Abstract
The large size of raw 3D optical coherence tomography (OCT) volumes poses challenges for deep learning methods as it cannot be accommodated on a single GPU in its original resolution. The direct analysis of these volumes however, provides advantages such as circumventing the need for the segmentation of retinal structures. Previously, a deep learning (DL) approach was proposed for the detection of glaucoma directly from 3D OCT volumes, where the volumes were significantly downsampled first. In this paper, we propose an end-to-end DL model for the detection of glaucoma that doubles the number of input voxels of the previously proposed method, and also boasts an improved AUC = 0.973 over the results obtained using the previously proposed approach of AUC = 0.946. Furthermore, this paper also includes a quantitative analysis of the regions of the volume highlighted by grad-CAM visualization. Occlusion of these highlighted regions resulted in a drop in performance by 40%, indicating that the regions highlighted by gradient-weighted class activation maps (grad-CAM) are indeed crucial to the performance of the model.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
- 3D-CNN
- Glaucoma detection
- Optical coherence tomography
- Gradient-weighted class activation maps
- Visual explanations
1 Introduction
Glaucoma is the second leading cause of irreversible blindness worldwide. The number of worldwide glaucoma patients, aged 40–80 years, is estimated to be approximately 80 million in 2020 with about 20 million increase since 2010 [4]. This disease is characterised by optic nerve damage, the death of retinal ganglion cells [3], and the ultimate loss of vision. It is a slowly progressing disease, with a long asymptomatic phase, where patients do not notice the increasing loss of peripheral vision. Since glaucomatous damages are irreversible, early detection is crucial.
Spectral-domain OCT imaging provides clinicians with high-resolution images of the retinal structures, which are employed for diagnosing and monitoring retinal diseases, evaluating progression, and assessing response to therapy [5]. While the previous approaches around the detection of glaucoma have primarily depended on segmented features such as the thickness of the retinal nerve fibre layer (RNFL) and the ganglion cell layer (GCL), there have been limited efforts around evaluating the utility of deep learning (DL) models in improving the diagnostic accuracy and early detection of glaucoma using 3D OCT scans.
For example, in [2] and [16], DL networks were proposed to diagnose early glaucoma using retinal thickness features. Similarly in [11] and [1], pretrained models (trained on ImageNet [14]) were used for the detection task. In fact, none of these techniques use the raw volumes for DL training. Rather, they rely entirely on segmented features or measurements generated by the SD-OCT scanners. One limitation for this is the segmentation error propagation where failure rate increases with the disease severity and co-existing pathologies. This also does not allow the diagnosis model to learn other unknown features existing within the image data. The only end-to-end DL model which uses the 3D raw scans was proposed by Maetschke et al. [9] (referred to as CAM-3D-CNN). This approach utilised 3D convolutional layers in the CNN, but was forced to downsample the volumes by nearly a factor of 80 to enable CAM and train the model on a GPU (due to memory constraints on the GPU itself).
Another important aspect of DL is the clinical interpretability and transparency [12] of the models developed. Class activation mapping (CAM) [18] and gradient-weighted class activation maps (grad-CAM) [13] have been recently proposed to reveal insights into the decisions of deep learning models. Both of these techniques identify areas of the images that the networks relied on heavily to generate the classification. However, CAMs requires a specific network architecture, namely the use of a global average pooling layer prior to the output layer. Grad-CAM is a generalized form of CAM and can be used with any CNN-based architecture without any additional requirements. In this regard, the visualization of DL model for glaucoma detection has been studied in two papers [1, 9]. An et al. [1] identified pathologic regions in 2D thickness maps using grad-CAM [13], which have shown to be in agreement with the important decision making regions used by physicians. Similarly, Maetschke et al. [9] implemented 3D-CAM [18] to identify the important regions in 3D OCT volumes. The maps were however, in a coarse resolution that matched the downsampled input image. This method also employed specific architecture changes to accommodate the requirements of CAM generation. It is also noteworthy that neither of these approaches analysed the CAMs in any systematic fashion, and merely used the heat maps to validate findings in a small number of images that were qualitatively assessed.
In this paper, we propose an end-to-end 3D-CNN for glaucoma detection trained directly on 3D OCT volumes (gradCAM-3D-CNN). This approach continues to avoid the dependency on segmented structural thicknesses, but also improves on previously approached techniques by doubling the size of the input volumes [9], and also improves on the performance in a direct comparison between gradCAM-3D-CNN and CAM-3D-CNN models. The use of 3D grad-CAM [13] allows for the visualization of the important regions of the 3D OCT cubes in a higher resolution than was not available before. Crucially, we validate the grad-CAM heat maps in a quantitative fashion, by occluding regions identified in the heat maps and assessing the impact of this on the performance of the model.
