Abstract
Deep convolutional networks (ConvNets) have achieved the state-of-the-art performance and become the de facto standard for solving a wide variety of medical image analysis tasks. However, the learned models tend to present degraded performance when being applied to a new target domain, which is different from the source domain where the model is trained on. This chapter presents unsupervised domain adaptation methods using adversarial learning, to generalize the ConvNets for medical image segmentation tasks. Specifically, we present solutions from two different perspectives, i.e., feature-level adaptation and pixel-level adaptation. The first is to utilize feature alignment in latent space, and has been applied to cross-modality (MRI/CT) cardiac image segmentation. The second is to use image-to-image transformation in appearance space, and has been applied to cross-cohort X-ray images for lung segmentation. Experimental results have validated the effectiveness of these unsupervised domain adaptation methods with promising performance on the challenging task.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
1 Introduction
Deep convolutional networks (ConvNets) have made wide success in a variety of automatic medical image analysis tasks, such as anatomical structure segmentation [1, 2], lesion detection [3, 4], cancer diagnosis [5, 6], attributing to the network’s learned highly representative features. In typical practice, the deep ConvNets are trained and tested on datasets where all the images come from the same dataset, i.e., samples are drawn from the same data distribution. However, it has been frequently observed that domain shift can bring about performance degradation. The ConvNets tend to present poor results when being applied to new target data, which are acquired using different protocols, scanners, or modalities [7, 8]. It is crucial to close the performance gap, for large-scale study or deployment of deep learning models in real-world clinical practice.
Domain adaptation has been a long-standing topic in machine learning. It is a very common challenge to investigate the generalization capability of the learning systems. In medical imaging, some traditional automatic methods also suffer from similar poor generalization problem. For example, Philipsen et al. [9] have studied the influence of data distribution variations across chest radiography datasets on segmentation methods based on k-nearest neighbor classification and active shape modeling. In recent years, the study of adapting ConvNets have gradually attracted more attention. In the concept of domain adaptation, the domain of labeled training data is termed as source domain, and the unseen test data is termed as target domain. One straightforward solution is transfer learning, i.e., fine-tuning the ConvNets learned on source domain with extra labeled data from the target domain. Remarkably, Ghafoorian et al. [7] investigated on the number of fine-tuned layers to reduce the required amount of annotations for brain lesion segmentation across MRI datasets. However, the way of supervised transfer learning (STL) still relies on extra labeled data, which is quite expensive or sometimes even infeasible to obtain in the medical field.
Instead, the unsupervised domain adaptation (UDA) methods are more appealing and feasible, since these scenarios transfer knowledge across domains without using additional target domain labels. Generally speaking, existing literatures tackle the unsupervised domain adaptation task based on adversarial learning [10] from two directions: (1) feature-level adaptation with latent space alignment; (2) pixel-level adaptation with image-to-image translation. More specifically, for feature-level adaptation, the source and target inputs are mapped into a shared latent feature space, such that a classifier learned based on this common space can work for both domains. For pixel-level adaptation, the images from target domain are transformed into the appearance of source domain, such that ConvNets trained on source domain can be used for target images, or vice versa. Detailed literatures within these two solution directions are described in the next section.
Illustration of performance degradation of deep learning models on medical images. a ConvNet trained on source chest X-ray images can perform well on source data (left) but get poor results on unseen target data (right). b ConvNet trained on cardiac MRI images (left) receives a complete failure when tested on cardiac CT images (right)
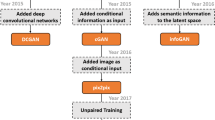
In this chapter, we focus on demonstrating how to conduct unsupervised domain adaptation of ConvNets on medical image segmentation tasks, with two case studies as illustrated in Fig. 5.1. One is using feature space alignment for adapting ConvNets between different modalities of images (i.e., CT and MRI) for cardiac segmentation. The other is employing pixel space transformation for adapting ConvNets between different cohorts of chest X-ray images for lung segmentation. Our works related to this chapter have been published in [11, 12].
2 Related Works
Domain adaptation aims to recover the performance degradation caused by any distribution change occurred after learning a classifier. For deep learning models, this situation also applies, and it has been an active and fruitful research topic in recent investigations of deep neural networks. In this section, we review the literatures of unsupervised domain adaptation methods proposed from two different perspectives, i.e., feature-level adaptation and pixel-level adaptation.
2.1 Feature-Level Adaptation
One group of prior studies on unsupervised domain adaptation focused on aligning the distributions between domains in the feature space, by minimizing measures of distance between features extracted from the source and target domains. Pioneer works tried to minimize the distance between domain statistics. For example, the maximum mean discrepancy (MMD) was minimized together with a task-specific loss to learn the domain-invariant and semantic-meaningful features in [13, 14]. The correlations of layer activations between the domains were aligned in the study of [15]. Later on, [16] pioneered adversarial feature adaptation where a domain discriminator aims to classify the source and target representations while a feature generator competes with the discriminator to produce domain-invariant features. The [17] introduced a more flexible adversarial learning method with untied weight sharing, which helps effective learning in the presence of larger domain shifts. Recent studies [18, 19] proposed to apply the adversarial learning in other lower dimensional spaces instead of the high-dimensional feature space for more effective feature alignment.
Effectiveness of the adversarial framework for feature adaptation has also been validated in medical applications. Kamnitsas et al. [20] made the earliest attempt to align feature distributions in cross-protocol MRI images with adversarial loss. The adversarial framework was further extended to cross-modality cardiac segmentation in [11, 21]. Most recently, the adversarial loss was combined with a shape prior to improve domain adaptation performance for left atrium segmentation across ultrasound datasets [22]. In [23], the adaptation for whole-slide images was achieved through the adversarial training between domains along with a Siamese architecture on the target domain to add a regularization. Dong et al. [24] discriminated segmentation predictions of the heart on both source and target X-rays from those ground truth masks, based on the assumption that segmentation masks should be domain independent. Zhang et al. [25] proposed multi-view adversarial training for dataset-invariant left and right-ventricular coverage estimation in cardiac MRI.
2.2 Pixel-Level Adaptation
With the success of generative adversarial networks (GANs) [10] and its powerful extensions such as CycleGAN [26] for producing realistic images, there exists lines of researches performing adaptation in pixel-level through image-to-image transformation. Some methods first trained a ConvNet in source domain, and then transformed the target images into source-like ones, such that the transformed image can be tested using the pretrained source model [12, 27, 28]. Inversely, other methods tried to transform the source images into the appearance of target images [29,30,31]. The transformed target-like images were then used to train a task model which could perform well in the target domain. For pixel-level adaptation, it is important that the structural contents of original images are well preserved in the generated images. For example, Shrivastava et al. [29] used an L1 reconstruction loss to ensure the contents similarity between the generated target images and original images. Bousmalis et al. [30] proposed a content similarity loss to force the generated image to preserve original contents.
