Abstract
Fluorescence lifetime imaging (FLIM) is increasingly recognized as a valuable approach for its ability not only to assess structural information, but also to follow certain biochemical processes in living organisms. Particularly, a measurement of fluorescence lifetimes of endogenous cofactors reduced nicotinamide adenine dinucleotide (phosphate) NAD(P)H and oxidized flavin adenine dinucleotide FAD and flavin mononucleotide FMN provides insight into cellular metabolism on a label-free basis. NAD(P)H and flavin cofactors are primarily involved in cellular bioenergetics pathways, in which they participate in the states with distinct fluorescence lifetimes. In recent years, metabolic FLIM has received increased attention in cancer studies. On the one hand, metabolic abnormalities that support the higher proliferative rate of cancer cells may serve as a basis for new diagnostic methods and targeted therapies. On the other hand, detection of early metabolic changes in response to anticancer therapy is a new way to personalized medicine. Currently available FLIM systems allow metabolic visualization from a micro- to a macroscopic scale with excellent spatial resolution and high molecular specificity. This chapter describes three main techniques on the basis of fluorescence time-resolved measurements—microscopic, spectroscopic, and macroscopic, and provides multiple examples of their applications in probing metabolism in cancer.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
1 Introduction
Multiple alterations in cancer metabolism are now evident. The key underlying reasons for metabolic changes accompanying neoplastic transformation are the high bioenergetic and biosynthetic demands of growing and proliferating cells and adaptation to limited nutrient and oxygen supply and to an acidic environment.
One of the primary metabolic features of cancer is enhanced rate of glycolysis. While it is natural to switch to glycolysis in hypoxic conditions, which occur in many advanced tumors, cancer cells actively use glycolysis even in the presence of oxygen—a phenomenon known as the Warburg effect—or aerobic glycolysis [1]. Aerobic glycolysis makes cancer cells insensitive to fluctuations of oxygen in cellular environment and gives them several other important advantages. Specifically, it provides a constant supply of metabolic intermediates to support tumor growth, reduces level of reactive oxygen species (ROS), thus, protecting cells against ROS-mediated cell death, generates adenosine triphosphate (ATP) more rapidly than oxidative phosphorylation (OXPHOS), and produces lactate that has an important functional role in promoting tumorigenesis [2, 3].
Mitochondrial respiration is frequently impaired in cancer cells due to dysfunctional mitochondria. Meanwhile, there is a growing body of evidence that some cancer cells preserve mitochondrial function and rely mainly on OXPHOS for energy production [4, 5].
The fate of lactate secreted by glycolytic cancer cells remains to be clarified. Recent findings suggest that adjacent oxidative cancer cells can consume and use it as a source for the tricarboxylic acid (TCA) cycle intermediates and for ATP production [6]. Moreover, cancer-associated fibroblasts , being in a metabolic crosstalk with cancer cells, also can extrude lactate to directly feed cancer cells [7].
Glucose is a major, but not the only fuel source for cancer cells. Many cancer cells have been shown to rely heavily upon an exogenous supply of glutamine [8]. Glutamine is converted to glutamate, which is further metabolized to α-ketoglutarate for utilization through the TCA cycle. Another important energy source in cancer cells is fatty acids , catabolized by the fatty acid oxidation (FAO; also known as β-oxidation) pathway [9]. FAO produces a pool of the reduced electron carriers NADH and FADH2, which are oxidized in the mitochondrial electron transport chain for ATP production, and acetyl CoA, which together with oxaloacetate is transformed in the TCA cycle into citrate, which on export to the cytoplasm, can enter NADPH-producing reactions.
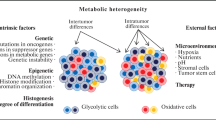
There are number of factors that collectively determine metabolic phenotype of a tumor, including the tissue of origin, the underlying cancer genetics and subtype, the tumor microenvironment, and other variables such as diet and host physiology, which lead to a vast intertumor heterogeneity in metabolism. In addition, individual cells within a tumor can explore distinct metabolic programs. The emerging view on cancer metabolism is that malignant cells have an ability to switch between different biochemical pathways in order to satisfy their immediate biosynthetic and energetic demands and to adopt to varying microenvironment [10]. However, the mechanisms by which cancer cells coordinate their metabolic activities and the conditions upon which cancer cells upregulate specific metabolic pathways are poorly defined.
Multiple studies suggest that cellular metabolism has an impact not only on biological behavior of tumors, but also on treatment response. The links between metabolic processes and response to cancer therapeutics are complex and not fully understood. On the one side, metabolism and its derangement are involved in the initiation and execution of cell death at multiple levels and in regulation of cell survival [11, 12]. A hallmark of cancer cells is the ability to evade apoptosis. On the other side, increased aerobic glycolysis and glutamine metabolism under suppression of mitochondrial respiration in cancer cells contribute to resistance to various therapies by providing higher intracellular ATP and NADPH levels and extracellular acidification, to name a few proposed mechanisms [13]. Significant intertumor and intratumor heterogeneity and metabolic plasticity of tumors impose additional difficulties for cancer treatment.
To reach a deeper understanding of cancer metabolism, highly sensitive, quantitative, high-resolution metabolic assays are critically needed. Established methods to characterize cellular metabolic activity include both solely laboratory techniques, e.g. mass spectrometry, routine analytical methods, molecular and genetic analyses, optical biosensors, and clinical imaging, e.g. positron emission tomography (PET) with 18F-labeled fluoro-2-deoxyglucose, 13C magnetic resonance spectroscopy (MRS). All these methods, however, either destructive or require the use of exogenous labeling of molecules of interests. Time-resolved optical imaging of endogenous fluorophores has emerged as a powerful approach for non-invasive, label-free assessment of metabolic state of living cells in vitro and tissues in vivo.
The book chapter is organized as following. Section 3.2 gives biochemical basis of optical metabolic imaging by introducing fluorescent metabolic cofactors. In Sect. 3.3 we address theoretical background for time-correlated single photon counting (TSCPC) technique as all the described systems are based on it. Fluorescence lifetime imaging on a microscopic scale together with the examples of metabolic imaging in cancer cells, spheroids, and tumors is presented in Sect. 3.4. Section 3.5 reports about the developed system for fiber-based fluorescence lifetime measurements and its applications for interrogation of metabolic state of a tumor in vivo. Section 3.6 describes fluorescence lifetime imaging on a macroscale and its applications in cancer studies. Finally, the conclusions of the book chapter are presented in the Summary section.
2 Fluorescence Lifetime of Metabolic Cofactors as an Indicator of a Metabolic State
There are two groups of fluorescent molecules involved in various metabolic pathways in cells (1) reduced nicotinamide adenine dinucleotide NADH and its phosphorylated derivative NADPH (collectively denoted NAD(P)H) and (2) oxidized flavin adenine dinucleotide FAD and flavin mononucleotide FMN . These molecules function as coenzymes, being linked with various dehydrogenases. There is a constant interchange between the reduced and the oxidized states of these molecules, which jointly constitute a redox couple (NAD+/NADH, NADP+/NADPH, FAD/FADH2, FMN/FMNH2). The primary role of these cofactors, except NADPH, is the transfer of electrons across the respiratory chain of mitochondria. Reduced electron carriers NADH and FADH2 are produced in catabolic pathways by transferring electrons from the substrate to NAD+ and FAD. Subsequently, NADH feeds electrons into complex I and FADH2—into complex II of the mitochondrial electron transport chain (ETC). ETC and oxidative phosphorylation (OXPHOS) reoxidize NADH and FADH2 and trap the released energy in the form of adenosine triphosphate (ATP). FMN, a prosthetic group of NADH-ubiquinone oxidoreductase (complex I), accepts electrons from NADH and passes to FeS centers (iron-sulfur clusters). NADPH acts as a reducing agent during the synthesis of nucleic acids and lipids and plays a central role in defense against oxidative damage and detoxification of reactive oxygen species.
NAD(P) H, FAD and FMN possess intrinsic fluorescence, detection of which is the basis for a group of methods referred as “optical metabolic imaging.” NADH and NADPH are spectrally identical—excitation peaks at 260 nm and 340 nm, emission peak at 460 nm. Flavin cofactors absorb light maximally at ~450 nm and emit at ~520 nm, with minor variations in spectra of FAD and FMN.
While fluorescence of NAD(P)H is derived from both mitochondria and cytosol (mitochondrial NAD(P)H fluorescence is dominant), flavin fluorescence originates predominantly from mitochondrial flavoproteins [14], although additional subcellular localizations (e.g., nucleus) cannot be ruled out.
The first approach to probe metabolism using fluorescence of the metabolic cofactors was developed in 1970th by B. Chance, who measured oxidation-reduction states of mitochondria taking the ratio of the fluorescence intensities of oxidized flavoproteins to reduced NADH [15, 16].
