Abstract
An abdominal aortic aneurysm (AAA) is a permanent and irreversible dilation of the lower aortic region. The current clinical rupture risk indicator for AAA repair is an anterior-posterior AAA diameter exceeding 5.5 cm. This is an inadequate rupture risk indicator given that 60% of AAAs with larger diameters than 5.5 cm often remain stable for the patient’s lifetime while 20% of smaller AAAs have ruptured. A more robust predictor of rupture risk is therefore crucial to save lives and reduce medical costs worldwide. Rupture is a local failure of the wall that occurs when local mechanical stress exceeds local wall strength. A comparison of the AAA tension and stretch during the cardiac cycle will provide the indication of wall structural integrity necessary for reliable rupture risk stratification. Employing engineering logic, mismatches between tension and stretch are likely to indicate localized wall weakening and the likelihood of rupture (e.g. a high stretch resulting from a low tension). Biomechanics based Prediction of Aneurysm Rupture Risk (BioPARR) is an AAA analysis software application that currently only determines aneurysm wall tension. This study seeks to investigate the feasibility of determining surface stretches within the AAA wall using methods compatible with clinical practices. It additionally aims to create and validate a new procedure for AAA rupture risk stratification.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
- Abdominal aortic aneurysm
- Rupture
- Computed tomography angiography
- Time-resolved
- Four-dimensional
- Synthetic
- Tension
- Stretch
1 Introduction
An abdominal aortic aneurysm (AAA) is a permanent and irreversible dilation of the lower aortic region. The condition is usually symptomless and is typically detected during an unrelated procedure. If left untreated, the aneurysm can dissect or rupture with the high mortality rates of approximately 80–90% [1]. Considering the dangers and expenses related to the surgical treatment, rupture risk classification is essential. If this rupture risk outweighs the risk of surgery, the patient will be considered for endovascular (EVAR) or open repair surgery.
The current clinical rupture risk indicator for repair is an anterior-posterior AAA diameter exceeding 5.5 cm or a diameter growth rate greater than 1 cm/year [2]. This is an inadequate rupture risk indicator given 60% of AAAs with larger diameters than 5.5 cm often remain stable for the patient’s lifetime [3] while 20% of smaller AAAs have ruptured [4]. Additionally, AAA rupture has been linked to other risk factors, including: genetic history, smoking, high mean arterial pressure (MAP), gender, vessel asymmetry, growth of intraluminal thrombus (ILT) and increased metabolic activity [5, 6]. Simplistic conclusions based on diameter alone are thus inadequate. A more robust and reliable predictor of rupture risk is therefore crucial to save lives and reduce medical costs worldwide.
Many researchers believe that a patient specific biomechanics-based approach is a promising alternative that could significantly improve the clinical management of AAA patients. With recent advancements in medical imaging and analysis software, geometrically accurate patient specific AAA three-dimensional (3D) models can now be constructed for the purpose of computer simulations that calculate wall stress. Studies have demonstrated that peak wall stress is a better indicator of individual rupture risk compared to aortic diameter [7]. Stress alone, however, will not provide an accurate estimation of rupture risk as mechanical failure of the wall is dependent on both local wall stress and local wall strength. Vande Geest et al. derived a statistical model for the non-invasive estimation of wall strength [8]. This strength model, however, is population-based, not patient specific and moreover not localized.
Many studies have utilized displacement tracking algorithms on time-resolved (4D) ultrasound scans to investigate local AAA wall deformations [9]. High local strains alone, however, cannot provide an indication of wall strength, as they may be generated by high local wall tensions.
AAA rupture is a local failure of the wall that occurs when local mechanical stress exceeds local wall strength [10]. This study proposes that a comparison of AAA tension with stretch during the cardiac cycle will provide the indication of wall structural integrity necessary for reliable rupture risk stratification. It is hypothesized that mismatches between local tension and resulting tangential stretch, such as high stretch with low tension, indicate localised wall weakening and the likelihood of rupture.
