
Note that, even though the indexes we presented have been developed for data series, they are equally applicable to and extremely efficient in the case of general high-dimensional vectors [23, 24]. This opens up several exciting application opportunities, including in deep learning analysis pipelines, where we often need to perform similarity search in high-dimensional vector embeddings.
5 Conclusions
In this work, we discussed the evolution of the iSAX family of indexes, which represent the current state-of-the-art in several variations of the problem of indexing for similarity search in very large data series collections. We reviewed the basic design decisions behind these indexes, and contrasted their strong points. The presentation (for the first time together) of all these indexes contributes to the better understanding of which particular problem each one solves, how their features could be combined, and what the opportunities for future work are.
References
ADHD-200 (2011). http://fcon\(\_\)1000.projects.nitrc.org/indi/adhd200/
Sloan Digital Sky Survey (2015). https://www.sdss3.org/dr10/data_access/volume.php
Agrawal, R., Faloutsos, C., Swami, A.: Efficient similarity search in sequence databases. In: Lomet, D.B. (ed.) FODO 1993. LNCS, vol. 730, pp. 69–84. Springer, Heidelberg (1993). https://doi.org/10.1007/3-540-57301-1_5
An, N., Kothuri, R.K.V., Ravada, S.: Improving performance with bulk-inserts in Oracle R-trees. In: VLDB, pp. 948–951. VLDB Endowment (2003)
Assent, I., Krieger, R., Afschari, F., Seidl, T.: The TS-tree: efficient time series search and retrieval. In: EDBT (2008)
Aßfalg, J., Kriegel, H.-P., Kröger, P., Kunath, P., Pryakhin, A., Renz, M.: Similarity search on time series based on threshold queries. In: Ioannidis, Y., et al. (eds.) EDBT 2006. LNCS, vol. 3896, pp. 276–294. Springer, Heidelberg (2006). https://doi.org/10.1007/11687238_19
Bagnall, A.J., Cole, R.L., Palpanas, T., Zoumpatianos, K.: Data series management (Dagstuhl seminar 19282). Dagstuhl Rep. 9(7), 24–39 (2019)
Bentley, J.L.: Multidimensional binary search trees used for associative searching. Commun. ACM 18(9), 509–517 (1975)
Berchtold, S., Keim, D.A., Kriegel, H.-P.: The X-tree: an index structure for high-dimensional data. In: VLDB, pp. 28–39 (1996)
Berndt, D.J, Clifford, J.: Using dynamic time warping to find patterns in time series. In: AAAIWS, pp. 359–370 (1994)
Bu, Y., Leung, T.W., Fu, A.W.C., Keogh, E., Pei, J., Meshkin, S.: WAT: finding top-k discords in time series database. In: SDM, pp. 449–454 (2007)
Camerra, A., Palpanas, T., Shieh, J., Keogh, E.: iSAX 2.0: indexing and mining one billion time series. In: ICDM (2010)
Camerra, A., Shieh, J., Palpanas, T., Rakthanmanon, T., Keogh, E.J.: Beyond one billion time series: indexing and mining very large time series collections with iSAX2+. KAIS 39(1), 123–151 (2014). https://doi.org/10.1007/s10115-012-0606-6
Chakrabarti, K., Keogh, E., Mehrotra, S.: Locally adaptive dimensionality reduction for indexing large time series databases. ACM Trans. Database Syst. (TODS) 27(2), 188–228 (2002)
Chakrabarti, K., Keogh, E., Mehrotra, S., Pazzani, M.: Locally adaptive dimensionality reduction for indexing large time series databases. In: SIGMOD (2002)
Chan, K.-P., Fu, A.-C.: Efficient time series matching by wavelets. In: ICDE (1999)
Chandola, V., Banerjee, A., Kumar, V.: Anomaly detection: a survey. ACM Comput. Surv. 41(3), 1–58 (2009)
Chen, Y., Nascimento, M.A., Ooi, B.C., Tung, A.K.H.: SpADe: on shape-based pattern detection in streaming time series. In: ICDE (2007)