2 Materials and Methods
2.1 Dataset
The dataset contained 1248 OCT scans from both eyes of 624 subjects, acquired on a Cirrus SD-OCT Scanner (Zeiss; Dublin, CA, USA). 138 scans with signal strength less than 7 were discarded. The final dataset contained 263 scans on healthy eyes and 847 scans with primary open angle glaucoma (POAG). The scans were centered on the optic nerve heard (ONH) and had 200 \(\times \) 200 \(\times \) 1024 (a-scans \(\times \) b-scans \(\times \) depth) voxels per cube covering an area of 6 \(\times \) 6 \(\times \) 2 mm\(^3\).
2.2 Network Architecture
The proposed CNN model receives input scans with a resolution of 256 \(\times \) 128 \(\times \) 128 (depth \(\times \) b-scans \(\times \) a-scans) to classify an OCT volume as healthy or glaucoma. The network consists of eight 3D-convolutional layers, where each is followed by ReLU activation [6], batch-normalization [8] and max-pooling in order. The 3D convolutional layers have incremental number of the filters of 16-16-32-32-32-32-32-32 with kernel sizes of 3-3-3-3-5-5-3-3 in order, and stride of 1 for all layers. Also, 3D max-pooling layers has size of 2 and stride of 2. Finally, two fully-connected layers connect all the activated neurons in the previous layer to the next layer with 64 and 2 units respectively.
2.3 Evaluation: Training and Testing
The 1110 OCT volumes were downsampled to size 256 \(\times \) 128 \(\times \) 128 and split into a training, validation and testing subsets, containing 889 (healthy: 219, POAG: 670), 111 (healthy: 23, POAG: 88) and 110 (healthy: 21, POAG: 89) scans, respectively. The proposed 3D-CNN model was trained using the RMSprop optimizer with a learning rate of \(1e^{-4}\). Training was performed with a batch of size four through 50 epochs. Data was stratified per epoch by down-sampling to obtain balanced training samples. After each epoch, the area under the curve (AUC) was computed for the validation set and the network is saved if an improvement in AUC is observed.
2.4 DL Visualization
We implemented 3D grad-CAM [13] for visual explanations of the proposed model. We do not use CAM as it requires adding the global average pooling (GAP) after the last convolutional layer (i.e. conv#8) which restricts the network architecture design. Further, CAM would generate visualization only for feature map of conv#8, which in our case has a size of 2 \(\times \) 1 \(\times \) 1. Hence, when resizing and overlaying on the original cube of size 200 \(\times \) 200 \(\times \) 1024 will not provide any meaningful results. In this paper, we calculated the heat map for each of the first 6 convolutional layers (conv#1-6) separately, following the explanation provided in [13]. We did not compute grad-CAM for conv #7 and #8 layers due to the very small size of the corresponding heat maps (conv#7: 8 \(\times \) 4 \(\times \) 4 and conv#8: 4 \(\times \) 2 \(\times \) 2). The generated heat maps have the same size as the feature map of the corresponding convolutional layer. Instead of clipping the negative values in the resulted heat maps, as performed in the grad-CAM paper [13], we used the absolute value. To get rid of noisy gradients and to highlight only the important decision regions, we clipped the smallest 30% values and then resize the heat map to the original cube size.
To validate the generated heat maps, we occluded the input volumes by zeroing the rows and columns with the highest weights. Specifically, we extracted a set of indices with the highest weights per each dimension. This was done by spatial dimension reduction using average pooling. For example, a heat map with size 1024 \(\times \) 200 \(\times \) 200 was reduced to a vector of size 1024 \(\times \) 1 \(\times \) 1 by averaging the values of each 200 \(\times \) 200 map to get a single value. The indices of the top highest values (top x) in the resulted vector represent the most important region for this dimension. We applied this process on the b-scans and depth dimensions with x values of 64 and 256 respectively, while we considered the 200 a-scan columns were all important. This means that a fixed region of size 256 \(\times \) 64 \(\times \) 200 was occluded for each volume. Finally, the network was examined by evaluating the performance using the test set and its occluded volumes (2 \(\times \) 110 scans).