Biomechanics based Prediction of Aneurysm Rupture Risk (BioPARR) is an existing, free and semi-automatic AAA analysis software application that currently only determines aneurysm wall tension [11]. This study seeks to investigate the feasibility of determining surface stretches within the AAA wall using methods compatible with clinical practice. It additionally aims to validate the approach of pairing surface stretches with tension as a measure of AAA rupture potential.
A variety of approaches have been utilized by researchers to obtain ground truth data for validation purposes. Most methods are inaccurate and inefficient due to the errors introduced by reference tracking algorithms, sparse location of reference markers and the bias introduced by these markers on the tracking problem. Additionally, fabrication of physical phantoms to simulate realistic physiological deformation is both challenging and expensive.
Synthetic data provides a valuable reference for assessing the accuracy of tracking algorithms due to knowledge of the exact deformation. In this case, the reference displacement field is unbiased by any motion estimation algorithm. Additionally, exact deformation is known at each voxel. Furthermore, a wide range of digital data can easily be created by researchers thus eliminating the requirement for complex experimental phantoms. The usefulness of synthetic data as a validation tool, however, is highly dependent on the degree of realism of the generated synthetic scans.
One method of creating synthetic datasets involves the use of algorithms that simulate the physics of the imaging process. Models of virtual patient anatomy can consequently be ‘imaged’ using these projection algorithms. Models of the patient anatomy are only simplified geometries that have been mathematically derived and are therefore largely unrealistic. Furthermore, the organs and substructures are modelled as homogenous with constant pixel intensity. Image artefacts introduced by the heterogenous tissues are not simulated [12]. Therefore, although these phantoms can be used for dosimetry studies, they are inadequate for reliably assessing techniques dependent on image quality.
In the pursuit of increasingly realistic synthetic data, new techniques use biomechanical models extracted from the segmentation of real patient anatomy. A single static real medical scan is then warped with the deformation field of this model [13]. The use of real scans enables more accurate synthetic data creation by accounting for the heterogeneous tissue voxel intensities. Exact and simple methods to achieve this have not been clearly outlined in the literature. Additionally, these methods have mainly been restricted to the modelling of cardiac motion using only echocardiography and MRI [13]. This study therefore additionally aims to extend the existing literature by developing and clearly outlining simple methods for the simulation of realistic CT images using open source software for the given application of AAA.
2 Methods
2.1 Synthetic Data
A simple method of creating a synthetic 4D CT dataset was developed. This was achieved by warping a static CT scan using the transformation matrices obtained after modelling the pulsatile motion of the abdominal aortic aneurysm geometry.
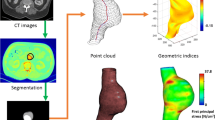
One abdominal aortic aneurysm computed tomography angiography DICOM scan was provided by Dr. Hozan Mufty of UZ Leuven academic hospital, Belgium. A 3D model of the AAA was created by segmenting the CT scan in 3D Slicer 4.10.1, a free open source medical image analysis and visualization software package.
The outer wall of the abdominal aortic aneurysm model was imported into Abaqus Explicit 2018. This was taken as the geometry that had been pre-loaded with the diastolic pressure. A linear tetrahedral element mesh was used due to its compatibility with Abaqus Explicit. The mesh contained approximately 4 × 106 nodes. The simulation consisted of a periodic loading cycle using an internal pulsatile pressure of 10 kPa. This represents a high pulse pressure that would realistically be observed in AAA patients. The upper and lower ends of the aneurysm were constrained in all directions using fixed boundary conditions (Fig. 1). Non-linear, hyper-elastic material properties were used to model the aneurysm tissue using the strain energy function presented by Raghavan and Vorp [14]. This strain energy function (W) shown below, was obtained by the researchers after examining the mechanical properties of excised AAA tissue.
a and b are the material properties and I1c is the first invariant of the right Cauchy-Green tensor. Most of the aneurysm tissue was modelled using a = 113.4 kPa, b = 9.2 kPa and a density of 1000 kg/m3 [15]. A randomly chosen local region of the aneurysm model was purposely weakened by halving each of these material parameters. In addition to location, the extent and range of weakening was arbitrarily selected. The local weakened and healthier tissue regions are indicated in Fig. 1 in red and green respectively.