The “optical redox ratio ” has been widely used as a simple, label-free metric of cellular redox states [16,17,18]. In a general case, the redox ratio is sensitive to the changes in the rates of glycolysis and mitochondrial respiration. NADH is generated during glycolysis, therefore enhanced glycolysis causes decrease in FAD/NADH ratio, which is often observed in cancer. Activation of oxidative metabolism increases the portion of oxidized and decreases the portion of reduced cofactors, which results in increased FAD/NADH ratio. However, the intensity-based redox ratio imaging and interpretation face some challenges associated with the fact that emission intensity is influenced by many factors besides concentration of the fluorophores, e.g. conformation or binding to proteins, light scattering and absorption—especially in tissues, excitation power, and participation of the same cofactors in various metabolic pathways.
Fluorescence lifetimes of NAD(P)H and flavoproteins provide an additional dimension to probing the cellular metabolic status.
For NAD(P)H, the fluorescence lifetime largely depends on binding to enzyme. This observation was first reported by Lakowicz et al. in 1992 [19]. Freely diffusing NAD(P)H has distinctly shorter fluorescence lifetime (~0.3–0.5 ns) due to the large rate of non-radiative relaxation mediated by small scale motion of the nicotinamide ring [20]. The fluorescence lifetime of protein-bound NADH is, at least, threefold higher and strongly vary—on average, in the range 1.2–2.5 ns in solutions [21, 22], when bound to different enzymes, which is presumably attributed to different levels of conformational restriction upon binding. Moreover, the fluorescence lifetime of enzyme-bound NADH may increase upon formation of a ternary complex with the specific substrate [23]. Protein-bound NADPH possesses a significantly larger fluorescence lifetime (~4.4 ns) than protein-bound NADH within the cellular environment [24]. Therefore, fluorescence lifetime of NAD(P)H depends on the composition and activity of NADH-binding enzymes and the amount of the phosphorylated form of NADH.
Currently, nonlinear curve-fitting analysis is the most common approach to analyze fluorescence lifetimes. With a large range of different enzymes to which NAD(P)H can bind, it is natural that its fluorescence is characterized by a complex multi-exponential decay. However, limited photon counts, acquisition and computational times, especially when measurements are made in live cells or tissues, allow to resolve most effectively only two lifetime components. When using a bi-exponential function for fitting NAD(P)H fluorescence decay, the first (short, t1) component is attributed to its free, and the second (long, t2) component to its protein-bound state, and their relative amplitudes (a1 ~ 85% and a2 ~ 15%) reflect the relative amounts of free and protein-bound NAD(P)H. The three-exponential model for fitting NAD(P)H fluorescence decay is appropriate for estimation of the contribution of protein-bound NADPH, as was suggested by Blacker et al. [24]. Three-exponential fitting requires larger photon numbers compared to single and bi-exponential fitting. Therefore, fixation of the one of the fluorescence lifetimes significantly relaxes the requirements on the minimum photon numbers and speeds up the computational times. This approach is more appropriate for the cells with increased NADPH level, for example, for stem cells undergoing adipogenic differentiation [25]. In general, NADPH contribution in FLIM of most cell and tissue types is not taken into account, because of its much lower concentration compared with NADH and insensitivity to perturbations in energy metabolism [26].
There is also possible to use four-exponential model for fitting NAD(P)H fluorescence decay curves bearing in mind that the free NADH has two decay components [27]. However, as in case with three-exponential fitting, one has to fix at least two lifetime components to be able to analyze the data with the limited photon budget.
We have to mention that phasor analysis has become recently a quite effective tool in the evaluation of the fluorescence lifetime data of NADH [28, 29]. However, the detailed description of it is out of scope of this book chapter.
In contrast to NAD(P)H, flavin cofactor in the form of FAD or FMN, more specifically termed a prosthetic group, is permanently linked to the protein [30]. Fluorescence of FAD decays multiexponentially with two major lifetime components—around 7 ps and 2.7 ns in aqueous solution [31]. The presence of two lifetime components is due to existence of FAD in two conformations—“closed” or stacked, in which the coplanar isoalloxazine and adenine rings interact through π-π interactions, resulting in very efficient fluorescence quenching, and “open” or unstacked, in which the two aromatic ring are separated from each other. The majority of FAD molecules are in a closed conformation, that determines low quantum yield of its fluorescence (0.033) [32]. FMN and riboflavin (vitamin B2), the precursor of flavin cofactors, exhibit monoexponential fluorescence decays with lifetime ~4.7 ns and ~5.1 ns, correspondingly [17].
Among flavins , FAD is the most abundant in tissues, while concentration of FMN is typically 3–5 times lower and riboflavin—40-400 times lower [33, 34], although in cultured cells riboflavin concentration can be comparable with FAD [35]. For flavoproteins, the emission maxima and quantum yield are dependent on the nature of the flavin-binding site [36].
It is common for fluorescence decay of the cellular flavins to be best fit by a bi-exponential model, where the short lifetime component (~0.3 to 0.4 ns, ~75 to 85%) is consistent with the quenched state of FAD and the long lifetime component (~2.5 to 2.8 ns, ~15 to 25%) contains contributions from unquenched FAD, FMN, and riboflavin. Given that the discrimination of different flavins within cells is problematic, biochemical interpretation of the fluorescence lifetime measurements remains a challenge.
Furthermore, in cancer cells and tumors with mitochondrial respiration malfunction fluorescence emission from flavins is often fairly weak and insufficient to collect the required number of photons for FLIM. Therefore, most of the metabolic studies with FLIM are concentrated on NAD(P)H.
3 Technical Basis of Time-Correlated Single Photon Counting
Time-correlated single photon counting (TCSPC) is a technique that allows to record photons, measure their time relative to the excitation pulse, and build up a histogram of the photon times [37]. The technique uses the fact that for low level, high-repetition-rate signals the probability to detect one photon per laser period is extremely low. That means that there are many signal periods that do not contain photons, and only few contain just one.
To measure fluorescence decay curves, one excites the sample with a train of short pulses (typically in the ps or fs range). Then emitted fluorescence photons are registered by a detector operating in a single photon counting mode, and their time with respect to the excitation pulse is recorded. With each pulse, more and more photons are recorded and the statistical distribution of the photons within the laser period is built up. The principle of TCSPC is shown in Fig. 3.1.
This method of decay curves measurements is very effective as all the registered photons contribute to the statistical distribution. The efficiency in this case is much higher than for recording techniques that shift a time-gate over the optical waveform [38].
Another advantage of using TCSPC for fluorescence decay measurements compared to other methods is that its time resolution is better than the width of the single-electron response of the detector, as the arrival times of the detector pulses can be measured with an accuracy much better than the width of the pulses. That in its turn means that the instrument response function determined by the transient time spread of the pulses at the detector can be much shorter than the single photon response of the detector. Finally, TCSPC technique is not sensitive to the waveform changes during the recording process as it measures an average waveform.
On the other side, if a time-gating approach is used, this may result in a distorted waveform in this case.
The photon distribution can be built up not only with respect to the time after the excitation pulse (one-dimensional TCSPC), but also with respect to other parameters (multi-dimensional TCSPC), like the spatial coordinates, the wavelength, the time from the start of experiment, polarization, etc. Depending on which and how many additional parameters are used, different photon distributions are obtained. The data that will be presented later in this book chapter is based on the TCSPC measurements recorded with one or two variable parameters. In the first case (one parameter-time after the excitation laser pulse) the experiments represent single point, or fluorescence lifetime spectroscopy measurements. In the latter case (two parameters, and that are the time after the excitation pulse in each laser period and the spatial coordinates within an image area), the measurements result in the building up fluorescence lifetime image, or FLIM.
4 FLIM-Microscopy
The principle of multi-dimensional TCSPC in laser scanning microscopy (LSM) is shown in Fig. 3.2.
In LSM , the sample is scanned with a focused beam of a pulsed laser (usually femtosecond or picosecond). Unlike one-dimensional TCSPC, the multi-parameter TCSPC involves determination of the laser beam position in addition to counting the number of photons in each period of laser pulses and estimation of the arrival times at the detector. The spatial coordinates are determined by transferring scan synchronization signals from the scanner to the single photon counting module: a “frame” pulse indicates the beginning of a new frame, a “line” pulse—the beginning of a new line, and a pixel pulse—the transition to the new pixel within the line. The spatial coordinates of the photons are determined by counting these pulses.
Based on the registered parameters (number of photons, photon arrival times, and spatial coordinates), spatial-temporal photon histograms which are in the basis of the time-resolved fluorescence microscopy, or fluorescence lifetime imaging are built up. If other photon parameters (e.g., wavelength) are registered in addition to spatial coordinates, this represents the three-parameter version of FLIM—spectral time-resolved fluorescence microscopy [39, 40].