Dallachiesa, M., Nushi, B., Mirylenka, K., Palpanas, T.: Uncertain time-series similarity: return to the basics. PVLDB 5(11), 1662–1673 (2012)
Dallachiesa, M., Palpanas, T., Ilyas, I.F.: Top-k nearest neighbor search in uncertain data series. PVLDB 8(1), 13–24 (2014)
Das, G., Gunopulos, D., Mannila, H.: Finding similar time series. In: Komorowski, J., Zytkow, J. (eds.) PKDD 1997. LNCS, vol. 1263, pp. 88–100. Springer, Heidelberg (1997). https://doi.org/10.1007/3-540-63223-9_109
Ding, H., Trajcevski, G., Scheuermann, P., Wang, X., Keogh, E.: Querying and mining of time series data: experimental comparison of representations and distance measures. In: PVLDB (2008)
Echihabi, K., Zoumpatianos, K., Palpanas, T., Benbrahim, H.: The Lernaean Hydra of data series similarity search: an experimental evaluation of the state of the art. PVLDB 12(2), 112–127 (2018)
Echihabi, K., Zoumpatianos, K., Palpanas, T., Benbrahim, H.: Return of the Lernaean Hydra: experimental evaluation of data series approximate similarity search. PVLDB 13, 403–420 (2019)
Soisalon-Soininen, E., Widmayer, P.: Single and bulk updates in stratified trees: an amortized andworst-case analysis. In: Klein, R., Six, H.-W., Wegner, L. (eds.) Computer Science in Perspective. LNCS, vol. 2598, pp. 278–292. Springer, Heidelberg (2003). https://doi.org/10.1007/3-540-36477-3_21
Fekete, J.-D., Primet, R.: Progressive analytics: a computation paradigm for exploratory data analysis. CoRR (2016)
Gogolou, A., Tsandilas, T., Palpanas, T., Bezerianos, A.: Comparing similarity perception in time series visualizations. IEEE TVCS 25(1), 523–533 (2019)
Gogolou, A., Tsandilas, T., Palpanas, T., Bezerianos, A.: Progressive similarity search on time series data. In: Workshops of the EDBT/ICDT (2019)
Guttman, A.: R-trees: a dynamic index structure for spatial searching. In: SIGMOD (1984)
Huijse, P., Estévez, P.A., Protopapas, P., Principe, J.C., Zegers, P.: Computational intelligence challenges and applications on large-scale astronomical time series databases. IEEE Comput. Int. Mag. 9(3), 27–39 (2014)
Jensen, S.K., Pedersen, T.B., Thomsen, C.: Time series management systems: a survey. IEEE Trans. Knowl. Data Eng. 29(11), 2581–2600 (2017)
Seeger, B., Van den Bercken, J.: An evaluation of generic bulk loading techniques. In: VLDB, pp. 461–470 (2001)
Widmayer, P., Van den Bercken, J., Seeger, B.: A generic approach to bulk loading multidimensional index structures. In: VLDB (1997)
Kashino, K., Smith, G., Murase, H.: Time-series active search for quick retrieval of audio and video. In: ICASSP (1999)
Kashyap, S., Karras, P.: Scalable KNN search on vertically stored time series. In: KDD (2011)
Keogh, E., Chakrabarti, K., Pazzani, M., Mehrotra, S.: Dimensionality reduction for fast similarity search in large time series databases. KAIS 3(3), 263–286 (2000). https://doi.org/10.1007/PL00011669
Keogh, E.J., Palpanas, T., Zordan, V.B., Gunopulos, D., Cardle, M.: Indexing large human-motion databases. In: VLDB, pp. 780–791 (2004)
Kondylakis, H., Dayan, N., Zoumpatianos, K., Palpanas, T.: Coconut: a scalable bottom-up approach for building data series indexes. In: PVLDB (2018)
Kondylakis, H., Dayan, N., Zoumpatianos, K., Palpanas, T.: Coconut palm: static and streaming data series exploration now in your palm. In: SIGMOD, pp. 1941–1944 (2019)
Kondylakis, H., Dayan, N., Zoumpatianos, K., Palpanas, T.: Coconut: sortable summarizations for scalable indexes over static and streaming data series. VLDBJ 28, 847–869 (2019). https://doi.org/10.1007/s00778-019-00573-w