Mesh nodal coordinates from five phases of the pulsating biomechanical model, between the two extremes of ‘diastole’ and ‘systole’, were extracted and exported from Abaqus to 3D Slicer. The transformation matrices, mapping each of the nodal coordinates from phase 0 to each of the respective phases, were obtained using the ‘Scattered Transform’ module [16]. The module interpolates displacements at nodes using a BSpline Algorithm. Once the transformation matrices were obtained, the 4D synthetic dataset was created using the ‘Data’ module. The initial CT scan was warped by each of these transformation matrices after dragging and dropping it onto the relevant transform. The new CT frames were then saved by hardening the transforms onto the volume. This resulted in a stack of synthetic CTs corresponding to each phase of the pulsating biomechanical model.
2.2 Voxel Displacement Tracking
As an alternative to producing an in-house code for the implementation of the displacement tracking techniques, open-source tools are available, such as those used for the registration of medical scans. Registration is the task of mapping one image to another image. This is typically used by clinicians to align scans of different modalities, or even align scans taken at different points in time such as for follow up procedures. Registration can therefore also be used to determine displacements of the aneurysm wall from scans at different points in time during the cardiac cycle.
Thirion proposed the Demons algorithm for non-rigid registration [17]. The Diffeomorphic Demons algorithm minimizes the sum of square differences of intensity, contains a smoothness constraint and additionally limits the transformation to be one-to-one. The Demons algorithm embodies a computationally efficient simplification of the optical flow problem.
The Demons Diffeomorphic Registration was implemented in 3D Slicer using the ‘BRAINSDemonWarp’ module. A course-to-fine pyramidal approach was utilized using 5 pyramid levels. A shrink factor of 16 and iteration count of 300, 50, 30, 20 and 15 for each respective pyramid level was employed. Linear interpolation and a Diffeomorphic Registration Filter were used. These parameter settings produced the most accurate results when visually compared with ground truth.
Each synthetic CT frame was registered to the initial frame. The outputs of these registrations were transformation matrices mapping points from one image to the next. The transformation matrices were then converted to displacement fields in the ‘Transforms’ module. Using the ‘Probe Volume’ module, the displacement field was then overlayed onto the surface of the segmented aneurysm geometry.
2.3 Determining Maximum Principal Stretch
The point coordinates of the AAA surface and the displacements at these nodes were read into MATLAB. An in-house modified moving least squares (MMLS) code was utilized in order to determine the deformation gradient from these nodal displacements [18]. The deformation gradient (F) was obtained by determining the derivative of the displacement vectors with respect to the undeformed configuration (X) and adding the identity matrix (I):
Additional code was added in order to determine the principal stretches. We computed the right Cauchy Green strain tensor: C=F TF. Eigenvalues of the right Cauchy Green strain tensor are the square of the principal stretches. The maximum principal tangential stretches and its directions were obtained after aligning the minimum eigenvectors with the surface normals. This is compatible with reality whereby the aorta wall will compress radially but stretch tangentially when it is inflated by the blood pressure.
2.4 Determining Maximum Principal Tension
The Maximum Principal Tension was determined via BioPARR utilizing the following inputs: a constant wall thickness of 1 mm, 16 kPa pressure applied to the interior AAA surface representing the patient’s mean arterial blood pressure and a ten-node tetrahedral hybrid element (C3D10H) mesh. The ‘no ILTP’ case was modelled. This case ignores the intraluminal thrombus and loads the interior surface of the AAA with blood pressure. This was done for simplicity and because the ILT was neglected when modelling the AAA motion.
2.5 New Rupture Risk Index
The MATLAB code was additionally updated to read-in the maximum principal tensions obtained from BioPARR. A structural integrity index (SII) was created by dividing the maximum principal tension by the largest maximum principal stretch during the cardiac cycle. A relative structural integrity index map (RSII) was created by dividing the SII map by the maximum structural integrity index over the AAA volume. This enables clear visualization of weakened areas by comparing all the structural integrity indices over the AAA volume with the strongest tissue present.
