Abstract
Salvia miltiorrhiza is an important medicinal crop in traditional Chinese medicine (TCM). Knowledge of its genetic foundation is limited. The first genetic linkage map of S. miltiorrhiza was constructed in 94 F1 individuals from an intraspecific cross, consisting of 53 SSR, 38 SRAP, and 2 ISSR loci in eight linkage groups. Another genetic map was constructed based on 111 genomic SSRs markers. Moreover, specific length amplified fragment sequencing (SLAF-seq) is a recently developed high-throughput strategy for large-scale SNP, which was used to construct a high-density genetic map for S. miltiorrhiza. The results not only provide a platform for mapping quantitative trait loci but also offer a critical new tool for S. miltiorrhiza genome and comparative genomics as well as a valuable reference for TCM studies. A total of six QTL have been identified through interval mapping. It includes three salvianolic acid B (SAB) content-related QTL on linkage group LG7, two lithospermic acid (LA) content markers on LG2 and LG4, and one rosmarinic acid (RA) content-linked QTL on LG7. These QTL could be used in marker-assisted breeding for phenolic acid contents of S. miltiorrhiza. The results provide important basis for QTL mapping, map-based cloning, and association studies for commercially important traits in S. miltiorrhiza.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
3.1 Introduction
Salvia miltiorrhiza Bunge, also known as Danshen in Chinese, is a typical herb plant used for traditional Chinese medicine (TCM). This plant has been extensively used for thousands of years to treat various diseases, particularly coronary heart disease and cerebrovascular diseases, in China and Japan and recently in the USA and many European countries (Zhou et al. 2005). Currently, more than 200 compounds have been isolated, of which diterpenoid quinones and hydrophilic phenolic acids are the major constituents (Ma et al. 2015). Moreover, recent studies have found several new bioactivities of Danshen constituents, such as antioxidant, antitumor, and protective effects on the kidney and liver. It suggests the potential for new applications. More than 320 Danshen preparations are produced by different pharmaceutical manufacturers.
There is constantly increasing demand for S. miltiorrhiza because of its varied and diverse pharmacologic properties. Currently, the annual demand for Danshen in China is approximately 10 million kg. Interest in biotechnology research on Danshen is increasing in many research groups. A large number of genes involved in the biosynthesis of phenolics and terpenoids have been identified through either molecular cloning or transcriptome-wide analysis (Gao et al. 2014; Luo et al. 2014; Xu et al. 2015). However, the genetic background and regulators of these two biosynthetic pathways, especially in the later steps, remain unknown. Danshen is of considerable research interest, and its superior genetic characteristics, such as its modest genome size, low number of chromosomes, vitality, short generation cycle, and mature tissue culture technology, have resulted in Danshen becoming an ideal model plant among TCM (Song et al. 2013).
At present, almost all of the Danshen preparations are primarily obtained by extraction from plants. An important and urgent task is to focus on improvements in Danshen for optimizing desirable traits, e.g., effective components, resistance, and yield. According to the literature on S. miltiorrhiza, several molecular markers, including amplified fragment length polymorphisms (AFLPs) (Yang et al. 2012), sequence-related amplified polymorphisms (SRAPs) (Song et al. 2010), EST-simple sequence repeats (SSRs), and inter-SSRs (ISSRs), have been used to analyze the genetic diversity of Danshen. Previous studies have shown that S. miltiorrhiza is a cross-pollinated plant with high differentiation of its germplasm (Song et al. 2010). These results provide an important basis for further construction of genetic maps with the aim of selecting parents and markers. Genetic linkage maps, particularly high-density genetic maps, are valuable tools in meeting the requirement of high-throughput superior trait selection among various germplasms, including plants and animals.
Secondary metabolites are usually synthesized in specific organs or tissues of plants with a relatively low level of contents, which were generally considered as complex agronomic traits, and controlled by multiple genes known as quantitative trait loci (QTL). QTL analysis based on linkage maps is useful for screening closely linked molecular markers of complex traits. It can effectively elucidate the genetic basis of complex agronomic traits, estimate gene actions and genetic parameters, and further accelerate the breeding progress through marker-assistant selection (MAS) in different species (Shehzad and Okuno 2016).
