Abstract
Missing data consists in the lack of information in a dataset and since it directly influences classification performance, neglecting it is not a valid option. Over the years, several studies presented alternative imputation strategies to deal with the three missing data mechanisms, Missing Completely At Random, Missing At Random and Missing Not At Random. However, there are no studies regarding the influence of all these three mechanisms on the latest high-performance Artificial Intelligence techniques, such as Deep Learning. The goal of this work is to perform a comparison study between state-of-the-art imputation techniques and a Stacked Denoising Autoencoders approach. To that end, the missing data mechanisms were synthetically generated in 6 different ways; 8 different imputation techniques were implemented; and finally, 33 complete datasets from different open source repositories were selected. The obtained results showed that Support Vector Machines imputation ensures the best classification performance while Multiple Imputation by Chained Equations performs better in terms of imputation quality.
Access provided by CONRICYT-eBooks. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Missing Data is a common problem that appears in real-world datasets and is an important issue since it affects the performance of classifiers [20]. Over the past decades, many methods have been proposed to impute the missing values. In the research community, three main missing mechanisms are recognised – Missing Completely At Random (MCAR), Missing At Random (MAR) and Missing Not At Random (MNAR) – and adjusting the imputation method to the missing mechanism is crucial, since an improper choice can bias the classification performance [23]. Deep Learning techniques are currently a hot topic in Machine Learning literature [2], although their application for imputation purposes remains an understudied topic.
This work analyses the appropriateness of Stacked Denoising Autoencoders (SDAE) to impute the different data mechanisms, considering univariate and multivariate scenarios. The performance of SDAE is then compared to the performance of state-of-the-art imputation techniques. To achieve that, we selected 33 complete datasets from different open source repositories and simulated the missing mechanisms using 6 different configurations. Then, 8 different imputation techniques are evaluated in terms of F-measure and Root Mean Squared Error (RMSE). Summing up, the contributions of this research are the following: (i) presenting a comparative study that considers several missing data mechanisms, imputation methods and missing rates (5, 10, 15, 20, 40%), (ii) proposing an imputation approach based on SDAE and (iii) simultaneously evaluating the quality of imputation (similarly to related work) and the benefits for classification performance (mostly overlooked in related work). Our experiments show that the imputation methods (and consequently the classification performance) are influenced by missing mechanisms and configurations. Furthermore, we conclude that SDAE do not show a significant advantage over other standard imputation algorithms: regarding the quality of imputation, Multiple Imputation by Chained Equations (MICE) seems to be a better approach while Support Vector Machines (SVM) provides the best imputation for the classification stage. This document is organised as follows: Sect. 2 presents several research works that considered different configurations to generate the missing mechanisms and studied well-know imputation techniques and some recent deep learning approaches. Then, Sect. 3 describes the different stages of the experimental setup while Sect. 4 discusses the obtained results. Finally, Sect. 5 concludes the paper and presents some possibilities for future work.
2 Background Knowledge and Related Work
In this section, we provide some background on missing data mechanisms and imputation methods, also including a thorough explanation on the procedure of SDAE. Along with some background information, we refer to previous work on both topics, highlighting their main objectives and conclusions.
2.1 Missing Mechanisms
There are three mechanisms under which missing data can occur [15]: MCAR, MAR and MNAR [10]. MCAR occurs when the reason why data is missing is unrelated to any observed or unobserved value from the dataset (e.g. a survey participant had a flat tire and misses his appointment). In the case of MAR, the cause of the missing data is unrelated to the missing values but it is related with observed values from the dataset (e.g. an investigator finds that women are less likely to reveal their weight) and finally, in the case of MNAR, the probability of a value to be missing is related to the value itself (e.g. obese subjects are less likely to reveal their weight).