From the point of view of biological studies, it is essential that the TCSPC-based FLIM allows identification of curves with complex (multi-exponent) profiles of fluorescence decay. It is also important that this method is insensitive to fluctuations in the laser power and fully compatible with fast scanning realized in modern LSM: scanning is continued until the photon statistics sufficient for the generation of reliable results of image analysis is accumulated. It should be mentioned that while photon statistics is not so essential in the standard fluorescence microscopy, the number of photons is a defining factor for accurate data analysis in the time-resolved fluorescence microscopy.
Apparently, the more complex the exponential descriptions of the decay curves are, the higher number of photons must be accumulated for correct estimation of fluorescence lifetimes. That is why the times for data accumulation in the TCSPC-based FLIM are longer than in the standard LSM .
Furthermore, the counting rate in the TCSPC-based systems is limited by the so-called accumulation effects [37]. The essence of these effects lies in the fact that if a very powerful laser radiation hits the sample and the rate of fluorescence photon emission is very high, the probability of the second photon arrival at the detector over one laser period (which means that this photon will be “ignored” by the electronics) increases, which can alter the decay curve profile. To avoid this, the rate of photon emission by the sample should not exceed 10% of the laser pulse repetition rate. This mode is realized in the majority of LSM setups to prevent the photodegradation of biological samples.
Laser scanning microscopes that allow to do FLIM experiments are typically equipped with either ps or fs lasers. While one-photon excitation with ps lasers makes a system cheaper, there are number of advantages of using two-photon excitation with fs lasers. First, multiphoton processes are excited only at the focal spot where the photon flux density is high enough for the nonlinear process to occur, eliminating the need for the confocal detector pinhole. Second, the illuminating laser is typically operates in the near infrared rather than in the visible or ultraviolet spectral regions. Using such long excitation wavelengths results in a deeper penetration due to lower scattering and causes less photodamage to the samples [41].
The first metabolic study with the use of multiphoton TSCPC-based FLIM-microscopy was reported by Bird et al. in 2005 [42]. The authors measured fluorescence lifetimes and ratio of free and protein-bound NADH in live cultured human breast cells upon chemical perturbations.
Multiphoton FLIM-microscopy of NADH was reported to detect the changes in metabolism with neoplasia development in vivo [43]. Using hamster cheek pouch model of carcinogenesis, the authors showed that the fluorescence lifetime of protein-bound NADH and its contribution to the fluorescence signal could serve as a marker that allowed differentiation between normal and pre-cancer tissue. Later the ability of FLIM-microscopy to distinguish cancer cells from normal ones on the basis of autofluorescence was demonstrated in different systems—from cultured cells of different origin to tumor samples. For example, this was shown for leukemia cells in vitro [44], breast cancer cells [21, 45], squamous carcinoma cells [46], head and neck squamous cell carcinoma [47] and for human lung cancer ex vivo [48], experimental glioma tissue [49], and mouse breast tumors [50]. Several of these studies [21, 44, 45, 47, 48] observed a shift towards shorter mean fluorescence lifetime and a higher ratio of free/bound NADH in malignant cells, which correlated with their glycolytic phenotype, while others reported on elongation of the NADH lifetime in cancer [45, 46, 49, 50], suggesting an important role of OXPHOS . Combination of metabolic FLIM-microscopy with phosphorescence lifetime measurements from oxygen-sensitive probes was shown to be promising approach to explore a relation between metabolic phenotypes and oxygen tension [51, 52].
A significant advantage of FLIM-microscopy over other FLIM modalities is the ability to produce a (sub)cellular resolution, which is essential in terms of metabolic heterogeneity at the cellular level.
As revealed by two-photon FLIM-microscopy , NAD(P)H exhibits a heterogeneous fluorescence lifetime (i.e., quantum yield) throughout living cells. The appreciable differences in NAD(P)H lifetime were shown between different cellular compartments; particularly, the relative contribution of the protein-bound species in mitochondria is larger than in cytosol or in nucleus [18, 21, 53]. Segmented cell analysis of FLIM images was proposed as a relevant approach to separate mitochondrial OXPHOS from cytosolic glycolysis in cancer cells [18].
Specifics of cellular metabolism enables FLIM-microscopy to distinguish between different subpopulations of cells that constitute the tumor and visualize the changes accompanying cellular interactions. For example, Heaster et al. demonstrated the feasibility of using the NAD(P)H and FAD fluorescence lifetimes and intensity-based redox ratio to discriminate proliferating, quiescent, and apoptotic cell populations in an acute myeloid leukemia model and generated classification algorithms based on a combination of all measured variables to improve separation between cell groups [54]. Using FLIM we also observed significant metabolic differences between proliferating cells of the outer layers and quiescent cells of the inner layers of multicellular spheroids (Fig. 3.3B) [55]. The common observation is that proliferating cancer cells use more glycolytic metabolism compared to quiescent cells and, consequently, display increased contribution of free NAD(P)H; other parameters can differ as well. Apoptotic cells show increased mean fluorescence lifetimes of NAD(P)H and FAD, increased lifetimes of the long components NAD(P)H and FAD, decreased contribution of free NAD(P)H, and decreased contribution of short lifetime FAD, indicating an elevated level of OXPHOS and binding cofactors to different enzymes [54, 56, 57].
Metabolic heterogeneity at the cellular level visualized by two-photon FLIM-microscopy . (A) The NAD(P)H-τm and FAD-τm FLIM images of co-culture of HeLa cancer cells and human skin fibroblasts. (B) The transmitted-light (upper) and NAD(P)H – a1% fluorescence lifetime (bottom) images of HeLa tumor spheroid. FLIM corresponds to the square area marked with a dashed white line. (C) The varying responses to chemotherapy with cisplatin in HeLa cells. The NAD(P)H-a1/a2 images are shown for the same field of view before treatment (control, left image) and at 6 h exposure (cisplatin, right image). Scale bar: 50 μm
Trinh et al. identified a significant shift towards longer NADH fluorescence lifetimes, suggesting an increase in the fraction of protein-bound NADH, in the invasive stem-like tumor-initiating cell subpopulation relative to the tumor mass-forming cell subpopulation of malignant gliomas [58].
The metabolic crosstalk between cancer cells and cancer-associated fibroblasts —the main stromal cell type of solid tumors—was examined in several studies with the use of FLIM. Specifically, it was shown that metabolic changes occurred in cancer cells and normal fibroblasts as a result of their interaction in a co-culture model; we observed a shift towards glycolysis in HeLa cancer cells, and from glycolysis to OXPHOS in fibroblasts (Fig. 3.3A) [59]. Walsh et al. found that isolated tumor-associated fibroblasts displayed decreased redox ratio NAD(P)H/FAD and increased mean fluorescence lifetimes of NAD(P)H and FAD compared to 3D organoids generated from primary pancreatic ductal adenocarcinoma [60]. Analysis of the metabolic status of cancer-associated and normal fibroblasts using FLIM revealed increased metabolic activity of fibroblasts derived from patient’s colon tumor with a shift to more oxidative metabolism compared to dermal fibroblasts [61].
Cellular-level metabolic heterogeneity was visualized by FLIM-microscopy in tumor models in vivo in a few studies. The elegant experiments by Szulczewski et al. demonstrate that at least two different sets of abundant cell types can be distinguished in a live mouse tumor by their endogenous fluorescence: cancer cells with high NAD(P)H fluorescence intensity and stromal cells with high FAD intensity. The latter were found predominantly outside of the tumor and concentrated mainly in the stroma, along collagen fibers, and verified to be tumor-associated macrophages. Notably, macrophages had significantly shorter τ mean of NAD(P)H with a greater fractional component of free NADH, indicating glycolytic-like metabolism [62]. In our previous study on human cervical carcinoma xenografts, we also detected a cellular population with greater contribution of free NAD(P)H in a collagen-rich zones compared to majority of cells comprising the tumor mass [63]. Yet, the interpretation of in vivo data remains challenging due to the difficulty of identifying specific cell types within tumor and a variety of factors that collectively determine the metabolic profile of cells.