Since the construction of genetic map of Artemisia annua L. and the identification of QTL affecting yields of artemisinin were reported (Graham et al. 2010), progresses have been obtained for various plant species, such as Bupleurum chinense DC., Siraitia grosvenorii, Eucommia ulmoides, and Dendrobium officinale (Zhan et al. 2010; Liu et al. 2011; Li et al. 2015; Lu et al. 2018). Although this opens a new door for genetic breeding researches of medicinal plant, genetic researches about active compound contents of medicinal or aromatic plants only conducted in a few species. The studies on medicinal or aromatic plants are behind far from other crops.
3.2 Molecular Linkage Map Construction
3.2.1 Map Strategy and Mapping Populations
An appropriate mapping population, which generally includes RIL, DH, F2 or backcrossed progeny, is very important in the construction of genetic maps (Grattapaglia and Sederoff 1994). However, similar to most medicinal species, it is very difficult to obtain a typical family-based population in S. miltiorrhiza due to its high heterozygosity resulting from a long history of natural cross-pollination and inbreeding depression. The double pseudo-testcross strategy was first proposed by Grattapaglia and Sederoff (1994) and was successfully applied to construct a genetic map of forest trees. In the pseudo-testcross, an F1 progeny is developed as a mapping population by hybridizing two unrelated and highly heterozygous individuals, where gene segregation patterns can be interpreted as a backcross. This strategy has been widely used in plant species that lack appropriate pedigrees (Myburg et al. 2003; Feng et al. 2013; Sudarshini et al. 2014).
Because of its cross-pollinated habit and lack of basic genetic and breeding study, it is difficult to construct the F2 and RIL populations in S. miltiorrhiza. In many fruit and forest trees with cross-pollination and asexual reproduction, the F1 mapping populations were used for genetic study of interested agronomic traits based on the principle of ‘pseudocrossing’, such as Mangifera indica L. (Luo et al. 2016) and Sesamum indicum L. (Zhang et al. 2013a, b). Because both parents were heterozygous, the genotype among different F1 progeny was different, which provides a feasible method for most medicinal plants. Many medicinal plants have a cross-pollinating habit. There is no doubt that this tendency provides favorable conditions for the selection of mapping parents. If there is a poor genetic basis and the lack of a typical family-based population, the pseudo-testcross is the most promising method for creating genetic maps of medicinal plants at the present time.
Study route for genetic mapping and QTL location of S. miltiorrhiza showed on Fig. 3.1. The first step is to select female and male parents with significant different characteristics, which is the guarantee for the analysis of QTL for important trait. A considerable difference between the parents was required to generate large polymorphic markers. The next step is to create F1 interspecific hybrid population, which usually contains at least 100 F1 lines. The third step is the main part of work, which includes two parallel elements. One is DNA extraction, genotyping by various molecular markers and further genetic map construction. Another is to investigate agronomic traits. The fourth step is association of agronomic traits with genotyping by software to identify the location, number of loci, and genetic effect of QTL. The final aim is to identify molecular markers closely linked to important trait, providing important basis for marker-assisted selection (MAS) and map-based gene cloning.
3.2.2 F1 Genotyping, Data Analysis and Map Construction Method
The marker analysis was carried out according to the isolation types of parents and F1 population. Marker segregation types can be divided into three categories: parental heterozygous markers, maternal-specific markers, and paternal-specific markers, including ab × cd, ef × eg, hk × hk, lm × ll, and nn × np (Fig. 3.2). There are four different amplification bands in the ab × cd segregation type with ab derived from maternal parent and cd from paternal parent, which produce four types including ac, ad, bc, and bd with same ratio of 1:1:1:1 in their F1 population. For the ef × eg type, the “e” amplification band was a common one from two parents, but “f” and “g” were the unique markers from maternal parent and paternal parent, respectively. There will appear four types including ee, eg, fe, and fg in F1 population with the ratio of 1:1:1:1. The two parents have the same bands type in the hk × hk type, and the F1 population show hh:hk:kk with the ratio of 1:2:1. There are two different amplification bands in lm × ll type or nn × np type, with “l” or “n” as a common band of two parents, “m” and “p” as unique band in maternal parent and paternal parent, respectively. Therefore, their F1 population displays two types with same ratio of 1:1.