These mechanisms could be generated in various ways and several different examples could be found in the literature [9, 12, 18, 23, 26, 28]. Twala et al. [23] investigated the robustness and accuracy of techniques for handling incomplete data for different mechanisms of missing data. Three suites of data were created corresponding to MCAR, MAR and MNAR. For each of them, univariate (one feature only) and multivariate (several features) generation of missing data was performed using 21 datasets. These approaches were implemented for 3 missing rates (15, 30 and 50%). Rieger et al. [18] performed an extensive study covering both classification and regression problems and a variety of missing data mechanisms. Four different types of MAR generation are proposed as well as a mechanism for MCAR generation. Garciarena et al. [12] studied the interaction between missing data mechanisms, imputation methods and supervised classification algorithms. The authors generated missing values for the three different mechanisms and present two different versions of MNAR. In total, 4 missing data configurations are created for 6 different missing rates (5, 10, 20, 30, 40 and 50%) on 10 datasets from UCI Machine Learning Repository.
2.2 Imputation Algorithms
Imputation methods aim to find a plausible value to replace one that is missing and are mainly divided into statistical-based or machine learning-based methods [11]. Statistical methods consist in substituting the missing observations with the most similar ones among the training data, without the need of constructing a predictive model to evaluate their ”similarity” (e.g. Mean imputation – Meanimp, MICE, Expectation-Maximization – EM). Machine learning-based techniques, construct a predictive model with the complete available data to estimate values for substituting those that are missing (e.g. k-Nearest Neighbours imputation – kNNimp, SVM imputation – SVMimp, DAE imputation).
Garciarena et al. [12] compared the performance of 8 different imputation techniques including MICE, Meanimp and EM. The classification results (evaluated with F-measure) showed that MICE was the best technique. García-Laencina et al. [9] proposed an approach that achieves a balance between classification and imputation by using Multi-Task Learning perceptrons. This approach is compared with 4 well-known imputation methods (including kNNimp) using classification accuracy. The results show that the proposed method outperforms the other well-known techniques. Twala et al. [23] studied the effect of different imputation techniques in classification accuracy of a decision tree. The authors used 7 imputation methods including EM and Meanimp. The results show that EM works well on small datasets, particularly for numeric attributes. Xia et al. [26] compared their proposed algorithm with 5 imputation methods, including Meanimp and kNNimp. They used accuracy and Area Under the ROC Curve (AUC) as evaluation metric for the classification process (using a Random Forest classifier).
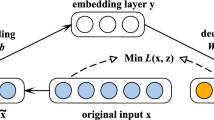
General neural network-based methods have been increasingly used for missing data imputation; however, deep learning architectures especially designed for missing data imputation has not yet been explored to its full potential. Denoising Autoencoders (DAE) [24] are an example of deep architectures that are designed to recover noisy data (\(\tilde{\mathbf{x }}\)), which can exist due to data corruption via some additive mechanism or by missing data. DAE are a variant of autoencoders (AE) – Fig. 1 – which is a type of neural network that uses backpropagation to learn a representation for a set of data. Each autoencoder is composed by three layers (input, hidden and output layer) which can be divided into two parts: encoder (from the input layer to the output of the hidden layer) and decoder (from the hidden layer to the output of the output layer). The encoder part maps an input vector x to a hidden representation y, through a nonlinear transformation \(f_\theta (\mathbf x )=s(\mathbf xW ^T+\mathbf b )\) where \(\theta \) represents W (weight matrix) and b (bias vector) parameters. The resulting y representation is then mapped back to a vector z which have the same shape of x, where z is equal to \(g_\theta '(\mathbf y )=s(\mathbf W'y +\mathbf b' )\). The train of an autoencoder consists in optimising the model parameters (W, W’, b and b’), minimising the reconstruction error between x and z. Vincent et al. [25] proposed a strategy to build deep networks by stacking layers of Denoising Autoencoders – SDAE. The results have shown that stacking DAE improves the performance over the standard DAE. In two recent works, Gondara et al. studied the appropriateness of SDAE for multiple imputation [13] and their application to imputation in clinical health records (recovering loss to followup information) [14]. In these works, the proposed algorithm is compared with MICE using the Predictive Mean Matching method. In the first work, authors consider only MCAR and MNAR mechanisms. The imputation results of both mechanisms are compared using sum of Root Mean Squared