Abstract
In the last two decades, several methods for airway segmentation from chest CT images have been proposed. The following natural step is the development of a tool to accurately assess the morphology of the bronchial system in all its aspects to help physicians better diagnosis and prognosis complex pulmonary diseases such as COPD, chronic bronchitis and bronchiectasis. Traditional methods for the assessment of airway morphology usually focus on lumen and wall thickness and are often limited due to resolution and artifacts of the CT image. Airway wall cartilage is an important characteristic related to airway integrity that has shown to be deteriorated during the airway disease process. In this paper, we propose the development of a Model-Based GAN Regressor (MBGR) that, thanks to a model-based GAN generator, generate synthetic airway samples with the morphological components necessary to resemble the appearance of real airways on CT at will and that simultaneously measures lumen, wall thickness, and amount of cartilage on pulmonary CT images. The method is evaluated by first computing the relative error on generated images to show that simulating the cartilage helps improve the morphological quantification of the airway structure. We then propose a cartilage index that summarizes the degree of cartilage of bronchial trees structures and perform an indirect validation with subjects with COPD. As shown by the results, the proposed approach paves the way for the use of CNNs to precisely and accurately measure small lung airways morphology, with the final goal to improve the diagnosis and prognosis of pulmonary diseases.
This work has been partially funded by the National Institutes of Health NHLBI awards R01HL116931, R01HL116473, and R21HL14042. We gratefully acknowledge the support of NVIDIA Corporation with the donation of the GPU used for this research.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
In the last two decades, several methods have been proposed to help physicians accurately locate small pulmonary bronchi on chest CT images, that otherwise requires a tedious and time-consuming analysis of the data.
Once the structures are identified, an equally important step is represented by a quantitative measurement and morphological analysis of the bronchial tree, as this is the part that is commonly affected by inflammatory and infectious lung diseases. As an example, the smaller conducting airways are the structures most affected in patients with chronic obstructive pulmonary disease (COPD) [4], while emphysema has been related to a diminished presence of cartilage in the airway walls [15]. For this reason, a method that provides accurate and precise information about the airway cartilage amount may greatly improve the diagnosis and prognosis of lung diseases.
To the best of our knowledge, while methods have attempted to characterize only wall thickness and lumen size on CT images [10, 12], information about the cartilage amount is not provided. One of the reasons why prior work has not been proposed to quantify airway wall cartilage is the complexities to define a ground truth. Histological studies are limited, and it is extremely complex to have imaging data with a well-characterized histological correlate for each airway point. That being said, the histological model of an airway is well understood and can be easily parameterized. On the other hand, the imaging reconstruction process can also be modeled to some extent.
Deep learning [6] is quickly becoming the de-facto approach for solving various tasks in the medical imaging field. One of the advantages of deep learning approaches is the ability to learn from a given domain that can be synthetically generated. In our context, where properly labeled airway morphology data is highly complicated to obtain as a tedious and prone to error analysis of CT images is required, this learning paradigm from a synthetic model is appealing.
In this paper, we propose the development of a Model-Based GAN Regressor (MBGR) that automatically generates 2D patches of synthetic airways resembling real small bronchi (radius < 6.0 mm) on CT images with known morphometric characteristics of cartilage, wall thickness, and lumen size. Due to the generative nature of our approach, infinite training samples with varying imaging characteristics are available to train a regressor that can estimate those morphometric quantities.
To validate the method, we first compute the relative error (RE) obtained on synthetic patches and we demonstrate that generating a more accurate airway model that takes into account the cartilage improves the estimation of the airway morphology (wall thickness and lumen) with respect to a model without cartilage. Then, we perform an indirect validation that shows that in the cartilage assessment in a population of 547 smoking subjects with and without COPD, our measurements relates to disease severity in a consistent manner with prior histological studies [15].
2 Materials and Methods
Workflow of the model-based GAN regressor. MBG (green square) indicates the model-based generator that creates the synthetic airways (black box), G (purple box) is the image-to-image generator, D (yellow box) is the discriminator, and AR (blue box) represents the airway regressor that provides the 4 airway morphology measurements. In the big squares the schemes of the three neural networks used in this paper are presented. C indicates a 2D convolutional layer, FC a fully connected, MP stands for max pooling, and s indicates the layer’s strides. (Color figure online)
Figure 1 shows the flow diagram of the proposed MBGR that automatically generates synthetic CT airways, refines them to resemble real bronchi and simultaneously predicts cartilage amount, wall thickness, and lumen size. Although less clinically relevant, for completeness and to help the network better learn airway characteristics, the size of the cartilage wall thickness is also measured.
