Abstract
The ability to generate samples of the random effects from their conditional distributions is fundamental for inference in mixed effects models. Random walk Metropolis is widely used to conduct such sampling, but such a method can converge slowly for medium dimension problems, or when the joint structure of the distributions to sample is complex. We propose a Metropolis–Hastings (MH) algorithm based on a multidimensional Gaussian proposal that takes into account the joint conditional distribution of the random effects and does not require any tuning, in contrast with more sophisticated samplers such as the Metropolis Adjusted Langevin Algorithm or the No-U-Turn Sampler that involve costly tuning runs or intensive computation. Indeed, this distribution is automatically obtained thanks to a Laplace approximation of the original model. We show that such approximation is equivalent to linearizing the model in the case of continuous data. Numerical experiments based on real data highlight the very good performances of the proposed method for continuous data model.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Mixed effects models are reference models when the inter-individual variability that can exist within the same population is considered (see [9] and the references therein). Given a population of individuals, the probability distribution of the series of observations for each individual depends on a vector of individual parameters. For complex priors on these individual parameters or models, Monte Carlo methods must be used to approximate the conditional distribution of the individual parameters given the observations. Most often, direct sampling from this conditional distribution is impossible and it is necessary to have resort to a Markov chain Monte Carlo (MCMC) procedure.
Designing a fast mixing sampler is of utmost importance for several tasks in the complex process of model building. The most common MCMC method for nonlinear mixed effects models is the random walk Metropolis algorithm [9, 14, 15]. Despite its simplicity, it has been successfully used in many classical examples of pharmacometry, when the number of random effects is not too large. Nevertheless, maintaining an optimal acceptance rate (advocated in [15]) most often implies very small moves and therefore a very large number of iterations in medium and high dimensions since no information of the geometry of the target distribution is used.
To make better use of this geometry and in order to explore the space faster, the Metropolis-adjusted Langevin algorithm (MALA) uses evaluations of the gradient of the target density for proposing new states which are accepted or rejected using the Metropolis-Hastings algorithm [16, 18]. The No-U-Turn Sampler (NUTS) is an extension of the Hamiltonian Monte Carlo [11] that allows an automatic and optimal selection of some of the settings required by the algorithm, [3]. Nevertheless, these methods may be difficult to use in practice, and are computationally involved, in particular when the structural model is a complex ODE based model.
The algorithm we propose is a Metropolis-Hastings algorithm, but for which the proposal is a good approximation of the target distribution. For general data model (i.e. categorical, count or time-to-event data models or continuous data models), the Laplace approximation of the incomplete pdf \(\texttt {p}(y_i)\) leads to a Gaussian approximation of the conditional distribution \(\texttt {p}(\psi _i |y_i)\).
In the special case of continuous data, linearisation of the model leads, by definition, to a Gaussian linear model for which the conditional distribution of the individual parameter \(\psi _i\) given the data \(y_i\) is a multidimensional normal distribution that can be computed and we fall back on the results of [8].
2 Mixed Effect Models
2.1 Population Approach and Hierarchical Models
We will adopt a population approach in the sequel, where we consider N individuals and \(n_i\) observations for individual i. The set of observed data is \(y = (y_i, 1\le i \le N)\) where \(y_i = (y_{ij}, 1\le j \le n_i)\) are the observations for individual i. For the sake of clarity, we assume that each observation \(y_{ij}\) takes its values in some subset of \(\mathbb {R}\). The distribution of the \(n_i-\)vector of observations \(y_i\) depends on a vector of individual parameters \(\psi _i\) that takes its values in a subset of \(\mathbb {R}^{p}\). We assume that the pairs \((y_i,\psi _i)\) are mutually independent and consider a parametric framework: the joint distribution of \((y_i,\psi _i)\) is denoted by \(\texttt {p}(y_i,\psi _i;\theta )\), where \(\theta \) is the vector of fixed parameters of the model. A natural decomposition of this joint distribution writes \(\texttt {p}(y_i,\psi _i;\theta ) = \texttt {p}(y_i|\psi _i;\theta )\texttt {p}(\psi _i;\theta )\), where \(\texttt {p}(y_i|\psi _i;\theta )\) is the conditional distribution of the observations, given the individual parameters, and where \(\texttt {p}(\psi _i;\theta )\) is the so-called population distribution used to describe the distribution of the individual parameters within the population. A particular case of this general framework consists in describing each individual parameters \(\psi _i\) as a typical value \(\psi _\mathrm{pop}\), and a vector of individual random effects \(\eta _i\): \(\psi _i = \psi _\mathrm{pop}+\eta _i\). In the sequel, we will assume a multivariate Gaussian distribution for the random effects: \(\eta _i \mathop {\sim }_\mathrm{i.i.d.}\mathcal {N}(0,\Omega )\). Several extensions of this model are straightforward, considering for instance transformation of the normal distribution, or adding individual covariates in the model.