Error (\(RMSE_{sum}\)). Additionally, MNAR mechanism is also evaluated in terms of classification error, using a Random Forest classifier. In the second work, authors propose a SDAE model to handle imputation in healthcare data, using datasets under MCAR and MNAR mechanisms. The simulation results showed that their proposed approach surpassed the state-of-the-art methods. In both previous works, although authors prove the advantages of SDAE for imputation, a complete study under all missing mechanisms is not provided, since in both cases, MAR generation is completely disregarded. Furthermore, they only compare two imputation methods (MICE and SDAE) and the classification performance is only evaluated for one mechanism (MNAR). Beaulieu et al. [4] used SDAE to impute data in electronic health records. This approach is compared with five other imputation strategies (including Meanimp and kNNimp) and evaluated with RMSE. The results show that the proposed SDAE-based approach outperforms MICE. Duan et al. [7, 8] used SDAE for traffic data imputation. In the first work [7], the proposed approach is compared with another one that uses artificial neural networks with the same set of layers and nodes as the ones used in SDAE. In the second work [8] another imputation method is used (ARIMA – AutoRegressive Integrated Moving-Average) for comparison. To evaluate the imputation process authors used RMSE, Mean Absolute Error (MAE) and Mean Relative Error (MRE). Ning et al. [17] proposed an algorithm based on SDAE for dealing with big data of quality inspection. The proposed approach is compared with two other imputation algorithms (GBWKNN [19] and MkNNI [16]) that are both based on the k-nearest neighbour algorithm. The results are evaluated through \(d_2\) (the suitability between the imputed value and the actual value) and RMSE. The above-mentioned works show that the proposed imputation methods outperform the ones used for comparison, showing that deep learning based techniques are promising in the field of imputation. Sánchez-Morales et al. [22] proposed an imputation method that uses a SDAE. The main goal of their work was to understand how the proposed approach can improve the results obtained in the pre-imputation step. They used three state of the art methods for the pre-imputation: Zero Imputation, kNNimp and SVMimp. The results, for three datasets from UCI, are evaluated in terms of MSE. Authors concluded that the SDAE is capable of improving the final results for a pre-imputed dataset. To summarise, most of related work does not address all three missing data mechanisms and mostly evaluates the results in terms of quality of imputation rather than also evaluating the usefulness of an imputation method to generate quality data for classification. Furthermore, none of the reviewed works studies the effect of different missing data mechanisms on imputation techniques (including DAE) for several missing rates.
3 Experiments
We start our experiments by collecting 33 publicly available real-world datasets (UCI Machine Learning Repository, KEEL, STATLIB) to analyse the effect of different missing mechanisms (using different configurations) on imputation methods. Some of the original datasets were preprocessed in order to remove instances containing small amounts of missing values. In the case of multiclass datasets, they were modified in order to represent a binary problem. Afterwards, we perform the missing data generation, inserting missing values at five missing rates (5, 10, 15, 20 and 40%) following 6 different scenarios (\(MCAR_{univa}\), \(MCAR_{unifo}\), \(MAR_{univa}\), \(MAR_{unifo}\), \(MNAR_{univa}\) and \(MNAR_{unifo}\)) based on state-of-the-art generation methods. Five runs were performed for each missing generation, per dataset and missing rate. To provide a clear explanation of all the generation methods it is important to establish some basic notation. Therefore, let us assume a dataset X represented by a \(n\times p\) matrix, where \(i = 1, ..., n\) patterns and \(j = 1, ..., p\) attributes. The elements of X are denoted by \(x_{ij}\), each individual feature in X is denoted by \(x_{j}\) and each pattern is referred to as \(\mathbf x _i = [x_{i,1}, x_{i,2}, ..., x_{i,j}, ..., x_{i,p}]\). For the univariate configuration, univa, the feature that will have the missing values, \(x_{m}\), will always be the one most correlated with the class labels and the determining feature \(x_d\) is the one most correlated with \(x_m\). Regarding multivariate configurations, unifo, there are several alternatives to choose the missing values positions which will be detailed later.
Missing Completely at Random. For the univariate configuration of MCAR, \(MCAR_{univa}\), we consider the method proposed by Rieger et al. [18] and Xia et al. [26]. This configuration chooses random locations in \(x_{m}\) to be missing, i.e., random values of \(x_{i,m}\) are eliminated. The multivariate configuration of MCAR is proposed in the work of Garciarena et al. [12]. \(MCAR_{unifo}\) chooses random locations, \(x_{i,j}\), in the dataset to be missing until the desired MR is reached.