The model-based generator, MBG, is characterized by an airway geometric model that creates 2D images of 32 \(\times \) 32 pixels on the reformatted axial plane, a reasonable simulation as in in-vivo CT images this plane can be extracted along the axis given by the first eigenvector of the Hessian matrix. The geometric model consists of two bright ellipses (inner and outer walls) and a central dark area (lumen) with tangent vessels represented by bright ellipses randomly rotated around the airway.
To simulate the presence of cartilage, two ellipses are created at the center of the airway walls with higher intensity and varying wall thickness. As reported in [15], the percentage of cartilage varies depending both on lumen size (based on Weibel dichotomous morphometry [16]) and amount of emphysema. Therefore, for each airway a random percentage of cartilage is selected between minimum and maximum values of each bronchial order and level of emphysema, in accordance to Table 1 in [15]. Based on the selected cartilage percentage, equally-spaced sections of the brighter ellipses are removed, as shown in Fig. 2a, using the area of the elliptical sector:
where \(\theta \) is the angle inside the sector and a and b represent the main axis of the ellipse. The cartilage wall thickness is chosen as a random percentage of the total wall thickness in the range [0.01 * wall thickness, 0.6 * wall thickness] mm, while the cartilage radius is given by:
where LR indicates the lumen radius size, WT the wall thickness, and CWT stands for cartilage wall thickness. All parameters to generate the geometry model of the airway were randomly chosen based on physiological values as described in Table 1.
An example of generated synthetic airway patch with cartilage mimicking (lumen radius = 3.5 mm, wall thickness = 2.55 mm, cartilage amount = 38%). The geometry model (a) is generated at a super resolution of 0.05 mm/pixel in a sampling grid of 640 \(\times \) 640 pixels. A point spread function (PSF) is then simulated to obtain the final resolution of 0.5 mm/pixel, the patch is cropped to a 32 \(\times \) 32 pixels grid (b), and the GAN makes it more realistic (c). An example of airway from a CT is shown in (d).
To mimic the structure of the parenchyma, first a Gaussian smoothing (with a standard deviation of 5) was applied to Gaussian distributed noise, to create some broadly correlated noise, which made a texture of multiple structures. Then, the correlated noise was altered to have a mean intensity of −900 HU and a standard deviation of 150. All values were empirically chosen. A super-resolution of 0.05 mm/pixel in a sampling grid of 640 \(\times \) 640 pixels that is then down-sampled to a resolution of 0.5 mm/pixel is used to create the initial patches (Fig. 2b). Next, a PSF is simulated under the assumption that the PSF can be approximated by means of a spatially locally invariant Gaussian function [13].
The standard deviation of the Gaussian filter was randomly chosen in an empirically determined range of 0.4 to 0.9 mm to simulate the differences in the PSF across CT scanners and manufacturers, and a spatially correlated Gaussian noise was added to the image based on Gaussian distributed random noise smoothed with a Gaussian filter (standard deviation of 2), with the empirically determined mean of zero and random standard deviation in a range [0, 40]. As a last step, the image is cropped to a 32 \(\times \) 32 pixels grid, a size chosen to include enough neighborhood information for big structures, without losing specificity for small and thin features.
The generated patch is then passed to an neural generator that is meant to improve the realism of the synthetic airway while preserving its main characteristics by comparing it to unlabeled real airways extracted from clinical CTs. To this end, a generative adversarial network (GAN) approach is leveraged to add realism.
A pixel-to-pixel fully convolutional neural network with ResNet blocks is used as generator, G (purple box in Fig. 1), which is trained with an adversarial loss with self-regularization (originally introduced in [14]) to preserve the patch global structure. By minimizing the generator’s loss function, G tries to “fool” a discriminator network, D (yellow box in Fig. 1), into classifying the refined airways as real. This is modeled as a minimax game, where the weights of G and D (optimized through a binary cross-entropy loss) are updated alternately.
Following the approach presented in [14], the receptive field of D is also limited to local regions, so that multiple local adversarial losses per image are considered. Also, to improve the stability of the network, a mini-batch of refined images (randomly selected from a buffer of refined images generated on previous iterations) are included into the training batch. An example of the refining step result is shown in Fig. 2c.
To train this model-based GAN, we extracted 273,600 real airways patches of \(32\times 32\) pixels and a resolution of 0.5 mm on the reformatted axial plane of 30 clinical CTs, using the multi-resolution particles method described in [5], initialized with the feature enhancement technique of [9]. We then trained the adversarial network for 10,000 steps using a batch size of 512, a stochastic gradient descent update and learning rate of 0.001, until real and synthetic patches become indistinguishable by D.