2.2 Continuous Data Models
A regression model is used to express the link between continuous observations and individual parameters:
where \(y_{ij}\) is the j-th observation for individual i measured at time \(t_{ij}\), \(\varepsilon _{ij}\) is the residual error, f is the structural model assumed to be a twice differentiable function of \(\psi _i\). We start by assuming that the residual errors are independent and normally distributed with zero-mean and a constant variance \(\sigma ^2\). Let \(t_i=(t_{ij}, 1\le n_i)\) be the vector of observation times for individual i. Then, the model for the observations rewrites \( y_i|\psi _i \sim \mathcal {N}(f_i(\psi _i),\sigma ^2\texttt {Id}_{n_i\times n_i})\;, \) where \(f_i(\psi _i) = (f(t_{i,1},\psi _i), \ldots , f(t_{i,n_i},\psi _i))\). If we assume that \(\psi _i \mathop {\sim }_\mathrm{i.i.d.}\mathcal {N}(\psi _\mathrm{pop},\Omega ) \), then the parameters of the model are \(\theta = (\psi _\mathrm{pop}, \Omega , \sigma ^2)\).
3 Sampling from Conditional Distributions
The conditional distribution \(\texttt {p}(\psi _i | y_i ; \theta )\) plays a crucial role in most methods used for inference in nonlinear mixed effects models.
One of the main task to perform is to compute the maximum likelihood (ML) estimate of \(\theta \), \(\hat{\theta }_\mathrm{ML}= \arg \max \limits _{\theta \in \Theta } \mathcal{L}(\theta , y)\), where \(\mathcal{L}(\theta , y) \triangleq \log \texttt {p}(y;\theta )\). The stochastic approximation version of EM [7] is an iterative procedure for ML estimation that requires to generate one or several realisations of this conditional distribution at each iteration of the algorithm.
Metropolis-Hasting algorithm is a powerful MCMC procedure widely used for sampling from a complex distribution [4]. To simplify the notations, we remove the dependency on \(\theta \). For a given individual i, the MH algorithm, to sample from the conditional distribution \(\texttt {p}(\psi _i | y_i)\), is described in Algorithm 1.

Current implementations of the MCMC algorithm, to which we will compare our new method, in Monolix [5], saemix (R package) [6], nlmefitsa (Matlab) and NONMEM [2] mainly use the same combination of proposals. The first proposal is an independent Metropolis-Hasting algorithm which consists in sampling the candidate state directly from the marginal distribution of the individual parameter \(\psi _i\). The other proposals are component-wise and block-wise random walk procedures [10] that update different components of \(\psi _i\) using univariate and multivariate Gaussian proposal distributions. Nevertheless, those proposals fail to take into account the nonlinear dependence structure of the individual parameters. A way to alleviate these problems is to use a proposal distribution derived from a discretised Langevin diffusion whose drift term is the gradient of the logarithm of the target density leading to the Metropolis Adjusted Langevin Algorithm (MALA) [16, 18]. The MALA proposal is given by:
where \(\gamma \) is a positive stepsize. These methods still do not take into consideration the multidimensional structure of the individual parameters. Recent works include efforts in that direction, such as the Anisotropic MALA for which the covariance matrix of the proposal depends on the gradient of the target measure [1]. The MALA algorithm is a special instance of the Hybrid Monte Carlo (HMC), introduced in [11]; see [4] and the references therein, and consists in augmenting the state space with an auxiliary variable p, known as the velocity in Hamiltonian dynamics.