Multiple studies demonstrate that anti-tumor therapies induce changes in the optical redox ratio and fluorescence lifetimes of NAD(P)H and FAD, and the extent of these changes correlate with responsiveness to the treatment. For example, in the study by Shah et al. head and neck squamous cell carcinoma cell lines were treated with cetuximab (anti-EGFR antibody), BGT226 (PI3K/mTOR inhibitor), or cisplatin (chemotherapy) [47]. Results showed a decreased redox ratio NAD(P)H/FAD and decreased contribution of free NADH-a1 with BGT226 and cisplatin treatment and decreased FAD-a1 with cisplatin treatment, which agreed with decreased proliferation and glycolytic rates. Treatment with cetuximab had no effects on the metabolic imaging variables neither on proliferation nor on glycolysis rate. Similar changes in redox ratio and NAD(P)H lifetime were detected in our study on human cervical cancer cells and xenografts after therapy with cisplatin, which correlated with inhibition of cancer cells growth [64]. Walsh et al. detected decreased redox ratio NAD(P)H/FAD and decreased mean fluorescence lifetime (τmean) NAD(P)H and FAD in HER2-overexpressing human breast cancer xenografts in mice treated with trastuzumab (herceptin, HER2 inhibitor). This response was confirmed with tumor growth curves and stains for Ki67 and cleaved caspase-3 [45]. In general, targeted therapy promoted more complex metabolic rearrangements, not easily interpreted, due to alterations at the level of signaling pathways and genes involved in regulation of cellular metabolism. In a recent work, we observed increase in the NAD(P)H τmean m and decrease in the free/bound NAD(P)H ratio in mouse colorectal tumors after chemotherapy with each of three drugs—cisplatin, paclitaxel, and irinotecan [65]. We should note that the treatment with different chemotherapeutic agents caused similar changes in the optical metabolic parameters. These changes serve as an indicator of the switch to a more oxidative/less glycolytic metabolism, which is rational for cells with disrupted cell division and decreased proliferative capacity.
It is important to mention that the changes in cellular metabolism, resolved by FLIM, precede manifestations of cell death and tumor size reduction, which allows to consider autofluorescence parameters as early indicators of drug efficacy.
With FLIM-microscopy longitudinal tracking of cells can be performed in the course of treatment, and subpopulations with different drug sensitivities can be identified on the basis of optical metabolic metrics. In our previous study, we monitored the metabolic activity of the same cervical cancer cells using fluorescence intensity and lifetime measurements of NAD(P)H and FAD in response to paclitaxel or cisplatin treatment (Fig. 3.3C) [55]. Among the optical metrics (the fluorescence intensity-based redox ratio FAD/NAD(P)H, and the fluorescence lifetimes of NAD(P)H and FAD), the fluorescence lifetime of NAD(P)H was the most sensitive to resolve heterogeneous drug response. It was found that metabolic changes developed faster in the more responsive (dying) cells. In the recent studies, the measurements of NAD(P)H, FAD, and tryptophan (Trp) lifetimes and their enzyme-bound fractions in individual cancer cells allowed to map heterogeneity in response to doxorubicin treatment [66, 67]. By measuring the optical redox ratio NAD(P)H/FAD and fluorescence lifetimes of NAD(P)H and FAD, Shah et al. observed increased metabolic heterogeneity in FaDu tumor xenografts of mice treated with cetuximab or cisplatin compared with untreated control [68]. To quantify the level of cellular metabolic heterogeneity and make quantitative comparisons across treatment groups, they developed a heterogeneity index that incorporates the number of subpopulations, evenness between subpopulations, and distance between subpopulations in frequency histograms for each optical metabolic imaging variable.
A series of studies was performed by Melissa Skala’ group to validate microscopic metabolic imaging of primary tumor organoid cultures to accurately predict drug response [60, 69, 70]. The possibility to detect early, heterogeneous treatment response, on a label-free basis with cellular resolution makes FLIM-microscopy an attractive platform for testing drugs on patient-derived cancer cells for individualized treatment planning.
5 Time-Resolved Fluorescence Spectroscopy
The measurement of autofluorescence signals from the coenzymes for comprehensive tumor analysis in vivo is technically challenging. While the signals from the surface of the tumors can be analyzed rather easily, the analysis of the luminescence from the volume of the tumor is hindered by strong absorption and scattering in the UV and blue spectral region. Even in red and near-infrared regions scattering in the tissue still deteriorate the signals. Thus, luminescence from tissues can be retrieved only within 1 mm from a surface irrespective of a wavelength and makes the quantitative analysis of NAD(P)H/FAD fluorescence rather difficult.
Fiber-optic based spectroscopic sensing and imaging have been previously employed for chemical quantification of the tissues, including FLIM and Raman spectroscopy [71, 72]. Fiber-optic probes have also been used for in vivo depth-resolved neuron-activity mapping, both with two-photon and one-photon configurations [73, 74]. One of the first applications of time-resolved spectroscopy for skin cancer based on analysis of autofluorescence from NAD(P)H and using a fiber was described in the work of P.A.A. de Beule et al. from P. French group [75]. The authors presented an approach based on analysis of both fluorescence spectrum and fluorescence lifetime of endogenous fluorophores that allowed to discriminate basal cell carcinoma skin cancer from healthy skin tissue in ex vivo human skin lesions.
An optical fiber integrated in a thin needle that is inserted into a tumor is a promising approach for the delivery and collection of the luminescence signal from the internal layers of the tissue. Such an attempt has been demonstrated for two-photon probing of xenograft tumors in mice using MCA 207 cells expressing green fluorescent protein (GFP) [76]. Measuring the fluorescence of intrinsic fluorophores poses a few challenges. The quantum yields of NADH and FAD are low, and so are the intensities [77]. A second problem is fluorescence from the optical fibers. The excitation wavelength is in the ultraviolet or blue range of the optical spectrum. Fluorescence from the fibers at these wavelengths can easily be stronger than that from the sample. Two-photon excitation may be employed to overcome the difficulties generated by autofluorescence from fibers, but at the expense of costs and bulkiness of the setups. Alternatively, one may use hollow waveguide (HW) [78] or an air-core photonic crystal fibers (PCF) [79]. In both cases, the excitation beams are guided through the air-core, thus, the background signals originating from the core material are not inherently generated. However, the HW has a disadvantage of a low numerical aperture (NA), which reduce the collection efficiency of the signal. Moreover, the air holes of the HW and PCF should not be directly in contact with the liquids to avoid damage due to the capillary action. Finally, there are no readily available PC fibers in the UV region and, in general, they are very expensive.
The problems associated with the time-resolved fluorescence measurements with a fiber have been addressed in our recent paper [80]. The authors proposed a fiber-optic configuration that allowed to overcome the problem with background autofluorescence from the fiber core and enable measurement of weak fluorescence from NAD(P)H and phosphorescence from oxygen sensors from the deep layers of a tumor in vivo. The generalized scheme of the experimental setup is presented in Fig. 3.4.
Experimental setup for fluorescence/phosphorescence spectroscopy measurements. MMF1: Multimode fiber (core diameter: 50 μm, cladding diameter: 65 μm); MMF2: Multimode fiber (core diameter: 200 μm, cladding diameter: 220 μm); C: miniature connector; P: exchangeable probe with a multimode fiber (core diameter: 300 μm, cladding diameter: 350 μm) in a needle G26) (see Visualization 1); FC-FC connector; D1 and D2: HPM-100-40 detector. F1/2: emission filters 510LP and 632/90 BP. F3/4: 405LP and 450/60BP. DM: 510 LP dichroic mirror. (Reprinted with permission from [80])
Using this system, spectroscopic measurements of NAD(P)H fluorescence lifetime were performed in model experiment in solutions and in colorectal tumors in mice in vivo. The mean fluorescence lifetime in tumors was considerably shorter, and the relative contribution of free NAD(P)H in tumors was greater than in muscle, indicating a shift to more glycolytic metabolism in tumor tissue as compared to muscle. Simultaneous measurements of phosphorescence lifetime of oxygen-sensitive probe administrated into the tissue showed increased values in tumor reflecting reduced oxygenation, which correlated with its glycolytic state.
In a recent study, we have used fiber-optic-based spectroscopy to investigate metabolic response of the tumors to chemotherapy with irinotecan. In vivo fluorescence time-resolved sensing of NAD(P)H showed decreased contribution from the unbound NAD(P)H fraction in the treated tumors in mice compare with untreated control (Fig. 3.5), which is in agreement with our studies using FLIM-microscopy.
In vivo one-photon fluorescence time-resolved spectroscopy of NAD(P)H in mouse colorectal tumors in response to irinotecan. (A) Fluorescence decay curves of NAD(P)H in the specific spots in subcutaneous tumors. (B) Relative amplitude of free NAD(P)H (a1%) calculated from the in vivo decay curves. Mean ± SD, n = 3 tumors, 5–7 measurements from each tumor. Measurements were performed on the 14th day of tumor growth
Although the examples of time-resolved measurements of intrinsic fluorescence using optical fiber-based systems are rather few, this approach has promises for development of clinical FLIM endoscopy.
6 FLIM-Macroscopy
Extending FLIM from microscopy to macroscopy is an extremely important issue in cancer studies. While it is essential to have a subcellular resolution to follow metabolic processes on a single cell level, it is also critical to understand metabolic status of a living system at the large scale. Intratumoral metabolic heterogeneity at the cellular level is, to a considerable degree, determined by heterogeneous microenvironment within a tumor, namely, the architecture of extracellular matrix, organization of vasculature, infiltration of immune cells and fibroblasts, and interaction with surroundings. As a result, heterogeneity of the metabolic picture on the macroscopic level can be even more pronounced than on the cellular level.