In the field of medical image analysis using deep learning, pixel-level domain adaptation has been more and more frequently explored to generalize learned models across domains. Zhao et al. [32] combined the annotated vessel structures with target image style to generate target-like retinal fundus data, then used the synthetic dataset to train a target domain model. Some CycleGAN-based methods have been proposed to tackle the cross-cohort or cross-modality domain shift. For the X-ray segmentation, both [12, 28] translated target X-ray images to resemble the source images, and directly applied the established source model to segment the generated source-like images. In [33], a two-stage approach was proposed to first translate CT images to appear like MRI using CycleGAN, and then used both generated MRI and a few real MRI for semi-supervised tumor segmentation. In [34], an end-to-end synthetic segmentation network was applied for MRI and CT images adaptation, which combined CycleGAN with a segmentation network.
3 Feature-Level Adaptation with Latent Space Alignment
In this section, we present a feature-level unsupervised domain adaptation framework with adversarial learning, applied to cross-modality cardiac image segmentations. To transfer the established ConvNet from source domain (MRI) to target domain (CT), we design a plug-and-play domain adaptation module (DAM) which implicitly maps the target input data to the feature space of source domain. Furthermore, we construct a discriminator which is also a ConvNet termed as domain critic module (DCM) to differentiate the feature distributions of two domains. Adversarial loss is derived to train the entire domain adaptation framework in an unsupervised manner, by placing the DAM and DCM into a minimax two-player game. Figure 5.2 presents overview of our method. The details of network architecture, adaptation method, adversarial loss, training strategies, and experimental results are elaborated in the followings.
3.1 Method
3.1.1 ConvNet Segmenter Architecture
Given a set of \(N^s\) labeled samples \(\{x_i^s,y_i^s\}_{i=1}^{N^s}\) from the source domain \(X^s\), we conduct supervised learning to establish a mapping from the input image to the label space \(Y^s\). In our setting, the \(x_i^s\) represents the sample (pixel or patch) of medical images and \(y_i^s\) is the category of anatomical structures. For the ease of denotation, we omit the index i in the following, and directly use \(x^s\) and \(y^s\) to represent the samples and labels from the source domain.
A segmentation ConvNet is established to implicitly learn the mapping \(M^s\) from input to the label space. The backbone of our segmenter is residual network for pixel-wise prediction of biomedical images. We employ the dilated residual blocks [35] to extract representative features from a large receptive field while preserving the spatial acuity of feature maps. This is for the considerations of our network design for feature space alignment, because short cut connections are not expected in our model. More specifically, the image is first input to a Conv layer, then forwarded to three residual modules (termed as RM, each consisting of two stacked residual blocks) and downsampled by a factor of 8. Next, another three RMs and one dilated RM are stacked to form a deep network. To enlarge receptive field for extracting global semantic features, four dilated convolutional layers are used in RM7 with a dilation factor of 2. For dense predictions in our segmentation task, we conduct upsamling at layer Conv10, which is followed by \(5 \! \times \! 5\) convolutions to smooth out the feature maps. Finally, a softmax layer is used for probability predictions of the pixels.
The segmentation ConvNet is optimized with labeled data from the source domain by minimizing the hybrid loss \(\mathcal {L}_\text {seg}\) composed of the multi-class cross-entropy loss and the Dice coefficient loss [36]. Formally, we denote \(y_{i,c}^s\) for binary label regarding class \(c \! \in \! C\) in sample \(x_i^s\), its probability prediction is \(\hat{p}_{i,c}^s\), and the label prediction is \(\hat{y}_{i,c}^s\), the source domain segmenter loss function is as follows:
where the first term is the cross-entropy loss for pixel-wise classification, with \(w_{c}^s\) being a weighting factor to cope with the issue of class imbalance. The second term is the Dice loss for multiple cardiac structures, which is commonly employed in biomedical image segmentation problems. We combine the two complementary loss functions to tackle the challenging cardiac segmentation task. In practice, we also tried to use only one type of loss, but the performance was not quite high.
3.1.2 Plug-and-Play Domain Adaptation Module
After obtaining the ConvNet learned on the source domain, our goal is to generalize it to a target domain. In transfer learning, the last several layers of the network are usually fine-tuned for a new task with new label space. The supporting assumption is that early layers in the network extract low-level features (such as edge filters and color blobs) which are common for vision tasks. Those upper layers are more task-specific and learn high-level features for the classifier [37, 38]. In this case, labeled data from target domain are required to supervise the learning process. Differently, we use unlabeled data from the target domain, given that labeling dataset is time consuming and expensive. This is critical in clinical practice where radiologists are willing to perform image computing on cross-modality data with as less extra annotation cost as possible. Hence, we propose to adapt the ConvNet with unsupervised learning.
In our segmenter, the source domain mapping \(M^s\) is layer-wise feature extractors composing stacked transformations of \(\{M^s_{l_1},\ldots ,M^s_{l_n}\}\), with the l denoting the network layer index. Formally, the predictions of labels are obtained by
For domain adaptation, the source and target domains share the same label space, i.e., we segment the same anatomical structures from medical MRI/CT data. Our hypothesis is that the distribution changes between the cross-modality domains are primarily low-level characteristics (e.g., gray scale values) rather than high-level (e.g., geometric structures). The higher layers (such as \(M^s_{l_n}\)) are closely in correlation with the class labels which can be shared across different domains. In this regard, we propose to reuse the feature extractors learned in higher layers of the ConvNet, whereas the earlier layers are updated to conduct distribution mappings in feature space for our unsupervised domain adaptation.
To perform segmentation on target images \(x^t\), we propose a domain adaptation module \(\mathcal {M}\) that maps \(x^t\) to the feature space of the source domain. We denote the adaptation depth by d, i.e., the layers earlier than and including \(l_d\) are replaced by DAM when processing the target domain images. In the meanwhile, the source model’s upper layers are frozen during domain adaptation learning and reused for target inference. Formally, the predictions for target domain is
where \(\mathcal {M}(x^t) = \mathcal {M}_{l_1:l_d}(x^t) = \mathcal {M}_{l_d} \circ \cdots \circ \mathcal {M}_{l_1} (x^t)\) represents the DAM which is also a stacked ConvNet. Overall, we form a flexible plug-and-play domain adaptation framework. During the test inference, the DAM directly replaces the early d layers of the model trained on source domain. The images of target domain are processed and mapped to deep learning feature space of source domain via the DAM. These adapted features are robust to the cross-modality domain shift, and can be mapped to the label space using those high-level layers established on source domain. In practice, the ConvNet configuration of the DAM is identical to \(\{M^s_{l_1},\ldots ,M^s_{l_d}\}\). We initialize the DAM with trained source domain model and fine-tune the parameters in an unsupervised manner with adversarial loss.