Once G is trained, 2,500,000 training and 1,000,000 validation synthetic patches are generated and automatically refined while training the airway regressor, AR, to learn airway and cartilage characteristics. To this end, a 9-layer 2D network consisting of seven convolutional layers and two fully-connected layers, (see the blue box in Fig. 1) is used. An Adam update (\(\beta _1\) = 0.9, \(\beta _2\) = 0.999, \(\epsilon = 1e^{-08}\), decay = 0.0), a learning rate of 0.001, and a customized loss function that combines accuracy and precision, as presented in [8], were used to train AR for 100 epochs with a batch size of 1,000:
where N is the total number of images, M is the number of augmented replicas for each original patch, and y and \(\widehat{y}\) indicate the true and the predicted values, respectively. In this work, M = 25 has been used.
Finally, in order to help the network focus more on geometry than intensity values, during training we applied a data augmentation that in addition to randomly inverting intensity values inside the patches it also randomly shifts and flips the images.
All networks were trained on a NVIDIA Titan X GPU machine, using the deep learning framework Keras [2] on top of TensorFlow [1].
2.1 Experimental Setup
To evaluate the proposed approach, we used both synthetic and in-vivo experiments. To demonstrate that having a more accurate airway geometric model helps the neural network better measure the structures of interest, we first generated a dataset of 200,000 synthetic patches and we compared the mean relative error (RE) obtained with the proposed MBGR and with the same network trained with a model without the cartilage.
For validation on clinical cases, since a reliable ground-truth cannot be obtained in an accurate way, we performed an indirect validation that leverages a physiological evaluation and demonstrates the clinical relevance of the proposed technique.
We analyzed the associations between the amount of cartilage in bronchi of various orders and the presence of COPD in 547 smoking subjects with and without COPD. To this end, we propose a cartilage index defined by the regression coefficient in a random sample consensus (RANSAC) regression [3] between the airway lumen and cartilage percentage across airway points with a lumen radius \(l_{r} \in [1.19, 3.95]\) mm, similar to what was proposed for the Pi10 calculation [7]. Subjects whose regression fitting was poor (\(R^2<0.8\)) were excluded from the analysis. The total number of subjects after exclusion was 390. An example of regression for one case is shown in Fig. 3a.
3 Results
The mean absolute RE obtained with MBGR for wall thickness, airway lumen, and cartilage percentage across the 200,000 generated patches was 6.19%, 1.73%, and 11.75%, respectively. If the geometric model is generated excluding the cartilage simulation (cartilage percentage set to 0%), the RE was 7.25% for wall thickness and 2.95 for lumen. When considering airways with a wall thickness of 1.0 mm, a mean absolute RE of 6.47% is obtained with the proposed MBGR and 7.93% with the same version without cartilage, whereas when the wall thickness is at the image resolution (0.5 mm) the mean absolute RE was at 12.90% for the proposed method and 13.09% if cartilage is not simulated. These REs are significantly lower than those previously reported in the literature for structures of similar sizes [10, 11].
Figure 3b shows the results of the association between the cartilage index and GOLD disease stage stratified by gender. A multi-factor analysis of variance (ANOVA) shows that both GOLD and gender are statistically significant factors (p < 0.0001 and p \(=\) 0.035 respectively) but not their interaction. A pair Tukey’s t-test analysis between GOLD stages shows that all pairs are statistically significant except GOLD 0–1, 2–3, and 3–4. This suggests a tendency of cartilage loss in moderate to severe COPD that is consistent in both males and females.
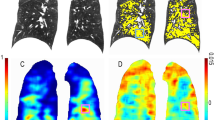
Finally, an example of airway segmentation color-coded by the amount of cartilage for five subjects (one for each GOLD level) is presented in Fig. 4. In this example, it is possible to notice a bigger amount of cartilage (around 50%) in peripheral airways of subjects with no or little COPD (GOLD 0 and 1) while this percentage decreases in subjects with more disease as suggested by our numerical results.
4 Discussion and Conclusion
In this paper, a novel method to automatically assess the amount of cartilage in small airways on chest CT images, while simultaneously measuring lumen radius and the wall thickness is proposed. A generator of a geometric model is used to create synthetic patches that are then automatically refined using a GAN refiner and used to train a regressor of the measures of interest. This is the first time that an automated approach is proposed for the assessment of in-vivo airway cartilage. Our results show that the cartilage area percentage had a small relative error. The indirect validation with subjects with and without COPD showed consistent results with results previously reported in histological studies. These findings indicate that the method here proposed may potentially be used to help physicians toward early diagnosis and prognosis of lung disorders.