2.6 Validation of Techniques
The technique was validated by correlating displacements and maximum principal stretches obtained from 4D CT registration with the ground truth values obtained from Abaqus. This was implemented for each phase of the cardiac cycle. A Pearson correlation test was conducted in Excel with significance evaluated using a p-value of 0.05. Similarity to ground truth was also observed by visualizing displacements and maximum principal stretches in Paraview, an open-source data analysis and visualization application.
This new rupture risk predictor was then validated by determining if the randomly located purposely weakened area of the model was detected. This was achieved by visualizing relative structural integrity indices below 0.15 using Paraview. This represents the weakest 15% of tissue within the AAA.
3 Results
3.1 Validation of Displacement Tracking
A high similarity was observed between the ground truth displacement fields obtained via Abaqus and that obtained from registration of the synthetic 4D CT scans. This is depicted in Fig. 2 which displays the tangential displacement fields of the abdominal aortic aneurysm model during one phase of the cardiac cycle. This is additionally indicated by the high Pearson’s correlation coefficients of displacement magnitudes (R = 0.986, 0.990, 0.993, 0.996, 0.998, p < 0.001) and directions for each of the respective phases analysed (Table 1).
3.2 Maximum Principal Stretches
A high similarity was also observed between maximum principal stretches obtained from registered synthetic 4D CT scans and ground truth stretches obtained via Abaqus. This is evident in Fig. 3, where for each of the phases analyzed, stretch magnitudes and patterns obtained via registration are comparable to ground truth.
3.3 Relative Structural Integrity Index (RSII)
The largest maximum principal stretch during the cardiac cycle was then paired with the maximum principal tension obtained from BioPARR to compute the relative structural integrity index (RSII). A correlation analysis between the ground truth and registered RSII distributions indicated that good agreement was obtained (R = 0.98, Pearson’s correlation, p < 0.001). As evident in Fig. 4, an illustration of the lowest 15% of RSII successfully identifies the purposely locally weakened tissue depicted in Fig. 1.
4 Discussion
This study has successfully developed a procedure to accurately determine surface stretches within the AAA wall using methods compatible with clinical practices.
Most researchers have focused on utilizing time-resolved ultrasound to determine deformation of AAAs. This study has highlighted the feasibility of using 4D CT as an alternative. This is compatible with clinical workflow due to the current practice of employing 3D CT angiography for preoperative imaging of the AAA. Unlike ultrasound, 4D CT additionally enables quick, repeatable acquisition of the full volume of the AAA.
The use of the Demons Diffeomorphic registration technique to track deformation during the cardiac cycle from 4D CT scans was validated. The obtained displacements and resulting stretches were highly accurate with strong correlation to ground truth.
This novel study has introduced a new and improved rupture risk metric. The RSII utilizes a holistic engineering approach by accounting for both local stretches and tensions to enable the characterization of tissue integrity local to the AAA. This enables a patient specific measure of wall strength that other procedures have not considered. Even if stresses are computed correctly, high stresses alone cannot be interpreted as a loss of wall structural integrity without knowledge of local wall strength. i.e. clearly high wall stress is not an issue if it is present in a strong wall. Similarly, methods utilizing only high stretch as a measure of tissue integrity are flawed. These local high stretches may be generated by local high tensions and may not be due to weakened tissue. The RSII was validated by illustrating that a randomly located, purposely weakened area of the model was detected with high accuracy. These findings have advanced the state of the art of AAA management.