3.1.3 Learning with Adversarial Loss
We propose to employ adversarial loss to train our domain adaptation framework in an unsupervised manner. The spirit of adversarial training roots in GAN, where a generator model and a discriminator model form a minimax two-player game. The generator learns to capture the real data distribution; and the discriminator estimates the probability that a sample comes from the real training data rather than the generated data. These two models are alternatively optimized and compete with each other, until the generator can produce real-like samples that the discriminator fails to differentiate. For our problem, we train the DAM, aiming that the ConvNet can generate source-like feature maps from target input. Hence, the ConvNet is equivalent to a generator from GAN’s perspective.
Considering that accurate segmentations come from high-level semantic features, which in turn rely on fine patterns extracted by early layers, we propose to align multiple levels of feature maps between source and target domains (see Fig. 5.2). In practice, we select several layers from the frozen higher layers, and refer their corresponding feature maps as the set of \(F_H (\cdot )\) where \(H \! = \! \{k,\ldots ,q\}\) being the set of selected layer indices. Similarly, we denote the selected feature maps of DAM by \(\mathcal {M}_A( \cdot )\) with the A being the selected layer set. In this way, the feature space of target domain is \((\mathcal {M}_A(x^t), F_H(x^t))\) and the \((M^s_A(x^s), F_H(x^s))\) is their counterpart for source domain. Given the distribution of \((\mathcal {M}_A(x^t), F_H(x^t)) \! \sim \! \mathbb {P}_g\), and that of \((M^s_A(x^s), F_H(x^s)) \! \sim \! \mathbb {P}_s\), the distance between these two domain distributions which needs to be minimized is represented as \(W(\mathbb {P}_s,\mathbb {P}_g)\). For stabilized training, we employ the Wassertein distance [39] between the two distributions as follows:
where \(\prod (\mathbb {P}_s, \mathbb {P}_g)\) represents the set of all joint distributions \(\gamma (\text {x},\text {y})\) whose marginals are respectively \(\mathbb {P}_s\) and \(\mathbb {P}_g\).
In adversarial learning, the DAM is pitted against an adversary: a discriminative model that implicitly estimates the \(W(\mathbb {P}_s,\mathbb {P}_g)\). We refer our discriminator as domain critic module and denote it by \(\mathcal {D}\). Specifically, our constructed DCM consists of several stacked residual blocks, as illustrated in Fig. 5.2. In each block, the number of feature maps is doubled until it reaches 512, while their sizes are decreased. We concatenate the multiple levels of feature maps as input to the DCM. This discriminator would differentiate the complicated feature space between the source and target domains. In this way, our domain adaptation approach not only removes source-specific patterns in the beginning but also disallows their recovery at higher layers [20]. In unsupervised learning, we jointly optimize the generator \(\mathcal {M}\) (DAM) and the discriminator \(\mathcal {D}\) (DCM) via adversarial loss. Specifically, with \(X^t\) being target set, the loss for learning the DAM is
Then, with the \(X^s\) representing the set of source images, the DCM is optimized via
where K is a constant that applies Lipschitz constraint to \(\mathcal {D}\).
During the alternative updating of \(\mathcal {M}\) and \(\mathcal {D}\), the DCM outputs a more precise estimation of \(W(\mathbb {P}_s, \mathbb {P}_g)\) between distributions of the feature space from both domains. The updated DAM is more effective to generate source-like feature maps for conducting cross-modality domain adaptation.
3.1.4 Training Strategies
In our setting, the source domain is biomedical cardiac MRI images and the target domain is CT data. All the volumetric MRI and CT images were resampled to the voxel spacing of \(1 \! \times \! 1 \! \times \! 1\) mm\(^3\) and cropped into the size of \(256 \! \times \! 256 \! \times \! 256\) centering at the heart region. In preprocessing, we conducted intensity standardization for each domain, respectively. Augmentations of rotation, zooming, and affine transformations were employed to combat over fitting. To leverage the spatial information existing in volumetric data, we sampled consecutive three slices along the coronal plane and input them to three channels. The label of the intermediate slice is utilized as the ground truth when training the 2D networks.
We first trained the segmenter on the source domain data in supervised manner with stochastic gradient descent. The Adam optimizer was employed with parameters as batch size of 5, learning rate of \(1 \! \times \! 10^{-3}\) and a stepped decay rate of 0.95 every 1500 iterations. After that, we alternatively optimized the DAM and DCM with the adversarial loss for unsupervised domain adaptation. Following the heuristic rules of training WGAN [39], we updated the DAM every 20 times when updating the DCM. In adversarial learning, we utilized the RMSProp optimizer with a learning rate of \(3\times 10^{-4}\) and a stepped decay rate of 0.98 every 100 joint updates, with weight clipping for the discriminator being 0.03.
3.2 Experimental Results
3.2.1 Dataset and Evaluation Metrics
We validated our proposed unsupervised domain adaptation method on the public dataset of MICCAI 2017 Multi-Modality Whole Heart Segmentation for cross-modality cardiac segmentation in MRI and CT images [40]. This dataset consists of unpaired 20 MRI and 20 CT images from 40 patients. The MRI and CT data were acquired in different clinical centers. The cardiac structures of the images were manually annotated by radiologists for both MRI and CT images. Our ConvNet segmenter aimed to automatically segment four cardiac structures including the ascending aorta (AA), the left atrium blood cavity (LA-blood), the left ventricle blood cavity (LV-blood), and the myocardium of the left ventricle (LV-myo) . For each modality, we randomly split the dataset into training (16 subjects) and testing (4 subjects) sets, which were fixed throughout all experiments.
For evaluation, we employed two commonly used metrics to quantitatively evaluate the segmentation performance of automatic methods [41]. The DICE coefficient \(([\%])\!\) was employed to assess the agreement between the predicted segmentation and ground truth for cardiac structures. We also calculated the average surface distance (ASD\([\text {voxel}]\)) to measure the segmentation performance from the perspective of the boundary. A higher Dice and lower ASD indicate better segmentation performance. Both metrics are presented in the format of mean±std, which shows the average performance as well as the cross-subject variations of the results (Table 5.1).