Markers with blurred or missing data are labeled as “−”. According to different types of markers, the data were analyzed by statistical analysis, and the obtained data were tested for segregation ratios using the Chi-squared (χ2) test. The distorted markers (p < 0.05) were assigned a “*” suffix. JoinMap 4.0 software was employed to establish the genetic linkage map at likelihood of odds (LOD) score at least 3.0 with a cross-pollinator (CP) population type. Map distances were calculated using Kosambi’s mapping function. MapChart 2.2 was used to draw the genetic map (Cui et al. 2015).
3.2.3 Genetic Linkage Map Construction of S. miltiorrhiza
In our group, we selected the lines ZH74, ZH23, and ZH105 as female parents and BH18 as male parent (Fig. 3.3) to create three F1 interspecific hybrid population of S. miltiorrhiza for marker genotyping and the construction of genetic linkage maps using the double pseudo-testcross mapping strategy. A total of four genetic linkage maps were constructed using the above three F1 populations.
3.2.3.1 The First Linkage Genetic Map and Another Map Based on Integration of Multiple Markers
The first genetic linkage map of S. miltiorrhiza was constructed in 94 F1 individuals from crossing ZH74 and BH18 by using simple sequence repeat (SSR), sequence-related amplified polymorphism (SRAP) and inter-simple sequence repeat (ISSR) markers. A total of 93 marker loci in the linkage map, consisting of 53 SSR, 38 SRAP, and 2 ISSR loci, were made up of eight linkage groups, covering a total length of 400.1 cm with an average distance of 4.3 cm per marker. The length of linkage groups varied from 3.3 to 132 cm and each of them included 2–23 markers, separately (Zong et al. 2015).
Another map was constructed in 138 F1 individuals from crossing ZH105 and BH18 based on integration of multiple markers. A set of 503 SSR primers, 100 SRAP primers, and 8 ISSR primers developed from S. miltiorrhiza were screened in the two parents. A total of 340 primers yielded amplification effectively, with 206 primers possess polymorphism. It contained 158 SSR primers, 46 SRAP primers, and 2 ISSR primers with the percentage to be 31.4, 46.0, and 25.0%, respectively. It included 9 linkage groups, contained 111 loci, and covered genome 857.4 cM with a mean marker interval of 7.7 cM. The length of linkage groups varied from 41.6 to 149.7 cM and each of them included 2–27 markers, respectively. Seventeen point one percent partially segregated markers distributed in the map were mainly located on LG1, LG2, and LG3 linkage groups.
3.2.3.2 The Genomic SSR Linkage Genetic Map
Genome SSR was developed by simplified genome sequencing of female parent ZH23. A total of 665 pairs of SSR primers were synthesized and used to analyze the polymorphism between ZH23 and BH18, of which 568 pairs of primers generated clear bands. A total of 138 pairs of genomic SSR primers that amplified 151 loci were polymorphic and stable among the parents and 206 F1 lines. Among 138 pairs of polymorphic SSR primers, a total of five separation types were produced. The separation types of ab × cd, ef × eg, hk × hk, lm × ll, and nn × np involved 1, 13, 0, 48, and 49 primer pairs, respectively. One hundred fifty-one polymorphic loci were subjected to the χ2 test to determine if they met the expected segregation ratio given by the JoinMap4.0 software program. The LOD value was selected to 3.0, and finally, 111 SSR markers were used to construct the genetic linkage map.