Although there have been a few systems developed for whole-body small animal FLIM, they are insensitive to tissue autofluorescence and have poor spatial resolution [81,82,83].
Recently, we have used a confocal macro-FLIM system to record fluorescence of NAD(P)H and to investigate heterogeneity of tumors on macroscale. The operation of the macroscanner is described in detail elsewhere [84, 85]. Briefly, the macroscopic imaging is performed by scanning of the objects placed directly in the image plane of a confocal scan head. To combine NAD(P)H imaging with observation of other spectrally distinct fluorophores, there are two excitation and two detection channels in the system.
The optical principle for time-resolved macroscopic scanning system is shown in Fig. 3.6. The image plane of the scan lens is brought in coincidence with the sample surface. As the galvo-mirrors change the beam angle the laser focus scans across the sample. Fluorescence signal produced by the sample is collimated by the scan lens, descanned by the galvo-mirrors, and separated from the excitation light by the main dichroic beamsplitter.
Schematics of the macroscanner for two-channel FLIM. D: HPM-100-40 detector (Becker & Hickl GmbH, Germany), F1, F3: long pass filters; F2, F4: bandpass filters; A: pinhole; L3: detector focusing lenses (f = 50 mm); L1&L2 telescope lenses (f1 = 7.5 mm; f2 = 45 mm); BC beam combiner mirror, MD main dichroic mirror, GM galvo-mirrors, SL scan lens (f = 40 mm); S sample. (Reprinted with permission from [85])
Usually, the signal further can be separated into two spectral or polarization channels, and focused into the pinholes. Light passing the pinholes is sent to detectors. The maximum diameter of the image area in the primary image plane of the scanner is about 18 mm. The size of the laser spot in the image plane is about 15 μm. While the numerical aperture of the excitation beam path is determined by the beam diameter (about 1.5 mm) and the focal length of the scan lens (40 mm), the detection path is determined by the scan lens and the aperture of the galvo-mirrors (3 mm). Since the incoming laser beam under-fills the lens aperture, the numerical aperture in excitation is smaller than in the detection.
Overall, collection efficiency of a macroscanner is considerably lower compared to the system consisting of the scanner combined with a microscope. However, in macroscopic imaging one can use much higher laser power, which compensates for low collection efficiency. The power is distributed over a large area so that photobleaching is not a problem. The acquisition time of this system depends on the number of pixels and the desired number of photons for the accurate evaluation of the fluorescence lifetimes.
The performance of metabolic FLIM on a macroscale was demonstrated in mouse tumors in vivo and rat glioma samples ex vivo.
In the in vivo study we visualized NAD(P)H fluorescence lifetime in a whole tumor inoculated subcutaneously in live mice. Since 375 nm wavelength is used for one-photon excitation of NAD(P)H, the skin flap was surgically opened to image tumor. As expected, a solid advanced tumor displayed essential spatial variations of the fluorescence lifetime parameters of NAD(P)H. Comparisons between tumor and muscles showed an increase in the contribution of the free NAD(P)H in tumor tissue, suggesting a shift to glycolytic metabolism, similarly to the above mentioned spectroscopic measurements [84].
A large field of view in the macro-FLIM opens the opportunity to explore metabolism not only in the whole tumor in animal, but also in intraoperational centimeter sized samples of patients’ tumors. The metabolic differences between cancer and adjacent normal tissue, if detected by FLIM at the macroscale, can serve for delineation of tumor margins, which is extremely important in glioma surgery. We have made an attempt to evaluate the capability of macroscopic FLIM to delineate glioma margins in rat models on the basis of NAD(P)H fluorescence lifetime parameters recently (Fig. 3.7) [86]. It was found that, in comparison with normal brain tissue, anaplastic astrocytoma had longer values of mean fluorescence lifetime and fluorescence lifetime of a bound fraction of NAD(P)H. Moreover, we have observed decreased contribution from a free NAD(P)H. This shows the potential of macro-FLIM for tumor delineation. The changes in the optical metabolic readouts in the brain tumors are consistent with studies that report on elevated levels of fatty acid oxidation and glutaminolysis in glioma cells [8, 87]. With further development, macro-FLIM can find use in the clinic as a sensitive and precise method for identifying the edges of tumors during surgery.
Macro-FLIM of glioma in rat brain. Representative fluorescence lifetime images of NAD(P)H in freshly excised brain with anaplastic astrocytoma. τmean is an amplitude weighted fluorescence lifetime. a1/a2 is free/bound ratio. In the histogram of the normalized pixel frequency red and black lines represent the distribution of free NAD(P)H in the tumor and in the adjacent tissue, respectively. The tumor is marked with a white dashed line. Collection time: 120 s. Scale bar: 5 mm
7 Summary
A significant progress in the field of cancer metabolism led to understanding that the changes in metabolism in cancer cells are not a simple consequence of the uncontrolled proliferation, but are rather a coordinated and complementary program that promote tumor growth. An increased interest in tumor metabolism in recent years has been driven by the idea to exploit the altered and heterogeneous metabolism of cancer cells to develop personalized therapeutic approaches and to find new targets for therapy. Novel imaging technologies that allow detection of intrinsic fluorescence and measurement of its lifetime open the possibility to monitor metabolic state of living cells and tissues non-invasively, in real time. FLIM based on TCSPC is one of such technologies. Metabolic imaging done by fluorescence lifetime and intensity measurements is often associated with Fluorescence Lifetime Imaging Microscopy. Consequently, it is often believed that FLIM of metabolism is related to the microscopic level. This is certainly not correct, because now TCSPC-based fluorescence lifetime measurements can be also realized on the macroscale. Recently developed macro-FLIM allows to interrogate metabolism on the scale of a whole tumor. It gives an opportunity not only to observe the heterogeneity of the tumor on a large scale, which is important from a fundamental point of view, but also to determine the surgical margins of tumors with high precision, and this is essential for clinical applications. Another method to probe tumor metabolism is not related to imaging option, but rather to time-resolved fluorescence single point measurement via a fiber. While this approach does not deliver a map of metabolic state of the tumor, it offers an opportunity to explore its deep layers, since strong scattering and absorption of an excitation light at the surface of the tissue is not an issue any more. Combination of all the modalities may give new insights in the complicated picture of tumor metabolism, not only being limited to experimental models of cancer but also extending to patient’s tumors. Moreover, correlation of metabolic measurements with other physiological parameters measured using spectrally distinct exogenous sensors, i.e. multi-parametric recordings, will enhance their value and provide more details of fundamental aspects of cancer biology.