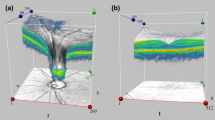
3D grad-CAM visualization results. Rows 1–4 show the b-scan slices #50, #100, #110, and #140 in order; \(5^{th}\) row displays the enface of the overlay of grad-CAM heat map on the original 3D cube; \(6^{th}\) row displays the enface of the occluded region (refer to Sect. 2.4 for the occlusion method). Note: scans are resized for display.
3 Experiments and Results
3.1 The Glaucoma Detection Model
The proposed gradCAM-3D-CNN model as well as the CAM-3D-CNN model described in [9] were implemented using Python, Keras with Tensorflow [7] and nuts-flow/ml [10] on a single K80 GPU. Performance of both models were evaluated using five statistical measures, namely, area under the curve (AUC), accuracy, Matthews correlation coefficient (MCC), recall, precision and F1-score. We computed the weighted average measures to avoid biased resulting from the class size imbalance in the data. The threshold with the highest validation F1-score was chosen for calculating the performance measures. The proposed model achieved an AUC of 0.973 for the test set (110 scans).
Further, to validate the performance of the proposed model (gradCAM-3D-CNN), we trained the CAM-3D-CNN architecture, proposed in [9] using same data split. Table 1 has the performance measures for each model using the same test set. The table shows that the proposed model outperforms the CAM-3D-CNN model with an increase of 3%, 5%, and 9% in the AUC, accuracy and F1-score respectively. We should note that CAM-3D-CNN has shown to outperform the conventional machine learning with an increase of 5% in the AUC measure [9].
Figure 1 visualizes grad-CAM heat maps for two convolutional layers: conv#1 and conv#4 for healthy and glaucoma cases. It is clear from the table that the last/deeper convolutional layers yield general and global important regions across all cubes, while the first convolutional layers give more detailed highlights which are comparable to the segmentation of retinal layers. The field of view of the deeper layers is larger than the more superficial layers, but the size of the heat maps are smaller in the deeper layers. This contributes to the generation of heat maps highlight larger swathes of the retina, but also lacks detail. For example, the heat map size for conv#1 is 256 \(\times \) 128 \(\times \) 128 while it is 16 \(\times \) 8 \(\times \) 8 for conv#6. The \(2^{nd}\) column in Table 2 shows the heat map size for each convolutional layer. In the heat maps generated from conv#4 we see that the optic disc region is highlighted, which is a region known to be affected by glaucoma.
3.2 Occlusion Experiment
To quantitatively assess different grad-CAM visualization results, we calculated the performance measure drops of gradCAM-3D-CNN model using the occluded set resulted from each convolutional layer. We also computed the occluded test set using CAM heat map generated from CAM-3D-CNN model. In total, 7 different occluded sets were generated and the corresponding performance measures drops are reported in Table 2. From the table, the highest drop in accuracy is 40%, achieved by gradCAM-conv#6 layer, followed by gradCAM-conv#4, gradCAM-conv#5, CAM, gradCAM-conv#1, gradCAM-conv#2, and gradCAM-conv#3 with accuracy drop of 39%, 39%, 35%, 35%, 34% and 33% in order. Further, heat map of gradCAM-conv#6 was also used to occlude the least important decision region, where the performance drop was only 4%, 3% and 4% in the AUC, accuracy and F1-score measures respectively. This confirms the effectiveness of grad-CAM for highlighting important decision regions.
Different variants of grad-CAM were implemented to enhance the heat maps by removing noisy gradients by either back propagating only positive gradients (relu-grads) [17] or positive gradients and positive input (guided-grads) [15]. Table 3 shows the impact of occlusion using different grad-CAM variants on the performance of the proposed model. We also investigated the impact of using different gradient modifiers, such as positive, negative, and absolute gradients, on the classification performance of the model. For example, in the case of absolute gradients we used the absolute values of the feature map gradients to compute the heat map. Table 3 shows that occlusion using grad-CAM without any modifier results in the highest drop in the performance.
4 Conclusion and Future Work
We present an end-to-end 3D CNN classification model that is able to effectively distinguish between healthy and glaucoma cases using 3D raw volumes. This approach improves on the accuracy of previously proposed methods [9], but also used an input that was double the size. This allowed for better CAMs to be generated using grad-CAM, which highlighted important regions of the retina. Further, grad-CAM heat maps were analyzed and quantitatively validated using the occlusion assessment method. In particular, the occlusion assessment method confirmed the effectiveness of grad-CAM in highlighting crucial decision regions. In the future, we will improve the evaluation using a cross-validation study, as well as extend this study to include other ocular diseases. We also plan to train the DL model on the important sub-volumes guided by grad-CAM results. We will also study the effect of fusing grad-CAM for different convolutional layers.