Missing at Random. The univariate configuration of MAR is based on ranks of \(x_d\): the probability of a pattern \(x_{i,m}\) to be missing is computed by dividing the rank of \(x_{i,m}\) in the determining feature \(x_d\) by the sum of all ranks for \(x_d\) – this configuration method is herein referred to as \(MAR_{univa}\). Then, the patterns to have missing values are sampled according to such probability, until the desired MR is reached [18, 26]. The multivariate configuration of MAR, \(MAR_{unifo}\), starts by defining pairs of features which include a determining and a missing feature {\(x_{d}\), \(x_{m}\)}. This pair selection was based on high correlations among all the features of the dataset. In the case of having an odd number of features, the unpaired feature may be added to the pair which contains its most correlated feature. For each pair of correlated features, the missing feature will be the one most correlated with the labels. In the case of having a triple of correlated features, there will be two missing features which will also be those most correlated with the class labels. \(x_{m}\) will be missing for the observations that are below the MR percentile in the determining feature \(x_{d}\). This means that the lowest observations of \(x_{d}\) will be deleted on \(x_{m}\).
Missing Not at Random. \(MNAR_{univa}\) was proposed by Twala et al. [23]: for this method the feature \(x_m\) itself is used as determining feature, i.e., the MR percentile of \(x_m\) is determined and values of \(x_m\) lower than a cut-off value are removed. The multivariate configuration of MNAR, \(MNAR_{unifo}\), was also proposed by Twala et al. [23] and is called Missingness depending on unobserved Variables (MuOV), where each feature of the dataset has the same number of missing values for the same observations. The missing observations are randomly chosen.
Nine imputation methods were then applied to the incomplete data: Mean imputation (Meanimp), imputation with kNN (kNNimp), imputation with SVM (SVMimp), MICE, EM imputation and SDAE-based imputation. Meanimp imputes the missing values with the mean of the complete values in the respective feature [12, 23, 26], while kNNimp imputes the incomplete patterns according to the values of their k-nearest neighbours [9, 26]. For kNNimp we considered the euclidean distance and a set of closest neighbours (1, 3 and 5). SVMimp was implemented considering a gaussian kernel – Radial Basis Function (RBF) [11]: the incomplete feature is used as target, while the remaining features are used to fit the model. The search for optimal parameters C and \(\gamma \) of the kernel was performed through a grid search for each dataset (different ranges of values were tested: \(10^{-2}\) to \(10^{10}\) for C and \(10^{-9}\) to \(10^3\) for \(\gamma \), both ranges increasing by a factor of 10). MICE is a multiple imputation technique that specifies a separate conditional model for each feature with missing data [3]. For each model, all other features can be used as predictors [13, 14]. EM is a maximum-likelihood-based missing data imputation method which estimates parameters by maximising the expected log-likelihood of the complete data [6]. The above methods were applied using open-source python implementations: scikit-learn for SVMimp and Meanimp, fancyimpute for kNNimp and MICE and impyute for EM.
Regarding the SDAE, we propose a model based on stacked denoising autoencoders, for the complete reconstruction of missing data. It was implemented using Keras library with a Theano backend. SDAE require complete data for initialisation so missing patterns are pre-imputed using the well-known Mean/Mode imputation method. We also apply z-score standardisation to the input data in order to have a faster convergence. There are two types of representations for an autoencoder [5]: overcomplete, where the hidden layer has more nodes than input layer and undercomplete, where the hidden layer is smaller than the input layer.