References
Warburg, O., Wind, F., Negelein, E.: The metabolism of tumors in the body. J. Gen. Physiol. 8(6), 519–530 (1927)
Bensinger, S.J., Christofk, H.R.: New aspects of the Warburg effect in cancer cell biology. Semin. Cell Dev. Biol. 23(4), 352–361 (2012). https://doi.org/10.1016/j.semcdb.2012.02.003
Coller, H.A.: Is cancer a metabolic disease? Am. J. Pathol. 184(1), 4–17 (2014). https://doi.org/10.1016/j.ajpath.2013.07.035
Solaini, G., Sgarbi, G., Baracca, A.: Oxidative phosphorylation in cancer cells. Biochim. Biophys. Acta Bioenerg. 1807(6), 534–542 (2011). https://doi.org/10.1016/j.bbabio.2010.09.003
Viale, A., Corti, D., Draetta, G.F.: Tumors and mitochondrial respiration: a neglected connection. Cancer Res. 75(18), 3687 (2015). https://doi.org/10.1158/0008-5472.CAN-15-0491
Hui, S., Ghergurovich, J.M., Morscher, R.J., Jang, C., Teng, X., Lu, W., Esparza, L.A., Reya, T., Le, Z., Yanxiang Guo, J., White, E., Rabinowitz, J.D.: Glucose feeds the TCA cycle via circulating lactate. Nature. 551(7678), 115–118 (2017). https://doi.org/10.1038/nature24057
Pavlides, S., Whitaker-Menezes, D., Castello-Cros, R., Flomenberg, N., Witkiewicz, A.K., Frank, P.G., Casimiro, M.C., Wang, C., Fortina, P., Addya, S., Pestell, R.G., Martinez-Outschoorn, U.E., Sotgia, F., Lisanti, M.P.: The reverse Warburg effect: aerobic glycolysis in cancer associated fibroblasts and the tumor stroma. Cell Cycle. 8(23), 3984–4001 (2009). https://doi.org/10.4161/cc.8.23.10238
Cluntun, A.A., Lukey, M.J., Cerione, R.A., Locasale, J.W.: Glutamine metabolism in cancer: understanding the heterogeneity. Trends Cancer. 3(3), 169–180 (2017). https://doi.org/10.1016/j.trecan.2017.01.005
Carracedo, A., Cantley, L.C., Pandolfi, P.P.: Cancer metabolism: fatty acid oxidation in the limelight. Nat. Rev. Cancer. 13(4), 227–232 (2013). https://doi.org/10.1038/nrc3483
Potter, M., Newport, E., Morten, K.J.: The Warburg effect: 80 years on. Biochem. Soc. Trans. 44(5), 1499–1505 (2016). https://doi.org/10.1042/BST20160094
Altman, B.J., Rathmell, J.C.: Metabolic stress in autophagy and cell death pathways. Cold Spring Harb Perspect Biol. 4(9), 008763 (2012). https://doi.org/10.1101/cshperspect.a008763
Green, D.R., Galluzzi, L., Kroemer, G.: Cell biology. Metabolic control of cell death. Science. 345(6203), 1250256 (2014). https://doi.org/10.1126/science.1250256
Zaal, E.A., Berkers, C.R.: The influence of metabolism on drug response in cancer. Front. Oncol. 8, 500 (2018). https://doi.org/10.3389/fonc.2018.00500
Scholz, R., Thurman, R.G., Williamson, J.R., Chance, B., Bücher, T.: Flavin and pyridine nucleotide oxidation-reduction changes in perfused rat liver. I. Anoxia and subcellular localization of fluorescent flavoproteins. J Biol Chem. 244, 2317–2324 (1969)
Chance, B.: Spectrophotometry of intracellular respiratory pigments. Science. 120(3124), 767–775 (1954)
Chance, B., Schoener, B., Oshino, R., Itshak, F., Nakase, Y.: Oxidation-reduction ratio studies of mitochondria in freeze-trapped samples. NADH and flavoprotein fluorescence signals. J Biol Chem. 254(11), 4764–4771 (1979)
Nakabayashi, T.: In: Ghukasyan, V.V., Heikal, A.A. (eds.) Intracellular Autofluorescent Species: Structure, Spectroscopy, and Photophysics. CRC Press, Boca Raton (2014)
Wallrabe, H., Svindrych, Z., Alam, S.R., Siller, K.H., Wang, T., Kashatus, D., Hu, S., Periasamy, A.: Segmented cell analyses to measure redox states of autofluorescent NAD(P)H, FAD & Trp in cancer cells by FLIM. Sci Rep. 8(1), 79 (2018). https://doi.org/10.1038/s41598-017-18634-x
Lakowicz, J.R., Szmacinski, H., Nowaczyk, K., Johnson, M.L.: Fluorescence lifetime imaging of free and protein-bound NADH. Proc. Natl. Acad. Sci. U. S. A. 89(4), 1271–1275 (1992)
Blacker, T.S., Marsh, R.J., Duchen, M.R., Bain, A.J.: Activated barrier crossing dynamics in the non-radiative decay of NADH and NADPH. Chem. Phys. 422, 184–194 (2013). https://doi.org/10.1016/j.chemphys.2013.02.019
Yu, Q., Heikal, A.A.: Two-photon autofluorescence dynamics imaging reveals sensitivity of intracellular NADH concentration and conformation to cell physiology at the single-cell level. J. Photochem. Photobiol. B. 95(1), 46–57 (2009). https://doi.org/10.1016/j.jphotobiol.2008.12.010
Sharick, J.T., Favreau, P.F., Gillette, A.A., Sdao, S.M., Merrins, M.J., Skala, M.C.: Protein-bound NAD(P)H lifetime is sensitive to multiple fates of glucose carbon. Sci Rep. 8(1), 5456 (2018). https://doi.org/10.1038/s41598-018-23691-x
Ladokhin, A., Brand, L.: Evidence for an excited-state reaction contributing to NADH fluorescence. J. Fluoresc. 5(1), 99–106 (1995). https://doi.org/10.1007/BF00718787
Blacker, T.S., Mann, Z.F., Gale, J.E., Ziegler, M., Bain, A.J., Szabadkai, G., Duchen, M.R.: Separating NADH and NADPH fluorescence in live cells and tissues using FLIM. Nat. Commun. 5, 3936 (2014). https://doi.org/10.1038/ncomms4936
Meleshina, A.V., Dudenkova, V.V., Shirmanova, M.V., Shcheslavskiy, V.I., Becker, W., Bystrova, A.S., Cherkasova, E.I., Zagaynova, E.V.: Probing metabolic states of differentiating stem cells using two-photon FLIM. Sci. Rep. 6, 21853 (2016). https://doi.org/10.1038/srep21853
Liu, Z., Pouli, D., Alonzo, C.A., Varone, A., Karaliota, S., Quinn, K.P., Münger, K., Karalis, K.P., Georgakoudi, I.: Mapping metabolic changes by noninvasive, multiparametric, high-resolution imaging using endogenous contrast. Sci Adv. 4(3), 9302 (2018). https://doi.org/10.1126/sciadv.aap9302
Yaseen, M.A., Sakadžić, S., Wu, W., Becker, W., Kasischke, K.A., Boas, D.A.: In vivo imaging of cerebral energy metabolism with two-photon fluorescence lifetime microscopy of NADH. Biomed. Opt. Express. 4(2), 307–321 (2013). https://doi.org/10.1364/BOE.4.000307
Stringari, C., Cinquin, A., Cinquin, O., Digman, M.A., Donovan, P.J., Gratton, E.: Phasor approach to fluorescence lifetime microscopy distinguishes different metabolic states of germ cells in a live tissue. Proc Natl Acad Sci U S A. 108(33), 13582 (2011). https://doi.org/10.1073/pnas.1108161108
Ma, N., Digman, M.A., Malacrida, L., Gratton, E.: Measurements of absolute concentrations of NADH in cells using the phasor FLIM method. Biomed. Opt. Express. 7(7), 2441–2452 (2016). https://doi.org/10.1364/BOE.7.002441
Starbird, C.A., Maklashina, E., Cecchini, G., Iverson, T.M.: Flavoenzymes: covalent versus noncovalent. In: eLS. Wiley, Chichester (2015)
van den Berg, P.A.W., Feenstra, K.A., Mark, A.E., Berendsen, H.J.C., Visser, A.J.W.G.: Dynamic conformations of flavin adenine dinucleotide: simulated molecular dynamics of the flavin cofactor related to the time-resolved fluorescence characteristics. J. Phys. Chem. B. 106(34), 8858–8869 (2002). https://doi.org/10.1021/jp020356s
Islam, S.D.M., Susdorf, T., Penzkofer, A., Hegemann, P.: Fluorescence quenching of flavin adenine dinucleotide in aqueous solution by pH dependent isomerisation and photo-induced electron transfer. Chem. Phys. 295, 137–149 (2003). https://doi.org/10.1016/j.chemphys.2003.08.013
Rivlin, R.S., Hornibrook, R., Osnos, M.: Effects of riboflavin deficiency upon concentrations of riboflavin, flavin mononucleotide, and flavin adenine dinucleotide in Novikoff hepatoma in rats. Cancer Res. 33(11), 3019–3023 (1973)
Pinto, J., Huang, Y.P., Rivlin, R.S.: Inhibition of riboflavin metabolism in rat tissues by chlorpromazine, imipramine, and amitriptyline. J. Clin. Invest. 67(5), 1500–1506 (1981)