All those methods aim at finding the proposal q that accelerates the convergence of the chain. Unfortunately they are computationally involved and can be difficult to implement (stepsizes and numerical derivatives need to be tuned and implemented).
We see in the next section how to define a multivariate Gaussian proposal for both continuous and noncontinuous data models, that is easy to implement and that takes into account the multidimensional structure of the individual parameters in order to accelerate the MCMC procedure.
4 A Multivariate Gaussian Proposal
For a given parameter value \(\theta \), the MAP estimate, for individual i, of \(\psi _i\) is the one that maximises the conditional distribution \(\texttt {p}(\psi _i|y_i,\theta )\):
4.1 General Data Models
For both continuous and noncontinuous data models, the goal is to find a simple proposal, a multivariate Gaussian distribution in our case, that approximates the target distribution \(\texttt {p}(\psi _i|y_i)\). In our context, we can write the marginal pdf \(\texttt {p}(y_i)\) that we aim to approximate as \(\texttt {p}(y_i) = \int {e^{\log \texttt {p}(y_i,\psi _i)}\text {d}\psi _i}\). Then, the Taylor expansion of \(\log (\texttt {p}(y_i,\psi _i)\) around the MAP \(\hat{\psi }_i\) (that verifies by definition \(\nabla \log \texttt {p}(y_i,\hat{\psi }_i)=0\)) yields the Laplace approximation of \(-2\log (\texttt {p}(y_i))\) as follows:
We thus obtain the following approximation of \(\log \texttt {p}(\hat{\psi }_i|y_i)\):
which is precisely the log-pdf of a multivariate Gaussian distribution with mean \(\hat{\psi }_i\) and variance-covariance \(-\nabla ^2 \log \texttt {p}(y_i,\hat{\psi }_i)^{-1}\), evaluated at \(\hat{\psi }_i\).
Proposition 1
The Laplace approximation of the conditional distribution \(\psi _i|y_i\) is a multivariate Gaussian distribution with mean \(\hat{\psi }_i\) and variance-covariance
We shall now see another method to derive a Gaussian proposal distribution in the specific case of continuous data models.
4.2 Nonlinear Continuous Data Models
When the model is described by (1), the approximation of the target distribution can be done twofold: either by using the Laplace approximation, as explained above, or by linearizing the structural model \(f_i\) for any individual i of the population. Once the MAP estimate \(\hat{\psi }_i\) has been computed, using an optimisation procedure, the method is based on the linearisation of the structural model f around \(\hat{\psi }_i\):
where \(\text {J}_{f_i(\hat{\psi }_i)}\) is the Jacobian matrix of the vector \(f_i(\hat{\psi }_i)\). Defining \(z_i \triangleq y_i - f_i(\hat{\psi }_i) + \text {J}_{f_i(\hat{\psi }_i)} \hat{\psi }_i\) yields a linear model \(z_i = \text {J}_{f_i(\hat{\psi }_i)}\psi _i + \epsilon _i\) which tractable conditional distribution can be used for approximating \(\texttt {p}(\psi _i|y_i,\theta )\):
Proposition 2
Under this linear model, the conditional distribution \(\psi _i|y_i\) is a Gaussian distribution with mean \(\mu _i\) and variance-covariance \(\Gamma _i\) where
We can note that linearizing the structural model is equivalent to using the Laplace approximation with the expected information matrix. Indeed:
We then use this normal distribution as a proposal in Algorithm 1 for model (1).