3.2.2 Experimental Settings
We employed the MRI images as the source domain and the CT dataset as the target domain. We demonstrated the effectiveness of the proposed unsupervised cross-modality domain adaptation method with extensive experiments. We designed several experiment settings: (1) training and testing the ConvNet segmenter on source domain (referred as Seg-MRI); (2) training the segmenter from scratch on annotated target domain data (referred as Seg-CT); (3) fine-tuning the source domain segmenter with annotated target domain data, i.e., the supervised transfer learning (referred as Seg-CT-STL); (4) directly testing the source domain segmenter on target domain data (referred as Seg-CT-noDA); (5) our proposed unsupervised domain adaptation method (referred as Seg-CT-UDA). We also compared with a previous state-of-the-art heart segmentation method using ConvNets [42]. Last but not least, we conducted ablation studies to observe how the adaptation depth would affect the performance.
3.2.3 Results of UDA on Cross-Modality Cardiac Images
Table 5.1 reports the comparison results of different methods, where we can see that the proposed unsupervised domain adaptation method is effective by mapping the feature space of the target CT domain to that of the source MRI domain. Qualitative results of the segmentations for CT images are presented in Fig. 5.3.
Results of different methods for CT image segmentations. Each row presents one typical example, from left to right: a raw CT slices b ground truth labels c supervised transfer learning d ConvNets trained from scratch e directly applying MRI segmenter on CT data f our unsupervised cross-modality domain adaptation results. The structures of AA, LA-blood, LV-blood, and LV-myo are indicated by yellow, red, green, and blue colors, respectively (best viewed in color)
In the experiment setting Seg-MRI, we first evaluate the performance of the source domain model, which serves as the basis for subsequent domain adaptation procedures. Compared with [42], our ConvNet segmenter reached promising performance with exceeding Dice on LV-blood and LV-myo, as well as comparable Dice on AA and LA-blood. With this standard segmenter network architecture, we conducted following experiments to validate the effectiveness of our unsupervised domain adaptation framework.
To experimentally explore the potential upper bounds of the segmentation accuracy of the cardiac structures from CT data, we implemented two different settings, i.e., the Seg-CT and Seg-CT-STL. Generally, the segmenter fine-tuned from Seg-MRI achieved higher Dice and lower ASD than the model trained from scratch, proving the effectiveness of supervised transfer learning for adapting an established network to a related target domain using additional annotations. Meanwhile, these results are comparable to [42] on most of the four cardiac structures.
To demonstrate the severe domain shift inherent in cross-modality biomedical images, we directly applied the segmenter trained on MRI domain to the CT data without any domain adaptation procedure. Unsurprisingly, the network of Seg-MRI completely failed on CT images, with average Dice of merely 14.3% across the structures. As shown in Table 5.1, the Seg-CT-noDA only got a Dice of 0.8% for the LV-blood. The model did not even output any correct predictions for two of the four testing subjects on the structure of LV-blood (please refer to (e) in Fig. 5.3). This demonstrates that although the cardiac MRI and CT images share similar high-level representations and identical label space, the significant difference in their low-level characteristics makes it extremely difficult for MRI segmenter to extract effective features for CT.
With our proposed unsupervised domain adaptation method, a great improvement of the segmentation performance on the target CT data was achieved compared with the Seg-CT-noDA. More specifically, our Seg-CT-UDA (d = 21) model has increased the average Dice across four cardiac structures by 43.4%. As presented in Fig. 5.3, the predicted segmentation masks from Seg-CT-UDA can successfully localize the cardiac structures and further capture their anatomical shapes. The performance on segmenting AA is even close to that of Seg-CT-STL. This reflects that the distinct geometric pattern and the clear boundary of the AA have been successfully captured by the DCM. In turn, it supervises the DAM to generate similar activation patterns as the source feature space via adversarial learning. Looking at the other three cardiac structures (i.e., LA-blood, LV-blood, and LV-myo), the Seg-CT-UDA performances are not as high as that of AA. The reason is that these anatomical structures are more challenging, given that they come with either relatively irregular geometrics or limited intensity contrast with surrounding tissues. The deficiency focused on the unclear boundaries between neighboring structures or noise predictions on relatively homogeneous tissues away from the ROI. This is responsible for the high ASDs of Seg-CT-UDA, where boundaries are corrupted by noisy outputs. Nevertheless, by mapping the feature space of target domain to that of the source domain, we obtained greatly improved and promising segmentations against Seg-CT-noDA with zero data annotation effort.
3.2.4 Ablation Study on Adaptation Depth
We conduct ablation experiments to study the adaptation depth d, which is an important hyperparameter in our framework to determine how many layers to be replaced during the plug-and-play domain adaptation procedure. Intuitively, a shallower DAM (i.e., smaller d) might be less capable of learning effective feature mapping function \(\mathcal {M}\) across domains than a deeper DAM (i.e., larger d). This is due to the insufficient capacity of parameters in shallow DAM, as well as the huge domain shift in feature distributions. Conversely, with an increase in adaptation depth d, DAM becomes more powerful for feature mappings, but training a deeper DAM solely with adversarial gradients would be more challenging.
Comparison of results using Seg-CT-UDA with different adaptation depths (colors are the same with Fig. 5.3)
The overview of our unsupervised domain adaptation framework. Left: the segmentation DNN learned on source domain; Middle: the SeUDA where the paired generator and discriminator are indicated with the same color, the blue/green arrows illustrate the data flows from original images (\(x^t/x^s\)) to transformed images (\(x^{t\rightarrow s}/x^{s\rightarrow t}\)) then back to reconstructed images (\(\hat{x}^t/\hat{x}^s\)) in cycle-consistency loss, the orange part is the discriminator for the semantic-aware adversarial learning; Right: the inference process of SeUDA given a new target image for testing
To experimentally demonstrate how the performance would be affected by d and search for an optimal d, we repeated the experiments with domain adaptation from MRI to CT by varying the \(d=\{13, 21, 31\}\), while maintaining all the other settings the same. Viewing the examples in Fig. 5.4, Seg-CT-UDA (d=21) model obtained an approaching ground truth segmentation mask for ascending aorta. The other two models also produced inspiring results capturing the geometry and boundary characteristics of AA, validating the effectiveness of our unsupervised domain adaptation method. From Table 5.1, we can observe that DAM with a middle-level of adaptation depth (\(d=21\)) achieved the highest Dice on three of the four cardiac structures, exceeding the other two models by a significant margin. For the LA-blood, the three adaptation depths reached comparable segmentation Dice and ASD, and the \(d=31\) model was the best. Notably, the model of Seg-CT-UDA \((d=31)\) overall demonstrated superiority over the model with adaptation depth \(d=13\). This shows that enabling more layers learnable helps to improve the domain adaptation performance on cross-modality segmentations.