Huhner, J., Ingles-Prieto, A., Neususs, C., Lammerhofer, M., Janovjak, H.: Quantification of riboflavin, flavin mononucleotide, and flavin adenine dinucleotide in mammalian model cells by CE with LED-induced fluorescence detection. Electrophoresis. 36(4), 518–525 (2015). https://doi.org/10.1002/elps.201400451
Munro, A.W., Noble, M.A.: In: Chapman, S.K., Reid, G.A. (eds.) Fluorescence Analysis of Flavoproteins, pp. 25–48. Humana Press, Totowa (1999)
Becker, W.: In: Castleman, A.W., Toennies, J.P., Zinth, W. (eds.) Advanced Time-Correlated Single Photon Counting Techniques. Springer, Berlin (2005)
Philip, J., Carlsson, K.: Theoretical investigation of the signal-to-noise ratio in fluorescence lifetime imaging. J. Opt. Soc. Am. A Opt. Image Sci. Vis. 20(2), 368–379 (2003). https://doi.org/10.1364/JOSAA.20.000368
Becker, W., Bergmann, A., Biskup, C., Zimmer, T., Klöcker, N., Benndorf, K., Becker, A., Gmbh, H.: Multi-wavelength TCSPC lifetime imaging. Proc. SPIE. 4620, 470679 (2002). https://doi.org/10.1117/12.470679
Becker, W., Bergmann, A., Biskup, C.: Multispectral fluorescence lifetime imaging by TCSPC. Microsc. Res. Tech. 70(5), 403–409 (2007). https://doi.org/10.1002/jemt.20432
Cheong, W.F., Prahl, S.A., Welch, A.J.: A review of the optical properties of biological tissues. IEEE J. Quantum Electron. 26(12), 2166–2185 (1990). https://doi.org/10.1109/3.64354
Bird, D.K., Yan, L., Vrotsos, K.M., Eliceiri, K.W., Vaughan, E.M., Keely, P.J., White, J.G., Ramanujam, N.: Metabolic mapping of MCF10A human breast cells via multiphoton fluorescence lifetime imaging of the coenzyme NADH. Cancer Res. 65(19), 8766–8773 (2005). https://doi.org/10.1158/0008-5472.can-04-3922
Skala, M.C., Riching, K.M., Gendron-Fitzpatrick, A., Eickhoff, J., Eliceiri, K.W., White, J.G., Ramanujam, N.: In vivo multiphoton microscopy of NADH and FAD redox states, fluorescence lifetimes, and cellular morphology in precancerous epithelia. Proc. Natl. Acad. Sci. U. S. A. 104(49), 19494–19499 (2007). https://doi.org/10.1073/pnas.0708425104
Lee, D.H., Li, X., Ma, N., Digman, M.A., Lee, A.: Rapid and label-free identification of single leukemia cells from blood in a high-density microfluidic trapping array by fluorescence lifetime imaging microscopy. Lab Chip. 18(9), 1349–1358 (2018). https://doi.org/10.1039/C7LC01301A
Walsh, A.J., Cook, R.S., Manning, H.C., Hicks, D.J., Lafontant, A., Arteaga, C.L., Skala, M.C.: Optical metabolic imaging identifies glycolytic levels, subtypes, and early-treatment response in breast cancer. Cancer Res. 73(20), 6164–6174 (2013). https://doi.org/10.1158/0008-5472.can-13-0527
Ruck, A., Hauser, C., Mosch, S., Kalinina, S.: Spectrally resolved fluorescence lifetime imaging to investigate cell metabolism in malignant and nonmalignant oral mucosa cells. J Biomed Opt. 19(9), 96005 (2014). https://doi.org/10.1117/1.jbo.19.9.096005
Shah, A.T., Demory Beckler, M., Walsh, A.J., Jones, W.P., Pohlmann, P.R., Skala, M.C.: Optical metabolic imaging of treatment response in human head and neck squamous cell carcinoma. PLoS One. 9(3), 90746 (2014). https://doi.org/10.1371/journal.pone.0090746
Wang, M., Tang, F., Pan, X., Yao, L., Wang, X., Jing, Y., Ma, J., Wang, G., Mi, L.: Rapid diagnosis and intraoperative margin assessment of human lung cancer with fluorescence lifetime imaging microscopy. BBA Clin. 8, 7–13 (2017). https://doi.org/10.1016/j.bbacli.2017.04.002
Leppert, J., Krajewski, J., Kantelhardt, S.R., Schlaffer, S., Petkus, N., Reusche, E., Huttmann, G., Giese, A.: Multiphoton excitation of autofluorescence for microscopy of glioma tissue. Neurosurgery. 58(4), 759–767 (2006). https://doi.org/10.1227/01.neu.0000204885.45644.22
Conklin, M.W., Provenzano, P.P., Eliceiri, K.W., Sullivan, R., Keely, P.J.: Fluorescence lifetime imaging of endogenous fluorophores in histopathology sections reveals differences between normal and tumor epithelium in carcinoma in situ of the breast. Cell Biochem. Biophys. 53(3), 145–157 (2009). https://doi.org/10.1007/s12013-009-9046-7
Kalinina, S., Breymayer, J., Schafer, P., Calzia, E., Shcheslavskiy, V., Becker, W., Ruck, A.: Correlative NAD(P)H-FLIM and oxygen sensing-PLIM for metabolic mapping. J. Biophotonics. 9(8), 800–811 (2016). https://doi.org/10.1002/jbio.201500297
Shcheslavskiy, V.I., Neubauer, A., Bukowiecki, R., Dinter, F., Becker, W.: Combined fluorescence and phosphorescence lifetime imaging. Appl Phys Lett. 108(9), 091111 (2016). https://doi.org/10.1063/1.4943265
Li, D., Zheng, W., Qu, J.Y.: Time-resolved spectroscopic imaging reveals the fundamentals of cellular NADH fluorescence. Opt. Lett. 33(20), 2365–2367 (2008)
Heaster, T.M., Walsh, A.J., Zhao, Y., Hiebert, S.W., Skala, M.C.: Autofluorescence imaging identifies tumor cell-cycle status on a single-cell level. J Biophotonics. 11(1), e201600276 (2018). https://doi.org/10.1002/jbio.201600276
Lukina, M.M., Dudenkova, V.V., Ignatova, N.I., Druzhkova, I.N., Shimolina, L.E., Zagaynova, E.V., Shirmanova, M.V.: Metabolic cofactors NAD(P)H and FAD as potential indicators of cancer cell response to chemotherapy with paclitaxel. Biochim. Biophys. Acta Gen. Subj. 1862(8), 1693–1700 (2018). https://doi.org/10.1016/j.bbagen.2018.04.021
Sergeeva, T.F., Shirmanova, M.V., Zlobovskaya, O.A., Gavrina, A.I., Dudenkova, V.V., Lukina, M.M., Lukyanov, K.A., Zagaynova, E.V.: Relationship between intracellular pH, metabolic co-factors and caspase-3 activation in cancer cells during apoptosis. Biochim. Biophys. Acta, Mol. Cell Res. 1864(3), 604–611 (2017). https://doi.org/10.1016/j.bbamcr.2016.12.022
Shirmanova, M.V., Sergeeva, T.F., Gavrina, A.I., Dudenkova, V.V., Lukyanov, K.A., Zagaynova, E.V.: Multiparametric analysis of cisplatin-induced changes in cancer cells using FLIM. Proc. SPIE. 10498, 1049807 (2018). https://doi.org/10.1117/12.2293996
Trinh, A.L., Chen, H., Chen, Y., Hu, Y., Li, Z., Siegel, E.R., Linskey, M.E., Wang, P.H., Digman, M.A., Zhou, Y.H.: Tracking functional tumor cell subpopulations of malignant glioma by phasor fluorescence lifetime imaging microscopy of NADH. Cancers. 9(12), 168 (2017). https://doi.org/10.3390/cancers9120168
Druzhkova, I.N., Shirmanova, M.V., Lukina, M.M., Dudenkova, V.V., Mishina, N.M., Zagaynova, E.V.: The metabolic interaction of cancer cells and fibroblasts - coupling between NAD(P)H and FAD, intracellular pH and hydrogen peroxide. Cell Cycle. 15(9), 1257–1266 (2016). https://doi.org/10.1080/15384101.2016.1160974
Walsh, A.J., Castellanos, J.A., Nagathihalli, N.S., Merchant, N.B., Skala, M.C.: Optical imaging of drug-induced metabolism changes in murine and human pancreatic cancer organoids reveals heterogeneous drug response. Pancreas. 45(6), 863–869 (2016). https://doi.org/10.1097/mpa.0000000000000543
Druzhkova, I.N., Lukina, M.M., Dudenkova, V., Shimolina, L.E., Zagaynova, E.V., Shirmanova, M.V.: Insight into microenvironment of tumor on the microscopic level with a focus on cancer-associated fibroblasts. Proc. SPIE. 10685, 106852R (2018). https://doi.org/10.1117/12.2307096
Szulczewski, J.M., Inman, D.R., Entenberg, D., Ponik, S.M., Aguirre-Ghiso, J., Castracane, J., Condeelis, J., Eliceiri, K.W., Keely, P.J.: In vivo visualization of stromal macrophages via label-free FLIM-based metabolite imaging. Sci. Rep. 6, 25086 (2016). https://doi.org/10.1038/srep25086
Shirmanova, M., Sergeeva, T., Druzhkova, I., Meleshina, A., Lukina, M., Dudenkova, V., Shcheslavskiy, V., Becker, W., Belousov, V., Mishina, N., Zagaynova, E.: In: König, K. (ed.) Metabolic Shifts in Cell Proliferation and Differentiation: Applications in Biology and Medicine. De Gruyter, Berlin (2018)