Arge, L., Hinrichs, K., Vahrenhold, J., et al.: Efficient bulk operations on dynamic R-trees. Algorithmica 33(1), 104–128 (2002). https://doi.org/10.1007/s00453-001-0107-6
Levchenko, O., et al.: Distributed algorithms to find similar time series. In: ECML/PKDD (2019)
Li, C.-S., Yu, P., Castelli, V.: HierarchyScan: a hierarchical similarity search algorithm for databases of long sequences. In: ICDE (1996)
Lin, J., Keogh, E., Lonardi, S., Chiu, B.: A symbolic representation of time series, with implications for streaming algorithms. In: DMKD (2003)
Linardi, M., Palpanas, T.: Scalable, variable-length similarity search in data series: the ULISSE approach. PVLDB 11(13), 2236–2248 (2018)
Linardi, M., Palpanas, T.: ULISSE: ULtra compact index for variable-length similarity SEarch in data series. In: ICDE (2018)
Linardi, M., Palpanas, T.: Scalable data series subsequence matching with ULISSE. Technical Report (2020)
Linardi, M., Zhu, Y., Palpanas, T., Keogh, E.J.: Matrix profile X: VALMOD - scalable discovery of variable-length motifs in data series (2018)
Linardi, M., Zhu, Y., Palpanas, T., Keogh, E.J.: VALMOD: a suite for easy and exact detection of variable length motifs in data series. In: SIGMOD (2018)
Mirylenka, K., Dallachiesa, M., Palpanas, T.: Correlation-aware distance measures for data series. In: EDBT, pp. 502–505 (2017)
Mirylenka, K., Dallachiesa, M., Palpanas, T.: Data series similarity using correlation-aware measures. In: SSDBM (2017)
Morton, G.M.: A Computer Oriented Geodetic Data Base and a New Technique in File Sequencing. International Business Machines Company, Ottawa (1966)
Palpanas, T.: Data series management: the road to big sequence analytics. SIGMOD Rec. 44, 47–52 (2015)
Palpanas, T.: Big sequence management: a glimpse of the past, the present, and the future. In: Freivalds, R.M., Engels, G., Catania, B. (eds.) SOFSEM 2016. LNCS, vol. 9587, pp. 63–80. Springer, Heidelberg (2016). https://doi.org/10.1007/978-3-662-49192-8_6
Palpanas, T.: The parallel and distributed future of data series mining. In: High Performance Computing & Simulation (HPCS) (2017)
Palpanas, T., Beckmann, V.: Report on the first and second interdisciplinary time series analysis workshop (ITISA). ACM SIGMOD Rec. 48(3), 36–40 (2019)
Palpanas, T., Vlachos, M., Keogh, E.J., Gunopulos, D.: Streaming time series summarization using user-defined amnesic functions. IEEE Trans. Knowl. Data Eng. 20(7), 992–1006 (2008)
Peng, B., Fatourou, P., Palpanas, T.: Paris: the next destination for fast data series indexing and query answering. In: IEEE BigData, pp. 791–800 (2018)
Peng, B., Fatourou, P., Palpanas, T.: MESSI: in-memory data series indexing. In: ICDE (2020)
Peng, B., Fatourou, P., Palpanas, T.: Paris+: data series indexing on multi-core architectures. In: TKDE (2020)
Rafiei, D.: On similarity-based queries for time series data. In: ICDE (1999)
Rafiei, D., Mendelzon, A.: Similarity-based queries for time series data. In: SIGMOD (1997)
Rakthanmanon, T.: Searching and mining trillions of time series subsequences under dynamic time warping. In: KDD (2012)
Raza, U., Camerra, A., Murphy, A.L., Palpanas, T., Picco, G.P.: Practical data prediction for real-world wireless sensor networks. TKDE 27(8), 2231–2244 (2015)
Choubey, R., Chen, L., Rundensteiner, E.A.: GBI: a generalized R-tree bulk-insertion strategy. In: Güting, R.H., Papadias, D., Lochovsky, F. (eds.) SSD 1999. LNCS, vol. 1651, pp. 91–108. Springer, Heidelberg (1999). https://doi.org/10.1007/3-540-48482-5_8