References
An, G., et al.: Glaucoma diagnosis with machine learning based on optical coherence tomography and color fundus images. J. Healthc. Eng. 2019, 1–9 (2019)
Asaoka, R., et al.: Using deep learning and transfer learning to accurately diagnose early-onset glaucoma from macular optical coherence tomography images. Am. J. Ophthalmol. 198, 136–145 (2019)
Davis, B.M., Crawley, L., Pahlitzsch, M., Javaid, F., Cordeiro, M.F.: Glaucoma: the retina and beyond. Acta Neuropathol. 132(6), 807–826 (2016)
Flaxman, S.R., et al.: Global causes of blindness and distance vision impairment 1990–2020: a systematic review and meta-analysis. Lancet Glob. Health 5(12), e1221–e1234 (2017)
Fujimoto, J., Swanson, E.: The development, commercialization, and impact of optical coherence tomography. Invest. Ophthalmol. Vis. Sci. 57(9), OCT1–OCT13 (2016)
Glorot, X., Bordes, A., Bengio, Y.: Deep sparse rectifier neural networks. In: Proceedings of the Fourteenth International Conference on Artificial Intelligence and Statistics, pp. 315–323 (2011)
Gulli, A., Pal, S.: Deep Learning with Keras. Packt Publishing Ltd, Birmingham (2017)
Ioffe, S., Szegedy, C.: Batch normalization: accelerating deep network training by reducing internal covariate shift. arXiv preprint arXiv:1502.03167 (2015)
Maetschke, S., Antony, B., Ishikawa, H., Garvani, R.: A feature agnostic approach for glaucoma detection in OCT volumes. arXiv preprint arXiv:1807.04855 (2018)
Maetschke, S., Tennakoon, R., Vecchiola, C., Garnavi, R.: Nuts-flow/ml: data pre-processing for deep learning. arXiv preprint arXiv:1708.06046 (2017)
Muhammad, H., et al.: Hybrid deep learning on single wide-field optical coherence tomography scans accurately classifies glaucoma suspects. J. Glaucoma 26(12), 1086–1094 (2017)
Razzak, M.I., Naz, S., Zaib, A.: Deep learning for medical image processing: overview, challenges and the future. In: Dey, N., Ashour, A.S., Borra, S. (eds.) Classification in BioApps. LNCVB, vol. 26, pp. 323–350. Springer, Cham (2018). https://doi.org/10.1007/978-3-319-65981-7_12
Selvaraju, R.R., Cogswell, M., Das, A., Vedantam, R., Parikh, D., Batra, D.: Grad-CAM: visual explanations from deep networks via gradient-based localization. In: Proceedings of the IEEE International Conference on Computer Vision, pp. 618–626 (2017)
Simonyan, K., Zisserman, A.: Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:1409.1556 (2014)
Springenberg, J.T., Dosovitskiy, A., Brox, T., Riedmiller, M.: Striving for simplicity: the all convolutional net. arXiv preprint arXiv:1412.6806 (2014)
Wang, J., et al.: SD Net: joint segmentation and diagnosis revealing the diagnostic significance of using entire RNFL thickness in glaucoma. In: Conference on Medical Imaging with Deep Learning (MIDL) (2018)
Zeiler, M.D., Fergus, R.: Visualizing and understanding convolutional networks. In: Fleet, D., Pajdla, T., Schiele, B., Tuytelaars, T. (eds.) ECCV 2014. LNCS, vol. 8689, pp. 818–833. Springer, Cham (2014). https://doi.org/10.1007/978-3-319-10590-1_53
Zhou, B., Khosla, A., Lapedriza, A., Oliva, A., Torralba, A.: Learning deep features for discriminative localization. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 2921–2929 (2016)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Switzerland AG
About this paper
Cite this paper
George, Y., Antony, B., Ishikawa, H., Wollstein, G., Schuman, J., Garnavi, R. (2019). 3D-CNN for Glaucoma Detection Using Optical Coherence Tomography. In: Fu, H., Garvin, M., MacGillivray, T., Xu, Y., Zheng, Y. (eds) Ophthalmic Medical Image Analysis. OMIA 2019. Lecture Notes in Computer Science(), vol 11855. Springer, Cham. https://doi.org/10.1007/978-3-030-32956-3_7
Download citation
DOI: https://doi.org/10.1007/978-3-030-32956-3_7
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-32955-6
Online ISBN: 978-3-030-32956-3
eBook Packages: Computer ScienceComputer Science (R0)