5 A Pharmacokinetic Example
5.1 Data and Model
32 healthy volunteers received a 1.5 mg/kg single oral dose of warfarin, an anticoagulant normally used in the prevention of thrombosis [12], for who we measure warfarin plasmatic concentration at different times. We will consider a one-compartment pharmacokinetics (PK) model for oral administration, assuming first-order absorption and linear elimination processes:
where ka is the absorption rate constant, V the volume of distribution, k the elimination rate constant, and D the dose administered. Here, ka, V and k are PK parameters that can change from one individual to another. Then, let \( \psi _i=(ka_i, V_i, k_i)\) be the vector of individual PK parameters for individual i lognormally distributed. We will assume in this example that the residual errors are independent and normally distributed with mean 0 and variance \(\sigma ^2\). We can use the proposal given by Proposition 2 and based on a linearisation of the structural model f proposed in (7). For the method to be easily extended to any structural model, the gradient is calculated by automatic differentiation using the R package ‘Madness’ [13].
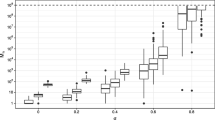
Modelling of the warfarin PK data: convergence of the empirical quantiles of order 0.1, 0.5 and 0.9 of \(\texttt {p}(\psi _i | y_i ; \theta )\) for a single individual. Our new MH algorithm is in red and dotted, the RWM is in black and solid, the MALA is in blue and dashed and the NUTS is in green and dashed
5.2 MCMC Convergence Diagnostic
We will consider one of the 32 individuals for this study and fix \(\theta \) to some arbitrary value, close to the Maximum Likelihood (ML) estimate obtained with SAEM (saemix R package [6]): \(ka_\mathrm{pop} =1\), \(V_\mathrm{pop}= 8\), \(k_\mathrm{pop}=0.01\), \(\omega _{ka}=0.5\), \(\omega _{V}=0.2\), \(\omega _{k}=0.3\) and \(\sigma ^2=0.5\). First, we compare our our nlme-IMH, which is a MH sampler using our new proposal, with the RWM, the MALA, which proposal, at iteration k, is defined by \(\psi _i^c \sim \mathcal {N}(\psi _i^{(k)}-\gamma _k \nabla \log \pi (\psi _i^{(k)}),2\gamma _k)\). The stepsize (\(\gamma = 10^{-2}\)) is constant and is tuned such that the optimal acceptance rate of 0.57 is reached [15]. \(20\,000\) iterations are run for each algorithm. Figure 1 highlights quantiles stabilisation using the MALA similar to our method for all orders and dimensions. The NUTS, implemented in rstan (R Package [17]), is fast and steady and presents similar, or even better convergence behaviors for some quantiles and dimension, than our method (see Fig. 1).
Then, we produce 100 independent runs of the RWM, the IMH using our proposal distribution (called the nlme-IMH algorithm), the MALA and the NUTS for 500 iterations. The boxplots of the samples drawn at a given iteration threshold are presented Fig. 2 against the ground truth (calculated running the NUTS for \(100\,000\) iterations) for the parameter ka.
For the three numbers of iteration considered in Fig. 2, the median of the nlme-IMH and NUTS samples are closer to the ground truth. Figure 2 also highlights that all those methods succeed in sampling from the whole distribution after 500 iterations. Similar comments can be made for the other parameters.
We decided to conduct a comparison between those sampling methods just in terms of number of iterations (one iteration is one transition of the Markov Chain). We acknowledge that the transition cost is not the same for each of those algorithms, though, our nmle-IMH algorithm, except the initialisation step that requires a MAP and a Jacobian computation, has the same iteration cost as RWM. The call to the structural model f being very costly in real applications (when the model is the solution of a complex ODE for instance), the MALA and the NUTS, computing its first order derivatives at each transition, are thus far computationally involved.
Since computational costs per transition are hard to accurately define for each sampling algorithm and since runtime depends on the actual implementation of those methods, comparisons are based on the number of iterations of the chain here.
6 Conclusion and Discussion
We presented in this article an independent Metropolis-Hastings procedure for sampling random effects from their conditional distributions in nonlinear mixed effects models. The numerical experiments that we have conducted show that the proposed sampler converges to the target distribution as fast as state-of-the-art samplers. This good practical behaviour is partly explained by the fact that the conditional mode of the random effects in the linearised model coincides with the conditional mode of the random effects in the original model. Initial experiments embedding this fast and easy-to-implement IMH algorithm within the SAEM algorithm [7], for Maximum Likelihood Estimation, indicate a faster convergence behavior.