Schäfer, P., Högqvist, M.: SFA: a symbolic fourier approximation and index for similarity search in high dimensional datasets. In: EDBT (2012)
Shasha, D.: Tuning time series queries in finance: case studies and recommendations. IEEE Data Eng. Bull. 22(2), 40–46 (1999)
Shieh, J., Keogh, E.: iSAX: indexing and mining terabyte sized time series. In: SIGKDD, pp. 623–631 (2008)
Shieh, J., Keogh, E.: iSAX: disk-aware mining and indexing of massive time series datasets. DMKD 19(1), 24–57 (2009). https://doi.org/10.1007/s10618-009-0125-6
Shieh, J., Keogh, E.J.: iSAX: indexing and mining terabyte sized time series. In: KDD, pp. 623–631 (2008)
Wang, X., Mueen, A., Ding, H., Trajcevski, G., Scheuermann, P., Keogh, E.: Experimental comparison of representation methods and distance measures for time series data. Data Min. Knowl. Discov. 26(2), 275–309 (2013)
Wang, Y., Wang, P., Pei, J., Wang, W., Huang, S.: A data-adaptive and dynamic segmentation index for whole matching on time series. PVLDB 6(10), 793–804 (2013)
Liao, T.W.: Clustering of time series data - a survey. Pattern Recogn. 38(11), 1857–1874 (2005)
Yagoubi, D.-E., Akbarinia, R., Masseglia, F., Palpanas, T.: DPiSAX: massively distributed partitioned iSAX. In: ICDM (2017)
Yagoubi, D.-E., Akbarinia, R., Masseglia, F., Palpanas, T.: Massively distributed time series indexing and querying. TKDE 32(1), 108–120 (2020)
Ye, L., Keogh, E.J.: Time series shapelets: a new primitive for data mining. In: KDD (2009)
Yi, B., Faloutsos, C.: Fast time sequence indexing for arbitrary Lp norms. In: VLDB (2000)
Zoumpatianos, K., Idreos, S., Palpanas, T.: Indexing for interactive exploration of big data series. In: SIGMOD (2014)
Zoumpatianos, K., Idreos, S., Palpanas, T.: RINSE: interactive data series exploration with ADS+. PVLDB 8(12), 1912–1923 (2015)
Zoumpatianos, K., Idreos, S., Palpanas, T.: ADS: the adaptive data series index. VLDB J. 25, 843–866 (2016). https://doi.org/10.1007/s00778-016-0442-5
Zoumpatianos, K., Lou, Y., Ileana, I., Palpanas, T., Gehrke, J.: Generating data series query workloads. VLDB J. 27(6), 823–846 (2018). https://doi.org/10.1007/s00778-018-0513-x
Zoumpatianos, K., Lou, Y., Palpanas, T., Gehrke, J.: Query workloads for data series indexes. In: KDD (2015)
Zoumpatianos, K., Palpanas, T.: Data series management: fulfilling the need for big sequence analytics. In: ICDE (2018)
Acknowledgements
I would like to thank my collaborators (in alphabetical order): R. Akbarinia, H. Benbrahim, A. Bezerianos, A. Camerra, M. Dallachiesa, N. Dayan, K. Echihabi, A. Gogolou, P. Fatourou, J. Gehrke, S. Idreos, I. Ilyas, E. Keogh, B. Kolev, H. Kondylakis, O. Levchenko, M. Linardi, Y. Lou, F. Masseglia, K. Mirylenka, B. Nushi, B. Peng, T. Rakthanmanon, D. Shasha, J. Shieh, T. Tsandilas, P. Valduriez, and D.-E. Yagoubi. Special thanks go to K. Zoumpatianos.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 Springer Nature Switzerland AG
About this paper
Cite this paper
Palpanas, T. (2020). Evolution of a Data Series Index. In: Flouris, G., Laurent, D., Plexousakis, D., Spyratos, N., Tanaka, Y. (eds) Information Search, Integration, and Personalization. ISIP 2019. Communications in Computer and Information Science, vol 1197. Springer, Cham. https://doi.org/10.1007/978-3-030-44900-1_5
Download citation
DOI: https://doi.org/10.1007/978-3-030-44900-1_5
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-44899-8
Online ISBN: 978-3-030-44900-1
eBook Packages: Computer ScienceComputer Science (R0)