The second genetic map of S. miltiorrhiza was constructed based on genomic SSRs. Simple sequence repeat (SSR) is one of the most popular and versatile marker type and is considered as an ideal co-dominant marker for plant genetic mapping (Morgantae and Olivieri 2010). It has several prior characteristics, such as neutrality, abundance and polymorphism, high information content, and mostly single-gene inheritance. Totally, 111 SSR markers were finally assigned into eight linkage groups, which corresponded to the haploid chromosome number of S. miltiorrhiza (2n = 16). The genetic distance spanned 397.5 cM in this maps accounting for about 92.7% of the entire genome of S. miltiorrhiza. The length of individual linkage group ranged from 24.8 cM (LG4) to 102.0 cM (LG1) with the number of markers on each group ranged from 6 to 24. Average distance between markers was 3.6 cM, with intervals between loci ranged from 1.8 to 9.7 cM (Feng et al. 2019).
3.2.3.3 The High-Density Linkage Genetic Map
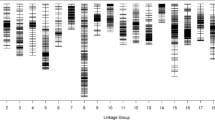
To construct a high-density genetic map, specific length amplified fragment sequencing (SLAF-seq) was used for the rapid discovery of SNPs in the F1 population. Analysis of the F1 mapping population indicated that 83,154 SLAFs were generated, with an average depth of 6.17-fold for each offspring. Among the 151,035 high-quality SLAFs, 62,834 were polymorphic, resulting in a polymorphism rate of 41.60%. Of the 62,834 polymorphic SLAFs, 47,701 were classified into eight segregation patterns. For the F1 population, five segregation patterns, including ab × cd, ef × eg, hk × hk, lm × ll, and nn × np, were used for genetic map construction, and a total of 5198 SLAFs were used for map construction (Fig. 3.4) (Liu et al. 2016).
Then, the high-density genetic map was constructed. After completing the data preparation, 5164 of the 5198 SLAFs were mapped onto the genetic map (i.e., a ratio of 99.34%). There were a total of 2966 BH18 (male) markers, 3038 ZH74 (female) markers, and 5164 SLAFs (7554 SNPs), which fell into 8 LGs, for the integrated map. The coverage of the markers was 75.67-fold in the female parent, 91.20-fold in the male parent, and 10.36-fold in each F1 individual (on average). The final map was 1516.43 cM in length, with an average inter-marker distance of 0.29 cM (Table 3.1). According to the formula for genetic linkage maps, the coverage ratio of the total length to the expected length was 99.83%, which is considerably stronger compared to the value of 84.4% obtained for the first map. The current linkage map covers nearly the entire genome with a resolution of 0.29 cM (Liu et al. 2016).
3.3 Quantitative Trait Loci Analysis of Important Agronomic Traits in S. miltiorrhiza
3.3.1 Quantitative Trait Loci (QTL) Mapping Method
Quantitative trait loci (QTL) mapping was performed using the MapQTL 6.0 software based on the interval mapping (IM) algorithm with a significance level of 0.05, 1000 times of permutation and walking speed of 1 cM. Quantitative trait loci (QTL) identified in this way were marked by the marker with the highest LOD score in the corresponding QTL region.
3.3.2 QTL Analysis for Phenolic Acid Content
The main peaks in the chromatograms of rosmarinic acid (RA), salvianolic acid (SAB), and lithospermic acid (LA) were identified by HPLC, based on their retention time and UV absorptions. Measurement of three phenolic acid contents showed significant differences between two parents. The variation coefficients of these traits varied from 33.64% (SAB content) to 53.03% (RA content), which showed a significant variation among F1 individuals. Skewness and peakedness test indicated that these traits were segregated continuously and controlled by multiple genes, suggesting their suitability for QTL analysis.