Results from the validation on synthetic patches showed a lower absolute relative error for bronchial lumen and wall thickness when the cartilage is included in the geometric model, indicating that creating a more accurate geometric model helps the network better regress the structures of interest.
For future work, we are planning on improving in-vivo validation by comparing the MGBR regression of the three structures to manual measurement, improve the generation of the synthetic model by reducing the level of approximation of the PSF and additive noise, and define a method to properly validate the refining process of the synthetic patches, which is currently done only by visual inspection.
References
Abadi, M., Agarwal, A., Barham, P., et al.: Tensorflow: Large-scale machine learning on heterogeneous distributed systems. arXiv preprint arXiv:1603.04467 (2016)
Chollet, F., et al.: Keras (2015). https://keras.io
Fischler, M.A., Bolles, R.C.: Random sample consensus: a paradigm for model fitting with applications to image analysis and automated cartography. Commun. ACM 24(6), 381–395 (1981)
Hogg, J.C., McDonough, J.E., Suzuki, M.: Small airway obstruction in COPD: new insights based on micro-CT imaging and MRI imaging. CHEST 143(5), 1436–1443 (2013)
Kindlmann, G.L., San José Estépar, R., Smith, S.M., Westin, C.F.: Sampling and visualizing creases with scale-space particles. IEEE TVCG 15(6), 1415–1424 (2009)
LeCun, Y., Bengio, Y., Hinton, G.: Deep learning. Nature 521(7553), 436 (2015)
Nakano, Y., Wong, J.C., de Jong, P.A., et al.: The prediction of small airway dimensions using computed tomography. AJRCCM 171(2), 142–146 (2005)
Nardelli, P., et al.: Accurate measurement of airway morphology on chest CT images. In: Stoyanov, D., et al. (eds.) RAMBO/BIA/TIA -2018. LNCS, vol. 11040, pp. 335–347. Springer, Cham (2018). https://doi.org/10.1007/978-3-030-00946-5_34
Nardelli, P., Ross, J.C., Estépar, R.S.J.: CT image enhancement for feature detection and localization. In: Descoteaux, M., Maier-Hein, L., Franz, A., Jannin, P., Collins, D.L., Duchesne, S. (eds.) MICCAI 2017. LNCS, vol. 10434, pp. 224–232. Springer, Cham (2017). https://doi.org/10.1007/978-3-319-66185-8_26
Reinhardt, J.M., D’Souza, N., Hoffman, E.A.: Accurate measurement of intrathoracic airways. IEEE TMI 16(6), 820–827 (1997)
Estépar, R.S.J., Washko, G.G., Silverman, E.K., Reilly, J.J., Kikinis, R., Westin, C.-F.: Accurate airway wall estimation using phase congruency. In: Larsen, R., Nielsen, M., Sporring, J. (eds.) MICCAI 2006. LNCS, vol. 4191, pp. 125–134. Springer, Heidelberg (2006). https://doi.org/10.1007/11866763_16
Schwab, R.J., Gefter, W.B., Pack, A.I., Hoffman, E.A.: Dynamic imaging of the upper airway during respiration in normal subjects. J. Appl. Physiol. 74(4), 1504–1514 (1993)
Schwarzband, G., Kiryati, N.: The point spread function of spiral CT. Phys. Med. Biol. 50(22), 5307 (2005)
Shrivastava, A., Pfister, T., Tuzel, O., Susskind, J., Wang, W., Webb, R.: Learning from simulated and unsupervised images through adversarial training. In: IEEE CVPR, vol. 3, p. 6 (2017)
Thurlbeck, W., Pun, R., Toth, J., Frazer, R.: Bronchial cartilage in chronic obstructive lung disease. Am. Rev. Respir. Dis. 109(1), 73–80 (1974)
Weibel, E.R., Cournand, A.F., Richards, D.W.: Morphometry of the Human Lung, vol. 1. Springer, Heidelberg (1963). https://doi.org/10.1007/978-3-642-87553-3
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Switzerland AG
About this paper
Cite this paper
Nardelli, P., Washko, G.R., San José Estépar, R. (2019). Bronchial Cartilage Assessment with Model-Based GAN Regressor. In: Shen, D., et al. Medical Image Computing and Computer Assisted Intervention – MICCAI 2019. MICCAI 2019. Lecture Notes in Computer Science(), vol 11769. Springer, Cham. https://doi.org/10.1007/978-3-030-32226-7_40
Download citation
DOI: https://doi.org/10.1007/978-3-030-32226-7_40
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-32225-0
Online ISBN: 978-3-030-32226-7
eBook Packages: Computer ScienceComputer Science (R0)