4 Pixel-Level Adaptation with Image-to-Image Translation
In this section, we present a pixel-level unsupervised domain adaptation framework with generative adversarial network, applied to cross-cohort X-ray lung segmentation. Different from feature-level adaptation method described in the last section, this pixel-level adaptation method detaches the segmentation ConvNets from the domain adaptation process. Given a test image, our framework conducts image-to-image transformation to generate a source-like image which is directly forwarded to the established source ConvNet. To enhance the preservation of structural information during image transformation, we improve CycleGAN with a novel semantic-aware loss by embedding a nested adversarial learning in semantic label space. Our method is named as SeUDA, standing for semantic-aware unsupervised domain adaptation, and Fig. 5.5 presents overview of it. Details of network configurations, adversarial losses and experimental results will be presented in the followings.
4.1 Method
With a set of the source domain images \(x^s \! \in \! \mathcal {X}^s\) and corresponding labels \(y^s \! \in \! \mathcal {Y}\), we train a ConvNet, denoted by \(f^s\), to segment the input images. For a new set of the target domain images \(x^t \! \in \! \mathcal {X}^t\), we aim to adapt the appearance of \(x^t\) to source image space \(\mathcal {X}^s\), so that the established \(f^s\) can be directly generalized to the transformed image.
4.1.1 ConvNet Segmenter Architecture
To establish a state-of-the-art segmentation network, we make complementary use of the residual connection, dilated convolution and multi-scale feature fusion. The backbone of our segmenter is modified ResNet-101. We replace the standard convolutional layers in the high-level residual blocks with the dilated convolutions. To leverage features with multi-scale receptive fields, we replace the last fully connected layer with four parallel \(3 \! \times \! 3\) dilated convolutional branches, with a dilation rate of {6, 12, 18, 24}, respectively. An upsampling layer is added in the end to produce dense predictions for the segmentation task. We start with 32 feature maps in the first layer and double the number of feature maps when the spatial size is halved or the dilation convolutions are utilized. The segmenter is optimized by minimizing the pixel-wise multi-class cross-entropy loss of the prediction \(f^s(x^s)\) and ground truth \(y^s\) with standard stochastic gradient descent.
4.1.2 Image Transformation with Semantic-Aware CycleGAN
With the source domain model \(f^s\) which maps the source input space \(\mathcal {X}^s\) to the semantic label space \(\mathcal {Y}\), our goal is to make it generally applicable to new target images. Given that annotating medical data is quite expensive, we conduct the domain adaptation in an unsupervised manner. Specifically, we map the target images toward the source image space. The generated new image \(x^{t\rightarrow s}\) appears to be drawn from \(\mathcal {X}^s\) while the content and semantic structures remain unchanged. In this way, we can directly apply the well-established model \(f^s\) on \(x^{t \rightarrow s}\) without retraining and get the segmentation result for \(x^t\).
To achieve this, we use generative adversarial networks [10], which have made a wide success for pixel-to-pixel image translation, by constructing a generator \(\mathcal {G}_{t \rightarrow s}\) and a discriminator \(\mathcal {D}_s\). The generator aims to produce realistic transformed image \(x^{t\rightarrow s} = \mathcal {G}_{t \rightarrow s}(x^t)\). The discriminator competes with the generator by trying to distinguish between the fake generated data \(x^{t \rightarrow s}\) and the real source data \(x^s\). The GAN corresponds to a minimax two-player game and is optimized via the following objective:
where the discriminator tries to maximize this objective to correctly classify the \(x^{t\rightarrow s}\) and \(x^s\), while the generator tries to minimize \(\text {log}(1-\mathcal {D}_s(\mathcal {G}_{t \rightarrow s}(x^t)))\) to learn the data distribution mapping from \(\mathcal {X}^t\) to \(\mathcal {X}^s\).
Cycle-consistency adversarial learning. To achieve domain adaptation with image transformation, it is crucial that the detailed contents in the original \(x^t\) are well preserved in the generated \(x^{t\rightarrow s}\). Inspired by the CycleGAN [26], we employ the cycle-consistency loss during the adversarial learning to maintain the contents with clinical clues of the target images.
We build a reverse source-to-target generator \(\mathcal {G}_{s \rightarrow t}\) and a target discriminator \(\mathcal {D}_t\), to bring the transformed image back to the original image. This pair of models are trained with a same way GAN loss \(\mathcal {L}_\text {GAN}(\mathcal {G}_{s\rightarrow t}, \mathcal {D}_t)\) following the Eq. (5.7). In this regard, we derive the cycle-consistency loss which encourages \(\mathcal {G}_{s \rightarrow t}(\mathcal {G}_{t \rightarrow s}(x^t)) \! \approx \! x^t\) and \(\mathcal {G}_{t \rightarrow s}(\mathcal {G}_{s \rightarrow t}(x^s)) \! \approx \! x^s\) in the transformation:
where the L1-Norm is employed for reducing blurs in the generated images. This loss imposes the pixel-level penalty on the distance between the cyclic transformation result and the input image.
Semantic-aware adversarial learning. The image quality of \(x^{t \rightarrow s}\) and the stability of \(\mathcal {G}_{t \rightarrow s}\) are crucial for the effectiveness of our method, since we apply the established \(f^s\) to \(x^{t \rightarrow s}\) which is obtained by inputting \(x^{t}\) to \(\mathcal {G}_{t\rightarrow s}\). Therefore, besides the cycle-consistency loss which composes both generators and constraints the cyclic input–output consistency, we further try to explicitly enhance the intermediate transformation result \(x^{t \rightarrow s}\). Specifically, for our segmentation domain adaptation task, we design a novel semantic-aware loss which aims to prevent the semantic distortion during the image transformation.
In our unsupervised learning scenario, we establish a nested adversarial learning module by adding another new discriminator \(\mathcal {D}_{m}\) into the system. It distinguishes between the source domain ground truth lung mask \(y^s\) and the predicted lung mask \(f^s(x^{t\rightarrow s})\) obtained by applying the segmenter on the source-like transformed image. Our underlying hypothesis is that the shape of anatomical structure is consistent across multicenter medical images. The prediction of \(f^s(x^{t\rightarrow s})\) should follow the regular semantic structures of the lung to fool the \(\mathcal {D}_{m}\), otherwise, the generator \(\mathcal {G}_{t\rightarrow s}\) would be penalized by the semantic-aware loss:
This loss imposes an explicit constraint on the intermediate result of the cyclic transformation. Its gradients can assist the update of the generator \(\mathcal {G}_{t\rightarrow s}\), which benefits the stability of the entire adversarial learning procedure.