Six QTL were identified for three phenolic acids content traits with phenotypic variance explained from 10.8 to 15.9%. Among them, 4 QTL was anchored on LG7, 1 QTL on LG2, and 1 QTL on LG4, respectively. Three QTL for SAB content were detected in a continuous interval between DSSR-540 and DSSR-98 markers on LG7 with a length of 27.2 cM (Fig. 3.5), in which, three continuous QTL explained 13.8, 12.9, and 13.6% of the phenotypic variation, respectively. Two QTL for LA content were located on LG2 and LG4, explained 10.8 and 15.9% of the phenotypic variation, respectively. These two QTL were localized with a single marker DSSR-140 and DSSR-77, respectively. Only one QTL for RA content was detected on LG7, which accounted for 12.4% of the phenotypic variation, and this locus was in the same region of QTL for SAB content.
Genetic map of the F1 population (SNDS1 × BH18) based on the simple sequence repeat (SSR) markers and distribution of quantitative trait loci (QTL) effecting phenolic acid concentration. Genetic linkage map of S. miltiorrhiza presented as eight pseudo-chromosomes. The names of loci are shown on the right, and the positions of the loci are shown, in Kosambi centiMorgan (cM), on the left. QTL are shown on the light blue boxes on LG2 and LG4 for two loci of lithospermic acid content. The light red box showed an important cluster consisting of four QTL, one for rosmarinic acid and three for salvianolic acid B content
3.3.3 QTL Analysis for Root Yield
Various characters in ZH105/BH18 F1 generation groups showed continuous distribution of the root traits of S. miltiorrhiza. These characters conformed the quantitative trait loci for genetic and normal distribution. Twenty-six loci were detected in the ten phenotypic traits of S. miltiorrhiza by the interval mapping method. Root diameter, root range, root fresh weight, and root number were detected QTL loci, explaining the phenotypic variation rate raged from 8.0 to 13.7%.
3.4 Discussion
Most of the genetic maps constructed in medicinal plants mainly use the universal markers (ISSR, SRAP, and AFLP), which indicates that the related genetic research of medicinal plants is still in its infancy, far behind that of food crops and other crops. The genetic map of Danshen, even though it is an emerging model medicinal plant, only recently begun to be constructed. The first map, which was constructed using SRAPs, ISSRs, and SSRs in the F1 population, includes 94 loci with an average interval distance of 4.3 cM. However, this unsaturated genetic map has limited future application. SNPs are the most abundant and stable form of genetic variation in most genomes and have become the marker type of choice in many genetic studies. Subsequently, a high-density genetic map of S. miltiorrhiza was constructed, which contained 5164 high-quality SLAFs and spanned 1516.43 cM, with an average marker interval of 0.29 cM. Due to different parents and different population, the genetic maps usually have poor versatility. In integration, the genomic SSR was essential because of the characteristics of co-dominance, easy to score, simple banding, and convenient identification (Wu et al. 2016; McCallum et al. 2016). Species-specific SSR markers need to be further developed and widely used in TCM. The methods used in this study for genetic mapping and for the development of markers provide a valuable reference for other medicinal plants.
Quantitative trait loci analysis based on linkage map can effectively elucidate the genetic basis of complex agronomic traits (Ma et al. 2014). A large number of QTL have been mapped in many crops related to various important agronomic traits, such as quantity (Tharanya et al. 2018), quality (Vallejo et al. 2015), plant adaptation and tolerance to stresses (Pilet-nayel et al. 2017), and some of which had been successfully utilized in breeding programs (Singh and Miklas 2015).
Phenolic acids are one of the main active components in S. miltiorrhiza root. Thus, improvement of phenolic acid contents has been an important objective of its genetic and breeding research. In our group, QTL analyses of phenolic acid contents in S. miltiorrhiza were firstly reported. It opened a new window for future MAS breeding of phenolic acids and identification of genes associated with phenolic acids biosynthesis.