References
Allassonniere, S., Kuhn, E.: Convergent stochastic expectation maximization algorithm with efficient sampling in high dimension. Application to Deformable Template Model Estimation. Comput. Stat. Data Anal. 91, 4–19 (2015)
Beal, S., Sheiner, L.: The NONMEM system. Am. Stat. 34(2), 118–119 (1980)
Betancourt, M.: A Conceptual Introduction to Hamiltonian Monte Carlo (2017). arXiv:1701.02434
Brooks, S., Gelman, A., Jones, G., Meng, X.-L.: Handbook of Markov Chain Monte Carlo. CRC Press (2011)
Chan, P.L.S., Jacqmin, P., Lavielle, M., McFadyen, L., Weatherley, B.: The use of the SAEM algorithm in MONOLIX software for estimation of population pharmacokinetic-pharmacodynamic-viral dynamics parameters of maraviroc in asymptomatic HIV subjects. J. Pharmacokinet. Pharmacodyn. 38(1), 41–61 (2011)
Comets, E., Lavenu, A., Lavielle, M.: Parameter estimation in nonlinear mixed effect models using saemix, an R implementation of the SAEM algorithm. J. Stat. Softw. 80(3), 1–42 (2017)
Delyon, B., Lavielle, M., Moulines, E.: Convergence of a stochastic approximation version of the EM algorithm. Ann. Stat. 27(1), 94–128 (1999)
Karimi, B., Lavielle, M., Moulines, E.: Non linear mixed effects models: bridging the gap between independent Metropolis-Hastings and variational inference. ICML 2017 Implicit Models Workshop (2017)
Lavielle, M.: Mixed Effects Models for The Population Approach: Models, Tasks. CRC Press, Methods and Tools (2014)
Metropolis, N., Rosenbluth, A.W., Rosenbluth, M.N., Teller, A.H., Teller, E.: Equation of state calculations by fast computing machines. J. Chem. Phys. 21(6), 1087–1092 (1953)
Neal, R.M.: MCMC using Hamiltonian dynamics. Handbook of Markov Chain Monte Carlo, vol. 2(11) (2011)
O’Reilly, R.A., Aggeler, P.M.: Studies on Coumarin anticoagulant drugs initiation of Warfarin therapy without a lading dose. Circulation 38(1), 169–177 (1968)
Pav, S.E.: Madness: A Package for Multivariate Automatic Differentiation (2016)
Robert, C.P., Casella, G.: Monte Carlo Statistical Methods. Springer Texts in Statistics (2004)
Roberts, G.O., Gelman, A., Gilks, W.R.: Weak convergence and optimal scaling of random walk Metropolis algorithms. Ann. Appl. Probab. 7(1), 110–120 (1997)
Roberts, G.O., Tweedie, R.L.: Exponential convergence of Langevin distributions and their discrete approximations. Bernoulli 2(4), 341–363 (1996)
Stan Development Team: RStan: the R interface to Stan. R Package Version 2.17.3 (2018)
Stramer, O., Tweedie, R.L.: Langevin-type models I: Diffusions with given stationary distributions and their discretizations. Methodol. Comput. Appl. Probab. 1(3), 283–306 (1999)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Switzerland AG
About this paper
Cite this paper
Karimi, B., Lavielle, M. (2019). Efficient Metropolis-Hastings Sampling for Nonlinear Mixed Effects Models. In: Argiento, R., Durante, D., Wade, S. (eds) Bayesian Statistics and New Generations. BAYSM 2018. Springer Proceedings in Mathematics & Statistics, vol 296. Springer, Cham. https://doi.org/10.1007/978-3-030-30611-3_9
Download citation
DOI: https://doi.org/10.1007/978-3-030-30611-3_9
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-30610-6
Online ISBN: 978-3-030-30611-3
eBook Packages: Mathematics and StatisticsMathematics and Statistics (R0)