4.1.3 Learning Procedure and Implementation Details
We follow the practice of [26] to configure the generators and discriminators. Specifically, both generators have the same architecture consisting of an encoder (three convolutions), a transformer (nine residual blocks), and a decoder (two deconvolutions and one convolution). All the three discriminators process \(70 \! \times \! 70\) patches and produce real/fake predictions via 3 stride-2 and 2 stride-1 convolutional layers. The overall objective for the generators and discriminators is as follows:
where the \(\{\alpha , \beta , \lambda \}\) denote trade-off hyperparameters adjusting the importance of each component, which is empirically set to be \(\{0.5, 10, 0.5\}\) in our experiments. The entire framework is optimized to obtain
The generators \(\{\mathcal {G}_{t\rightarrow s}, \mathcal {G}_{s\rightarrow t}\}\) and discriminators \(\{\mathcal {D}_s,\mathcal {D}_t,\mathcal {D}_m\}\) are optimized altogether and updated successively. Note that the segmenter \(f^s\) is not updated in the process of image transformation. In practice, when training the generative adversarial networks, we followed the strategies of [26] for reducing model oscillation. Specifically, the negative log likelihood in \(\mathcal {L}_{GAN}\) was replaced by a least-square loss to stabilize the training. The discriminator loss was calculated using one image from a collection of fifty previously generated images rather than the one produced in the latest training step. We used the Adam optimizer with an initial learning rate of 0.002, which was linearly decayed every 100 epochs. We implemented our proposed framework on the TensorFlow platform using an Nvidia Titan Xp GPU.
4.2 Experimental Results
4.2.1 Datasets and Evaluation Metrics
Our unsupervised domain adaptation method was validated on lung segmentations using two public Chest X-ray datasets, i.e., the Montgomery set (138 cases) [43] and the JSRT set (247 cases) [44]. Both the datasets are typical X-ray scans collected in clinical practice, but their image distributions are quite different in terms of the disease type, intensity, and contrast (see the first and fourth columns in Fig. 5.6a). The ground truth masks of left and right lungs are provided in both datasets. We randomly split each dataset into 7:1:2 for training, validation and test sets. All the images were resized to \(512 \! \times \! 512\), and rescaled to [0, 255]. The prediction masks were post-processed with the largest connected-component selection and hole filling.
To quantitatively evaluate our method, we utilized four common segmentation measurements, i.e., the Dice coefficient ([%]), recall ([%]), precision ([%]) and average surface distance (ASD)([mm]). The first three metrics are measured based on the pixel-wise classification accuracy. The ASD assesses the model performance at boundaries and a lower value indicates better segmentation performance.
4.2.2 Experimental Settings
In our experiments, the source domain is the Montgomery set and the target domain is the JSRT set. We first established the segmenter on source training data independently. Next, we test the segmenter under various settings: (1) testing on source domain (S-test); (2) directly testing on target data (T-noDA); (3) using histogram matching to adjust target images before testing (T-HistM); (4) aligning target features with the source domain as proposed in [20] (T-FeatDA); (5) fine-tuning the model on labeled target data before testing on JSRT (T-STL); In addition, we investigated the performance of our proposed domain adaptation method with and w/o the semantic-aware loss, i.e., SeUDA and CyUDA.
4.2.3 Results of UDA on Cross-Cohort Chest X-Ray Images
The comparison results of different methods are listed in Table 5.2. We can see that when directly applying the learned source domain segmenter to target data (T-noDA), the model performance significantly degraded, indicating that domain shift would severely impede the generalization performance of DNNs. Specifically, the average Dice over both lungs dropped from 95.61 to 79.47%, and the average ASD increased from 2.34 to 11.04 mm.
Typical results for the image transformation and lung segmentation. a Visualization of image transformation results, from left to right, are the target images in JSRT set, CyUDA transformation results, SeUDA transformation results, and the nearest neighbor of \(x^{t\rightarrow s}\) got from source set; each row corresponds to one patient. b Comparison of segmentation results between the ground truth, T-noDA, T-HistM, and our proposed SeUDA; each row corresponds to one patient
With our proposed method, we find a remarkable improvement by applying the source segmenter on transformed target images. Compared with T-noDA, our SeUDA increased the average Dice by 15.04%. Meanwhile, the ASDs for both lungs were reduced significantly. Also, our method outperforms the UDA baseline histogram matching T-HistM with the average dice increased by 3.97% and average ASD decreased from 5.19 to 3.18 mm. Compared with the feature-level domain adaptation method T-FeatDA, our SeUDA can not only obtain higher segmentation performance, but also provide intuitive visualization of how the adaptation is achieved. Notably, the performance of our unsupervised SeUDA is even comparable to the upper bound of supervised T-STL. In Table 5.2, the gaps of Dice are marginal, i.e., 1.32% for right lung and 2.42% for left lung.
The typical transformed target images can be visualized in Fig. 5.6a, demonstrating that SeUDA has successfully adapted the appearance of target data to look similar to source images. In addition, the positions, contents, semantic structures, and clinical clues are well preserved after transformation. In Fig. 5.6b, we can observe that without domain adaptation, the predicted lung masks are quite cluttered. With histogram matching, appreciable improvements are obtained but the transformed images cannot mimic the source images very well. With our SeUDA, the lung areas are accurately segmented attributing to the good target-to-source appearance transformation.
4.2.4 Effectiveness of Semantic-Aware Loss with Ablation Study
We conduct ablation experiments to investigate the contribution of our novel semantic-aware loss designed for segmentation domain adaptation. We implemented CyUDA by removing the semantic-aware loss from the SeUDA. One notorious problem of GANs is that their training would be unstable and sensitive to initialization states [30, 46]. In this study, we measured the standard deviation (std) of the CyUDA and SeUDA by running each model for 10 times under different initializations but with the same hyperparameters. We observed significant lower variability on the segmentation performance across the 10 SeUDA models than the 10 CyUDA models, i.e., Dice std: 0.25 versus 2.03%, ASD std: 0.16 versus 1.19 mm. Qualitatively, we observe that the CyUDA transformed images may suffer from distorted lung boundaries in some cases, see the third row in Fig. 5.6a. In contrast, adding the semantic-aware loss, the transformed images consistently present a high quality. This reveals that the novel semantic-aware loss contributes to stabilize the image transformation process and prevent the distortion in structural contents, and hence contributes to boost the performance of segmentation domain adaptation.
5 Discussion
This chapter introduces how to tackle domain adaptation problem in medical imaging from two different perspectives. This is an essential and urgent topic to study the generalization capability and robustness of ConvNets, given that deep learning nowadays has become the state of the art for solving image recognition tasks. Resolving this issue will help to promote deep learning studies based on large-scale real-world clinical dataset composing inhomogeneous images [47].