Quantitative trait loci for correlated traits often position on the same chromosomal regions. Consistently, QTL for rosmarinic acid (RA) and three QTL for salvianolic acid (SAB) content were located within the same region. This region can be considered as further target region for fine mapping and marker-assisted selection in S. miltiorrhiza breeding programs. Besides, SAB content also had a significantly high correlation with the other two secondary compounds contents. Selection of SAB content will have positive effects on other traits. The same phenomenon of overlaps in QTL regions was also reported in many plant species, such as Pyrus L. (Zhang et al. 2013a, b), Glycine max L. Merr. (Akond et al. 2014), and Cynodon dactylon var. dactylon (Guo et al. 2017a, b). The above findings may be due to two genes controlling different traits. These QTL are likely to be either within the same region or in adjacent locations on a chromosome or a single gene tended to have pleiotropic effect (Zhang et al. 2013a, b). A limited number of QTL for phenolic acid contents were detected due to the genetic maps with low density. Improvement in the marker density would increase the identification of QTL number. It is suggested that the density of the genetic map is a key to perform QTL fine mapping of interested agronomic traits.
In future breeding of S. miltiorrhiza, attention should be paid to the continuous recording and dynamic analysis of agronomic traits for many years. It will greatly improve the predictability and accuracy of the selection of good genotypes for quantitative traits and will effectively improve the MAS efficiency. In addition, expanding mapping population, increasing the number of markers in genetic map, constructing ultra-high density map, and integrating and locating physical map can effectively improve the establishment of closely linked QTL and lay an important foundation for molecular breeding of S. miltiorrhiza.
References
Akond M, Liu SM, Boney M, Kantartzi SK, Meksem K, Bellaloui N, Lightfoot DA, Kassem MA (2014) Identification of quantitative trait loci (QTL) underlying protein, oil, and five major fatty acids contents in soybean. Am J Plant Sci 5(1):158–167
Cui JJ, Cheng JW, Wang GP, Tang X, Wu ZM, Lin MB, Li LF, Hu KL (2015) QTL analysis of three flower-related traits based on an interspecific genetic map of Luffa. Euphytica 202(1):45–54
Feng YY, Guo LL, Jin H, Lin CC, Zhou CH, Fang XS, Wang JH, Song ZQ (2019) Quantitative trait loci analysis of phenolic acids contents in Salvia miltiorrhiza based on genomic simple sequence repeat markers. Industrial crops and products 133:365–372
Feng S, Zhao H, Lu J, Liu J, Shen B, Wang H (2013) Preliminary genetic linkage maps of chinese herb Dendrobium nobile and D. moniliforme. J Genet 92(2):205–212
Gao W, Sun HX, Xiao H, Cui G, Hillwig ML, Jackson A, Wang X, ShenY Zhao N, Zhang LX, Wang XJ, Peters RJ, Huang LQ (2014) Combining metabolomics and transcriptomics to characterize tanshinone biosynthesis in Salvia miltiorrhiza. BMC Genom 15(1):1–14
Graham IA, Besser K, Blumer S, Braniga, CA, Czechowski T, Elias L, Guterman I, Harvey D, Isaac PG, Khan AM, Larson TR, Li Y, Pawson T, Penfield T, Rae AM, Rathbone DA, Reid S, Ross J, Smallwood MF, Segura V, Townsend T, Vyas D, Winzer T, Bowles D (2010) The genetic map of Artemisia annua L. identifies loci affecting yield of the antimalarial drug artemisinin. Science 327(5963):328–331