This method of creating a synthetic 4D CT sequence has granted access to the required data to test the feasibility of determining surface stretches within the AAA wall, without reliance on a clinic. It additionally enabled accurate knowledge of ground truth values and thus the ability to reliably assess the novel techniques used. This essential validation step would not have been possible with real patient data where access to exact ground truth is unattainable. Synthetic data provides a reference displacement field that is unbiased to any motion estimation algorithm. This is unlike that required by intermodal registration reference methods and techniques relying on the tracking of implanted markers. Unlike previous methods that utilise sparsely located reference markers, the technique used in this study provides knowledge of exact deformation at each voxel. Furthermore, the simple, low cost computer-based biomechanical model is more realistic compared to other mock-ups such as complicated physical phantoms, due to easier control of material properties and pressures. This opens the door to the generation of a wide range of synthetic data, from normal to varying diseased states, as demonstrated by this AAA study. The usefulness of synthetic data as a validation tool, however, is highly dependent on the degree of realism of the generated sequence. Unlike synthetic datasets created using projection algorithms, this study uses methods that produce realistic synthetic data. This was achieved by using real scans to extract exact patient anatomy and to simulate the heterogenous voxel intensities of imaged tissue.
The simple and easily accessible methods developed in this study can similarly be used by other researchers to progress pilot studies without being impeded by clinical bureaucracy. Additionally, the flexibility offered by this simple technique provides a platform to optimize and validate emerging technologies and methods without being impeded by the multitude of external restrictions imposed by the other validation techniques discussed.
Limitations, however, do exist in the presented work. This method of synthetic CT creation does not completely take the physics of image acquisition into account. Instead it re-uses the same texture of the initial CT, which is warped according to the differences between the original scan and the simulated motion. Changes in the geometry of the moving organ, however, will alter the path length along which the radiation travels through the organ. This will cause variations in voxel intensity throughout the cardiac cycle. The change in voxel intensity during deformation is not reflected in the synthetic data creation technique discussed.
One method discussed in the literature partly accounts for this by using a template 4D DICOM dataset to partially increase the degree of realism of the generated synthetic sequence [19]. This is achieved by spatio-temporal alignment of the template sequence with the biomechanical model. In this method instead of warping a single static scan at the initial phase of the cardiac cycle, the template scan is warped by the biomechanical model at each of the respective phases. This partially accounts for the change in intensities that will be present as a result of deformation because it reduces the difference between the reference and deformed frames. The risk of unrealistic texture warping does, however, still exist with this method when the simulated motion of the model deviates too far from the template motion. That method, however, requires the presence of an initial 4D dataset. In novel studies such as this one, access to an initial 4D dataset is not always possible. A 4D CT protocol of the AAA is not yet utilized in the clinic. Once access to real data from this protocol is achieved, a future study can further validate the methods used by implementing this improved technique.
A basic assumption made using the Demons algorithm is that the intensity of voxels remains constant through time. The geometry of the aneurysm, however, will be changing during the cardiac cycle, which, as discussed, will alter voxel intensities. Since this synthetic data is slightly unrealistic in that the intensity of voxels remains constant despite motion, the methods used on this artificial dataset are acceptable. When using real data, however, this may not remain true. An option for dealing with this issue could be to not register each frame to the initial frame, as was done using this synthetic dataset. Instead one could register each frame to the previous frame but use the preceding transform as an initialization to the registration. This would enable the constant intensity assumption to hold true as the geometry between consecutive frames would not change significantly.
The next step required to progress this novel technique into normal clinical practice is an initial pilot study using real patient data. Further studies will need to establish the relationship between RSII and the progression of abdominal aortic aneurysms using follow up analyses.