Shirmanova, M.V., Druzhkova, I.N., Lukina, M.M., Dudenkova, V.V., Ignatova, N.I., Snopova, L.B., Shcheslavskiy, V.I., Belousov, V.V., Zagaynova, E.V.: Chemotherapy with cisplatin: insights into intracellular pH and metabolic landscape of cancer cells in vitro and in vivo. Sci. Rep. 7(1), 8911 (2017). https://doi.org/10.1038/s41598-017-09426-4
Lukina, M.M., Dudenkova, V.V., Shimolina, L.E., Snopova, L.B., Zagaynova, E.V., Shirmanova, M.V.: In vivo metabolic and SHG imaging for monitoring of tumor response to chemotherapy. Cytometry A. 95(1), 47–55 (2019). https://doi.org/10.1002/cyto.a.23607
Alam, S.R., Wallrabe, H., Svindrych, Z., Chaudhary, A.K., Christopher, K.G., Chandra, D., Periasamy, A.: Investigation of mitochondrial metabolic response to doxorubicin in prostate cancer cells: an NADH, FAD and tryptophan FLIM assay. Sci. Rep. 7(1), 10451 (2017). https://doi.org/10.1038/s41598-017-10856-3
Cao, R., Wallrabe, H., Siller, K., Rehman Alam, S., Periasamy, A.: Single-cell redox states analyzed by fluorescence lifetime metrics and tryptophan FRET interaction with NAD(P)H. Cytometry A. 95(1), 110–121 (2019). https://doi.org/10.1002/cyto.a.23711
Shah, A.T., Diggins, K.E., Walsh, A.J., Irish, J.M., Skala, M.C.: In vivo autofluorescence imaging of tumor heterogeneity in response to treatment. Neoplasia. 17(12), 862–870 (2015). https://doi.org/10.1016/j.neo.2015.11.006
Walsh, A.J., Cook, R.S., Sanders, M.E., Aurisicchio, L., Ciliberto, G., Arteaga, C.L., Skala, M.C.: Quantitative optical imaging of primary tumor organoid metabolism predicts drug response in breast cancer. Cancer Res. 74(18), 5184–5194 (2014). https://doi.org/10.1158/0008-5472.CAN-14-0663
Shah, A.T., Heaster, T.M., Skala, M.C.: Metabolic imaging of head and neck cancer organoids. PLoS One. 12(1), 0170415 (2017). https://doi.org/10.1371/journal.pone.0170415
Cheng, S., Rico-Jimenez, J.J., Jabbour, J., Malik, B., Maitland, K.C., Wright, J., Cheng, Y.-S.L., Jo, J.A.: Flexible endoscope for continuous in vivo multispectral fluorescence lifetime imaging. Opt. Lett. 38(9), 1515–1517 (2013). https://doi.org/10.1364/OL.38.001515
Dochow, S., Ma, D., Latka, I., Bocklitz, T., Hartl, B., Bec, J., Fatakdawala, H., Marple, E., Urmey, K., Wachsmann-Hogiu, S., Schmitt, M., Marcu, L., Popp, J.: Combined fiber probe for fluorescence lifetime and Raman spectroscopy. Anal. Bioanal. Chem. 407(27), 8291–8301 (2015). https://doi.org/10.1007/s00216-015-8800-5
Cui, G., Jun, S.B., Jin, X., Pham, M.D., Vogel, S.S., Lovinger, D.M., Costa, R.M.: Concurrent activation of striatal direct and indirect pathways during action initiation. Nature. 494(7436), 238–242 (2013). https://doi.org/10.1038/nature11846
Doronina-Amitonova, L.V., Fedotov, I.V., Ivashkina, O.I., Zots, M.A., Fedotov, A.B., Anokhin, K.V., Zheltikov, A.M.: Implantable fiber-optic interface for parallel multisite long-term optical dynamic brain interrogation in freely moving mice. Sci. Rep. 3, 3265 (2013). https://doi.org/10.1038/srep03265
De Beule, P.A., Dunsby, C., Galletly, N.P., Stamp, G.W., Chu, A.C., Anand, U., Anand, P., Benham, C.D., Naylor, A., French, P.M.: A hyperspectral fluorescence lifetime probe for skin cancer diagnosis. Rev. Sci. Instrum. 78(12), 123101 (2007). https://doi.org/10.1063/1.2818785
Thomas, T.P., Myaing, M.T., Ye, J.Y., Candido, K., Kotlyar, A., Beals, J., Cao, P., Keszler, B., Patri, A.K., Norris, T.B., Baker Jr., J.R.: Detection and analysis of tumor fluorescence using a two-photon optical fiber probe. Biophys. J. 86(6), 3959–3965 (2004). https://doi.org/10.1529/biophysj.103.034462
Lakowicz, J.R.: Principles of Fluorescence Spectroscopy, 3rd edn. Springer, Boston (2006)
Sun, Y., Chen, W.L., Lin, S.J., Jee, S.H., Chen, Y.F., Lin, L.C., So, P.T.C., Dong, C.Y.: Investigating mechanisms of collagen thermal denaturation by high resolution second-harmonic generation imaging. Biophys. J. 91(7), 2620–2625 (2006). https://doi.org/10.1529/biophysj.106.085902
Doronina-Amitonova, L.V., Fedotov, I.V., Ivashkina, O.I., Zots, M.A., Fedotov, A.B., Anokhin, K.V., Zheltikov, A.M.: Enhancing the locality of optical interrogation with photonic-crystal fibers. Appl Phys Lett. 101(2), 021114 (2012). https://doi.org/10.1063/1.4727901
Lukina, M., Orlova, A., Shirmanova, M., Shirokov, D., Pavlikov, A., Neubauer, A., Studier, H., Becker, W., Zagaynova, E., Yoshihara, T., Tobita, S., Shcheslavskiy, V.: Interrogation of metabolic and oxygen states of tumors with fiber-based luminescence lifetime spectroscopy. Opt. Lett. 42(4), 731–734 (2017). https://doi.org/10.1364/OL.42.000731
Bloch, S., Lesage, F., McIntosh, L., Gandjbakhche, A., Liang, K., Achilefu, S.: Whole-body fluorescence lifetime imaging of a tumor-targeted near-infrared molecular probe in mice. J. Biomed. Opt. 10(5), 054003 (2005). https://doi.org/10.1117/1.2070148
Rice, W.L., Shcherbakova, D.M., Verkhusha, V.V., Kumar, A.T.: In vivo tomographic imaging of deep-seated cancer using fluorescence lifetime contrast. Cancer Res. 75(7), 1236–1243 (2015). https://doi.org/10.1158/0008-5472.CAN-14-3001
Sinsuebphon, N., Rudkouskaya, A., Barroso, M., Intes, X.: Comparison of illumination geometry for lifetime-based measurements in whole-body preclinical imaging. J. Biophotonics. 11(10), 201800037 (2018). https://doi.org/10.1002/jbio.201800037
Shcheslavskiy, V.I., Shirmanova, M.V., Dudenkova, V.V., Lukyanov, K.A., Gavrina, A.I., Shumilova, A.V., Zagaynova, E., Becker, W.: Fluorescence time-resolved macroimaging. Opt. Lett. 43(13), 3152–3155 (2018). https://doi.org/10.1364/OL.43.003152
Zherdeva, V.V., Kazachkina, N.I., Shcheslavskiy, V.I., Savitsky, A.P.: Long-term fluorescence lifetime imaging of a genetically encoded sensor for caspase-3 activity in mouse tumor xenografts. J. Biomed. Opt. 23(3), 1–11 (2018). https://doi.org/10.1117/1.JBO.23.3.035002
Shirmanova, M.V., Maria, L., Kisileva, E.B., Fedoseeva, V.V., Dudenkova, V.V., Zagaynova, E.V., Wolfgang, B., Shcheslavskiy, V.I.: Interrogation of glioma metabolism on macroscale by FLIM. Proc. SPIE. 10882, 1088209 (2019). https://doi.org/10.1117/12.2511475
Strickland, M., Stoll, E.A.: Metabolic reprogramming in glioma. Front. Cell Dev. Biol. 5, 43 (2017). https://doi.org/10.3389/fcell.2017.00043
Acknowledgments
The authors are thankful to Dr. Varvara Dudenkova, Dr. Nadezhda Ignatova, Dr. Elena Kiseleva, Irina Druzhkova, Liubov Shimolina, Alena Gavrina and Dmitry Reunov from Privolzhsky Research Medical University for their kind assistance in these studies. The studies are supported by the Russian Science Foundation (project # 20-65-46018).
Author information
Authors and Affiliations
Corresponding authors
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 Springer Nature Switzerland AG
About this chapter
Cite this chapter
Shirmanova, M.V., Shcheslavskiy, V.I., Lukina, M.M., Becker, W., Zagaynova, E.V. (2020). Exploring Tumor Metabolism with Time-Resolved Fluorescence Methods: from Single Cells to a Whole Tumor. In: Tuchin, V.V., Popp, J., Zakharov, V. (eds) Multimodal Optical Diagnostics of Cancer. Springer, Cham. https://doi.org/10.1007/978-3-030-44594-2_3
Download citation
DOI: https://doi.org/10.1007/978-3-030-44594-2_3
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-44593-5
Online ISBN: 978-3-030-44594-2
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)