Grattapaglia D, Sederoff R (1994) Genetic linkage maps of Eucalyptus grandis and Eucalyptus urophylla using a pseudo-testcross: mapping strategy and RAPD markers. Genetics 137:1121–1137
Guo Q, Guo LL, Zhang LX, Zhang LX, Ma HL, Guo DL, Hou XG (2017a) Construction of a genetic linkage map in tree peony (Paeonia Sect. Moutan) using simple sequence repeat (SSR) markers. Sci Hortic 219:294–301
Guo Y, Wu Y, Anderson JA, Moss JQ, Zhu L, Fu J (2017b) SSR marker development, linkage mapping, and QTL analysis for establishment rate in common Bermuda grass. Plant Genome 10(1):1–11
Li Y, Wei JK, Li ZQ, Lei AN, Liu MH (2015) Quantitative trait locus analysis of leaf morphological characters, yield-related traits, and secondary metabolite contents in Eucommia ulmoides. Genet Mol Res 14(4):17871–17884
Liu LH, Ma XJ, Wei JH, Qin JM, Mo CM (2011) The first genetic linkage map of Luohanguo (Siraitia grosvenorii) based on ISSR and SRAP markers. Genome 54(1):19–25
Liu T, Guo LL, Pan YL, Zhao Q, Wang JH, Song ZQ (2016) Construction of the first high-density genetic linkage map of Salvia miltiorrhiza using specific length amplified fragment (SLAF) sequencing. Sci Rep 6:24070
Lu JJ, Liu YY, Xu J, Mei ZW, Shi YJ, Liu PL, He JB, Wang XT, Meng YJ, Feng SG, Shen CJ, Wang HZ (2018) High-density genetic map construction and stem total polysaccharide content-related QTL exploration for Chinese Endemic Dendrobium (Orchidaceae). Front Plant Sci 9:398
Luo C, Shu B, Yao QS, Wu HX, Xu WT, Wang SB (2016) Construction of a high-density genetic map based on large-scale marker development in mango using specific-locus amplified fragment sequencing (SLAF-seq). Front Plant Sci 7:1310
Luo H, Zhu Y, Song J, Xu L, Sun C, Zhang X, Xu Y, He L, Sun W, Xu H, Wang B, Li X, Li C, Liu J, Chen S (2014) Transcriptional data mining of Salvia miltiorrhiza in response to methyl jasmonate to examine the mechanism of bioactive compound biosynthesis and regulation. Physiol Plant 152(2):241–255
Ma JQ, Yao MZ, Ma CL, Wang XC, Jin JQ, Wang XM, Chen L (2014) Construction of a SSR-based genetic map and identification of QTLs for catechins content in tea plant (Camellia sinensis). Plos One 9(3):e93131
Ma PD, Liu JY, Osbourn A, Donga J, Liang ZS (2015) Regulation and metabolic engineering of tanshinone biosynthesis. RSC Adv 5:18137–18144
Mccallum S, Graham J, Jorgensen L, Rowland LJ, Bassil NV, Hancock JF, Wheeler EJ, Vining K, Poland JA, Olmstead JW, Buck E, Wiedow C, Jackson E, Brown A, Hackett CA (2016) Construction of a SNP and SSR linkage map in autotetraploid blueberry using genotyping by sequencing. Mol Breed 36(4):1–24
Morgantae M, Olivieri AM (2010) PCR-amplified microsatellites as markers in plant genetics. Plant J 3(1):175–182
Myburg AA, Griffin AR, Sederoff RR, Whetten RW (2003) Comparative genetic linkage maps of Eucalyptus grandis, Eucalyptus globulus and their F1 hybrid based on a double pseudo-back cross mapping approach. Theor Appl Genet 107:1028–1042
Pilet-Nayel ML, Moury B, Caffier V, Montarry J, Kerlan MC, Fournet S, Durel CE, Delourme R (2017) Quantitative resistance to plant pathogens in pyramiding strategies for durable crop protection. Front Plant Sci 8:1838
Shehzad T, Okuno K (2016) QTL mapping for yield and yield-contributing traits in sorghum (Sorghum bicolor (L.) Moench) with genome-based SSR markers. Euphytica 203(1):1–15
Singh SP, Miklas PN (2015) Breeding common bean for resistance to common blight: a review. Crop Sci 55(3):971
Song JY, Luo HM, Li CF, Sun C, Chen SL (2013) Salvia miltiorrhiza as medicinal model plant. Yao Xue Xue Bao 48(7):1099–1106