Fine-tuning the ConvNets with a set of new labeled images from the target domain can improve the model’s performance on target data. However, this straightforward supervised solution still requires extra efforts from clinicians for constructing the annotated fine-tune dataset. Unsupervised domain adaptation methods are more appealing and practical in the long-run, though it is technically challenging at current stage. Basically, the UDA requires to model and map the underlying distributions of different domains, either in latent feature space or appearance pixel space. The insights of adversarial networks fit well into this scope, as which can implicitly learn how to model, transform, and discriminate the data distributions via highly nonlinear networks. This forms the basis of the situation that adversarial learning has been frequently investigated for unsupervised domain adaptation tasks.
Feature-level adaptation and pixel-level adaptation are two independent ways to conduct unsupervised domain adaptation, with ideas from different perspectives. Feature-level adaptation aims to transform different data sources into a shared latent space with domain-invariant features, such that a shared classifier can be established in this common space. The advantage is that the classifier is learned in a high-quality homogeneous feature space, with reduced confounding factors from scanner effects. The disadvantage is that the obtained domain-invariant features are unclear for interpretation and intuitive visualization. Pixel-level adaptation aims to transform the image appearance from one domain to the other, and use the transformed images to train or test a model. The advantage for this stream of solution is that we can directly assess the quality of domain adaptation by observing the transformed images. The disadvantage is that there may still exist a domain gap between the synthetic images and real images. It is worth noting that these two independent manners of matching across domains can be complementary to each other. Jointly taking advantage of both is feasible and have good potential to present more appealing performance to narrow the domain gap.
6 Conclusion
In conclusion, this chapter presents unsupervised domain adaptation methods for medical image segmentation using adversarial learning. Solutions from two different perspectives are presented, i.e., feature-level adaptation and pixel-level adaptation. The feature-level adaptation method has been validated on cross-modality (MRI/CT) cardiac image segmentation. The pixel-level adaptation method has been validated on cross-cohort X-ray images for lung segmentation. Both application scenarios of unsupervised domain adaptation have demonstrated highly promising results on generalizing the ConvNets to the unseen target domain. The proposed frameworks are general and can be extended to other similar scenarios in medical image computing with domain shift issues.
References
Ronneberger O, Fischer P, Brox T (2015) U-net: convolutional networks for biomedical image segmentation. In: Navab N, Hornegger J, Wells WM, Frangi AF (eds) MICCAI 2015. LNCS. Springer, Munich, Germany, pp 234–241
Roth HR, Lu L, Farag A, Shin HC, Liu J, Turkbey EB, Summers RM (2015) Deeporgan: multi-level deep convolutional networks for automated pancreas segmentation. In: International conference on medical image computing and computer-assisted intervention. Springer, Berlin, pp 556–564
Roth HR, Lu L, Seff A, Cherry KM, Hoffman J, Wang S, Liu J, Turkbey E, Summers RM (2014) A new 2.5 d representation for lymph node detection using random sets of deep convolutional neural network observations. In: International conference on medical image computing and computer-assisted intervention. Springer, Berlin, pp 520–527
Dou Q, Chen H, Yueming J, Huangjing L, Jing Q, Heng P (2017) Automated pulmonary nodule detection via 3d convnets with online sample filtering and hybrid-loss residual learning. In: MICCAI, pp 630–638
Bejnordi BE, Veta M, Van Diest PJ, Van Ginneken B, Karssemeijer N, Litjens G, Van Der Laak JA, Hermsen M, Manson QF, Balkenhol M et al (2017) Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer. Jama 318(22):2199–2210
Esteva A, Kuprel B, Novoa RA, Ko J, Swetter SM, Blau HM, Thrun S (2017) Dermatologist-level classification of skin cancer with deep neural networks. Nature 542(7639):115
Ghafoorian M, Mehrtash A, Kapur T, Karssemeijer N, Marchiori E, Pesteie M, Guttmann CR, de Leeuw FE, Tempany CM, van Ginneken B et al (2017) Transfer learning for domain adaptation in mri: application in brain lesion segmentation. In: International conference on medical image computing and computer-assisted intervention. Springer, Berlin, pp 516–524
Gibson E, Hu Y, Ghavami N, Ahmed HU, Moore C, Emberton M, Huisman HJ, Barratt DC (2018) Inter-site variability in prostate segmentation accuracy using deep learning. In: International conference on medical image computing and computer-assisted intervention. Springer, Berlin, pp 506–514
Philipsen RH, Maduskar P, Hogeweg L, Melendez J, Sánchez CI, van Ginneken B (2015) Localized energy-based normalization of medical images: application to chest radiography. IEEE Trans Med Imaging 34(9):1965–1975
Goodfellow IJ, Pouget-Abadie J, Mirza M, Xu B et al (2014) Generative adversarial nets. In: Conference on neural information processing systems (NIPS), pp 2672–2680
Dou Q, Ouyang C, Chen C, Chen H, Heng PA (2018) Unsupervised cross-modality domain adaptation of convnets for biomedical image segmentations with adversarial loss. arXiv:180410916
Chen C, Dou Q, Chen H, Heng PA (2018) Semantic-aware generative adversarial nets for unsupervised domain adaptation in chest x-ray segmentation. arXiv:180600600
Tzeng E, Hoffman J, Zhang N, Saenko K, Darrell T (2014) Deep domain confusion: maximizing for domain invariance. arXiv:14123474
Long M, Cao Y, Wang J, Jordan MI (2015) Learning transferable features with deep adaptation networks. In: International conference on machine learning (ICML), pp 97–105
Sun B, Saenko K (2016) Deep coral: correlation alignment for deep domain adaptation. In: European conference on computer vision (ECCV) workshops, pp 443–450
Ganin Y, Ustinova E, Ajakan H, Germain P, Larochelle H, Laviolette F, Marchand M, Lempitsky V (2016) Domain-adversarial training of neural networks. J Mach Learn Res 17(1):2030–2096
Tzeng E, Hoffman J, Saenko K, Darrell T (2017) Adversarial discriminative domain adaptation. In: CVPR, pp 2962–2971