References
L.L. Hoornweg et al., Meta analysis on mortality of ruptured abdominal aortic aneurysms. Eur. J. Vasc. Endovasc. Surg. 35(5), 558–570 (2008)
B.J. Doyle et al., On the influence of patient-specific material properties in computational simulations: A case study of a large ruptured abdominal aortic aneurysm. Int. J. Numer. Methods Biomed. Eng. 29(2), 150–164 (2013)
D. Farotto et al., The role of biomechanics in aortic aneurysm management: Requirements, open problems and future prospects. J. Mech. Behav. Biomed. Mater. 77, 295–307 (2018)
S.C. Nicholls et al., Rupture in small abdominal aortic aneurysms. J. Vasc. Surg. 28(5), 884–888 (1998)
N. Sakalihasan et al., Positron emission tomography (PET) evaluation of abdominal aortic aneurysm (AAA). Eur. J. Vasc. Endovasc. Surg. 23(5), 431–436 (2002)
T.C. Gasser et al., A novel strategy to translate the biomechanical rupture risk of abdominal aortic aneurysms to their equivalent diameter risk: Method and retrospective validation. Eur. J. Vasc. Endovasc. Surg. 47(3), 288–295 (2014)
D.A. Vorp, Biomechanics of abdominal aortic aneurysm. J. Biomech. 40(9), 1887–1902 (2007)
J. Vande Geest et al., Towards a noninvasive method for determination of patient-specific wall strength distribution in abdominal aortic aneurysms. Ann. Biomed. Eng. 34(7), 1098–1106 (2006)
K. Karatolios et al., Method for aortic wall strain measurement with three-dimensional ultrasound speckle tracking and fitted finite element analysis. Ann. Thorac. Surg. 96(5), 1664–1671 (2013)
T.C. Gasser, Biomechanical rupture risk assessment: A consistent and objective decision-making tool for abdominal aortic aneurysm patients. Aorta J. 4(2), 42–60 (2016)
G. Joldes et al., BioPARR: A software system for estimating the rupture potential index for abdominal aortic aneurysms. Sci. Rep. 7(1), 4641–4641 (2017)
N. Lowther et al., Investigation of the XCAT phantom as a validation tool in cardiac MRI tracking algorithms. Phys. Med. 45, 44–51 (2018)
P. Clarysse et al., Simulation based evaluation of cardiac motion estimation methods in tagged-MR image sequences. J. Cardiovasc. Magn. Reson. 13(Suppl 1), P360 (2011)
M.L. Raghavan, D.A. Vorp, Toward a biomechanical tool to evaluate rupture potential of abdominal aortic aneurysm: Identification of a finite strain constitutive model and evaluation of its applicability. J. Biomech. 33(4), 475–482 (2000)
A. Satriano et al., In vivo strain assessment of the abdominal aortic aneurysm. J. Biomech. 48(2), 354–360 (2015)
G.R. Joldes et al., Performing Brain Image Warping Using the Deformation Field Predicted by a Biomechanical Model (Springer, New York, 2012), pp. 89–96
J.P. Thirion, Image matching as a diffusion process: An analogy with Maxwell’s demons. Med. Image Anal. 2(3), 243–260 (1998)
G.R. Joldes et al., Modified moving least squares with polynomial bases for scattered data approximation. Appl. Math. Comput. 266(C), 893–902 (2015)
N. Duchateau et al., Model-based generation of large databases of cardiac images: Synthesis of pathological cine MR sequences from real healthy cases. IEEE Trans. Med. Imaging 37(3), 755–766 (2018)
Acknowledgements
The authors wish to acknowledge help from is research was supported by Dr. Grand Roman Joldes, Dr. Mark Teh (Director of Training, Radiology, SCGH), Dr. Benjamin Khoo (Medical Physicist, SCGH), Dr. Toby Richards (HoD of Vascular Surgery, Fiona Stanley Hospital), Dr. Hozan Mufty (Vascular Surgeon, UZ Leuven academic hospital) and Dr. Abdul Maher (Vascular Surgeon, SCGH).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 Springer Nature Switzerland AG
About this paper
Cite this paper
Beinart, K.R., Bourantas, G.C., Miller, K. (2020). Towards Visualising and Understanding Patient-Specific Biomechanics of Abdominal Aortic Aneurysms. In: Miller, K., Wittek, A., Joldes, G., Nash, M., Nielsen, P. (eds) Computational Biomechanics for Medicine. MICCAI MICCAI 2019 2018. Springer, Cham. https://doi.org/10.1007/978-3-030-42428-2_7
Download citation
DOI: https://doi.org/10.1007/978-3-030-42428-2_7
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-42427-5
Online ISBN: 978-3-030-42428-2
eBook Packages: Physics and AstronomyPhysics and Astronomy (R0)