Song ZQ, Li XF, Wang HG, Wang JH (2010) Genetic diversity and population structure of Salvia miltiorrhiza Bge in China revealed by ISSR and SRAP. Genetica 138:241–249
Sudarshini KV, Padmakar B, Patil MS, Lakshman R, Aswath C (2014) The genetic linkage maps of Anthurium species based on RAPD, ISSR and SRAP marker. Sci Hortic 178:132–137
Tharanya M, Kholova J, Sivasakthi K, Seghal D, Hash CT, Raj B, Srivastav, RK, Baddam R, Thirunalasundari T, Yadav R, Vadez V (2018) Quantitative trait loci (QTL) for water use and crop production traits co-locate with major QTL for tolerance to water deficit in a fine-mapping population of pearl millet (Pennisetum glaucum L.R.Br.). Theor Appl Genet 131(7):1509–1529
Vallejo VA, Tychonievich J, Lin WK, Wangchu L, Barry CS, Warner RM (2015) Identification of QTL for crop timing and quality traits in an interspecific Petunia population. Mol Breed 35(1):2
Wu QH, Chen YX, Lin F, Zhou SH, Chen JJ, Zhao XJ, Zhang D, Ouyang SH, Wang ZZ, Li D, Wang GX, Zhang DY, Yuan CG, Wang LX, You MS, Han J, Liu ZY (2016) QTL mapping of flag leaf traits in common wheat using an integrated high-density SSR and SNP genetic linkage map. Euphytica 208(2):337–351
Xu Z, Peters RJ, Weirather J, Luo H, Liao B, Zhang X, Zhu Y, Ji A, Zhang B, Hu S, Au KF, Song J, Chen S (2015) Full-length transcriptome sequences and splice variants obtained by a combination of sequencing platforms applied to different root tissues of Salvia miltiorrhiza and tanshinone biosynthesis. Plant J 82:951–961
Yang D, Ma P, Liang X, Liang Z, Zhang M, Shen S, Liu H, Liu Y (2012) Metabolic profiles and cDNA-AFLP analysis of Salvia miltiorrhiza and Salvia castanea Diel f. tomentosa Stib. PLoS One 7:e29678
Zhan QQ, Sui C, Wei JH, Fan SC, Zhang J (2010) Construction of genetic linkage map of Bupleurum Chinense DC. Using ISSR and SSR markers. Yao Xue Xue Bao 45(4):517–529
Zhang RP, Wu J, Li XG, Khan, MA, Chen H, Korban SS, Zhang SL (2013a) An AFLP, SRAP, and SSR genetic linkage map and identification of QTL for fruit traits in Pear (Pyrus L.). Plant Mol Biol Rep 31(3):678–687
Zhang Y, Wang L, Xin H, Li D, Ma C, Ding X, Hong W (2013b) Construction of a high-density genetic map for sesame based on large scale marker development by specific length amplified fragment (SLAF) sequencing. BMC Plant Biol 13(1):141
Zhou L, Zuo Z, Chow MSS (2005) Danshen: an overview of its chemistry, pharmacology, pharmacokinetics, and clinical use. J Clin Pharmacol 45:1345–1359
Zong CK, Song ZQ, Chen HM, Liu C, Wang JH, Guo LL, Liu T, Pan YL (2015) Construction of the first genetic linkage map of Salvia miltiorrhiza Bge. Using SSR, SRAP and ISSR markers. Yao Xue Xue Bao 50(3):360–366
Acknowledgements
This work was supported by funds from the National Natural Science Foundation of China (81872949), the Natural Science Foundation of Shandong province of China (ZR2019HM081), and the key research and development plan of Shandong Province (2017CXGC1302).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Switzerland AG
About this chapter
Cite this chapter
Li, X., Wang, J., Song, Z. (2019). Molecular Maps and Mapping of Genes and QTLs of Salvia miltiorrhiza. In: Lu, S. (eds) The Salvia miltiorrhiza Genome. Compendium of Plant Genomes. Springer, Cham. https://doi.org/10.1007/978-3-030-24716-4_3
Download citation
DOI: https://doi.org/10.1007/978-3-030-24716-4_3
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-24715-7
Online ISBN: 978-3-030-24716-4
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)