Tsai Y, Hung W, Schulter S, Sohn K, Yang M, Chandraker M (2018) Learning to adapt structured output space for semantic segmentation. In: IEEE conference on computer vision and pattern recognition. CVPR, pp 7472–7481
Sankaranarayanan S, Balaji Y, Jain A, Lim SN, Chellappa R (2018) Learning from synthetic data: addressing domain shift for semantic segmentation. In: IEEE conference on computer vision and pattern recognition (CVPR), pp 3752–3761
Kamnitsas K et al (2017) Unsupervised domain adaptation in brain lesion segmentation with adversarial networks. In: IPMI. Springer, Berlin, pp 597–609
Joyce T, Chartsias A, Tsaftaris SA (2018) Deep multi-class segmentation without ground-truth labels. In: International conference on medical imaging with deep learning (MIDL)
Degel MA, Navab N, Albarqouni S (2018) Domain and geometry agnostic cnns for left atrium segmentation in 3d ultrasound. In: MICCAI, pp 630–637
Ren J, Hacihaliloglu I, Singer EA, Foran DJ, Qi X (2018) Adversarial domain adaptation for classification of prostate histopathology whole-slide images. In: MICCAI, pp 201–209
Dong N, Kampffmeyer M, Liang X, Wang Z, Dai W, Xing E (2018) Unsupervised domain adaptation for automatic estimation of cardiothoracic ratio. In: MICCAI. Springer, Berlin, pp 544–552
Zhang L, Pereañez M, Piechnik SK, Neubauer S, Petersen SE, Frangi AF (2018) Multi-input and dataset-invariant adversarial learning (mdal) for left and right-ventricular coverage estimation in cardiac mri. In: International conference on medical image computing and computer-assisted intervention. Springer, Berlin, pp 481–489
Zhu J, Park T, Isola P, Efros AA (2017) Unpaired image-to-image translation using cycle-consistent adversarial networks. In: ICCV, pp 2242–2251
Russo P, Carlucci FM, Tommasi T, Caputo B (2018) From source to target and back: Symmetric bi-directional adaptive GAN. In: IEEE conference on computer vision and pattern recognition. CVPR, pp 8099–8108
Zhang Y, Miao S, Mansi T, Liao R (2018) Task driven generative modeling for unsupervised domain adaptation: application to x-ray image segmentation. In: International conference on medical image computing and computer-assisted intervention (MICCAI), pp 599–607
Shrivastava A, Pfister T, Tuzel O, Susskind J, Wang W, Webb R (2017) Learning from simulated and unsupervised images through adversarial training. In: ieee conference on computer vision and pattern recognition. CVPR, pp 2242–2251
Bousmalis K, Silberman N, Dohan D, Erhan D, Krishnan D (2017) Unsupervised pixel-level domain adaptation with generative adversarial networks. In: IEEE conference on computer vision and pattern recognition. CVPR, pp 95–104
Hoffman J, Tzeng E, Park T, Zhu J, Isola P, Saenko K, Efros AA, Darrell T (2018) Cycada: Cycle-consistent adversarial domain adaptation. In: International conference on machine learning (ICML), pp 1994–2003
Zhao H, Li H, Maurer-Stroh S, Guo Y, Deng Q, Cheng L (2018) Supervised segmentation of un-annotated retinal fundus images by synthesis. IEEE Trans Med Imaging
Jiang J, Hu YC, Tyagi N, Zhang P, Rimner A, Mageras GS, Deasy JO, Veeraraghavan H (2018) Tumor-aware, adversarial domain adaptation from ct to mri for lung cancer segmentation. In: MICCAI. Springer, Berlin, pp 777–785
Huo Y, Xu Z, Moon H, Bao S, Assad A, Moyo TK, Savona MR, Abramson RG, Landman BA (2018) Synseg-net: synthetic segmentation without target modality ground truth. IEEE Trans Med Imaging
Yu F, Koltun V, Funkhouser T (2017) Dilated residual networks. In: CVPR, pp 636–644
Milletari F, Navab N, Ahmadi SA (2016) V-net: fully convolutional neural networks for volumetric medical image segmentation. In: 2016 fourth international conference on 3D vision (3DV). IEEE, pp 565–571
Zeiler MD, Fergus R (2014) Visualizing and understanding convolutional networks. In: ECCV. Springer, Berlin, pp 818–833
Yosinski J, Clune J, Bengio Y, Lipson H (2014) How transferable are features in deep neural networks? In: NIPS, pp 3320–3328
Arjovsky M, Chintala S, Bottou L (2017) Wasserstein gan. arXiv:170107875
Zhuang X, Shen J (2016) Multi-scale patch and multi-modality atlases for whole heart segmentation of mri. Med Image Anal 31:77–87
Dou Q, Yu L, Chen H, Jin Y, Yang X, Qin J, Heng PA (2017) 3d deeply supervised network for automated segmentation of volumetric medical images. Med Image Anal 41:40–54
Payer C, Štern D, Bischof H, Urschler M (2017) Multi-label whole heart segmentation using cnns and anatomical label configurations, pp 190–198
Jaeger S, Candemir S, Antani S, Wáng YXJ, Lu PX, Thoma G (2014) Two public chest x-ray datasets for computer-aided screening of pulmonary diseases. Quant Imaging Med Surg 4(6):475
Shiraishi J, Katsuragawa S, Ikezoe J, Matsumoto T, Kobayashi T, Komatsu Ki, Matsui M, Fujita H, Kodera Y, Doi K (2000) Development of a digital image database for chest radiographs with and without a lung nodule: receiver operating characteristic analysis of radiologists’ detection of pulmonary nodules. Am J Roentgenol 174(1):71–74
Wang L, Lai HM, Barker GJ, Miller DH, Tofts PS (1998) Correction for variations in mri scanner sensitivity in brain studies with histogram matching. Magn Reson Med 39(2):322–327
Salimans T, Goodfellow I, Zaremba W, Cheung V, Radford A, Chen X (2016) Improved techniques for training gans. In: Advances in neural information processing systems, pp 2234–2242
Wang X, Peng Y, Lu L, Lu Z, Bagheri M, Summers RM (2017) Chestx-ray8: Hospital-scale chest x-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases. In: Computer vision and pattern recognition (CVPR), pp 3462–3471
Acknowledgements
This work was supported by a research grant supported by the Hong Kong Innovation and Technology Commission under ITSP Tier 2 Platform Scheme (Project No. ITS/426/17FP) and a research grant from the Hong Kong Research Grants Council General Research Fund (Project No. 14225616).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Switzerland AG
About this chapter
Cite this chapter
Dou, Q., Chen, C., Ouyang, C., Chen, H., Heng, P.A. (2019). Unsupervised Domain Adaptation of ConvNets for Medical Image Segmentation via Adversarial Learning. In: Lu, L., Wang, X., Carneiro, G., Yang, L. (eds) Deep Learning and Convolutional Neural Networks for Medical Imaging and Clinical Informatics. Advances in Computer Vision and Pattern Recognition. Springer, Cham. https://doi.org/10.1007/978-3-030-13969-8_5
Download citation
DOI: https://doi.org/10.1007/978-3-030-13969-8_5
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-13968-1
Online ISBN: 978-3-030-13969-8
eBook Packages: Computer ScienceComputer Science (R0)