Our architecture is overcomplete, which means that the encoder block has more units in consecutive hidden layers than the input layer. This architecture of the SDAE is similar to the one proposed by Gondara et al. [13]. The model is composed by an input layer, 5 hidden layers and an output layer which form the encoder and the decoder (both constructed using regular densely-connected neural network layers). The number of nodes for each hidden layer was set to 7, as it has proven to obtain good results in related work [13]. For the encoding layers we chose hyperbolic tangent (tanh) as activation function due to its greater gradients [5]. Rectified Linear Units function (reLu) was used as activation function in the decoding layers. We have performed experiments with two different configurations for the training phase: the first one was adapted from Gondara et al. [13] while for the second one we have decided to study a different optimisation function – Adadelta optimisation algorithm – since it avoids the difficulties of defining a proper learning rate [27]. At the end, we have decided to use Adadelta since it proved to be most effective. Therefore, our final SDAE is trained with 100 epochs using Adadelta optimisation algorithm [27] and mean squared error as loss function. Our model has an input dropout ratio of 50%, which means that half of the network inputs are set to zero in each training batch. To prevent the training data from overfitting we add a regularization function named L2 [5]. Our imputation approach based on this SDAE considers the creation of three different models (for three different training sets), for which three runs will be made (multiple imputation). This approach is illustrated in Algorithm 1 and works as follows: (1) the instances of each dataset are divided in three equal-size sets; (2) each set is used as test set, while the remaining two are used to feed the SDAE in the training phase; (3) 3 multiple runs will be performed for each one of these models; (4) the output mean of the three models is used to impute the unknown values of the test set.

After the imputation step is concluded, we move towards the classification stage. We perform classification with a SVM with linear kernel (considering a value of \(C = 1\)) and considered two different metrics to evaluate two key performance requirements for imputation techniques: their efficiency on retrieving the true values in data (quality of imputation) [21] and their ability to provide quality data for classification [11]. The quality of imputation was assessed using RMSE, given by \(\sqrt{\frac{1}{n}\sum _{i=1}^{n}{({x}_i-\tilde{x}_i)^2}}\), where \(\tilde{x}\) are the imputed values of a feature, x are the corresponding original values and n is the number of missing values. The classification performance was assessed using F-measure which consists of an harmonic mean of precision and recall [1], defined as \(\text {F-measure} = \frac{2\times \text {precision} \times \text {recall}}{\text {precision} + \text {recall}}\).
4 Results and Discussion
Our work consists of a missing value generation phase followed by imputation and classification. Thus, we evaluate both the imputation quality and its impact on the classification performance. The results are divided by metric (F-measure and RMSE), missing mechanism (MCAR, MAR and MNAR), type of configuration (univariate and multivariate) and missing rate (5, 10, 15, 20 and 40%). Table 1 presents the average results obtained for all the datasets used in this study. As expected, the increase of missing rate leads to a decrease in the performance of classifiers (F-measure) and the quality of imputation (RMSE).
Quality of Imputation (RMSE). For univa configurations, MICE proved to be the best approach in most of the scenarios: for MNAR mechanism and a higher MR (40%), SDAE seems to be the best method. For the unifo configurations, MICE is the best imputation method for MCAR mechanism, regardless of the MR. Considering MAR mechanism, MICE is also the best method in most of scenarios, except for a higher MR (40%) – SDAE seems to be the best approach. In the case of MNAR mechanism and for lower MRs (5 and 10 %), MICE is also the best approach. However, for higher MRs, the SDAE-based approach guarantees a better imputation quality. Furthermore, there are some datasets where SDAE is the top winner, especially for MR 40% (dermatology, hcc-data-survival, hcc-data-mortality, lung-cancer, among others), although this is not generalisable for all datasets.
Impact on classification (F-measure). The results show that SVMimp seems to be the best imputation method in terms of classification performance, regardless the missing mechanism and configuration considered. This is shown in Table 1, where for the three highest MRs (15, 20 and 40%) SVMimp is the winner approach for most of the studied scenarios, although for MNAR univa configuration and under a MR of 40%, SDAE is the best approach. For the lowest MRs (5 and 10%) there is no standard, suggesting that small amounts of missing values have little influence on the quality of the dataset for classification purposes – there is an exception for the univa configurations under 5% of MR: in this case, SVM is the winner approach. Regarding the classification performance of the SDAE-based approach, it belongs to the top 3 imputation approaches for MNAR configurations under higher MRs (20 and 40%) .
We continue this section by referring to the results obtained by Gondara et al.[13], who used a similar benchmarking of datasets (although smaller, with only 15 datasets) and a SDAE approach. Gondara et al.[13] proposed a SDAE based model for imputation but only compare its results with MICE. Therefore, we also perform this comparison, only for unifo configuration, and present the respective results in Fig. 2. The SDAE seems to perform better than MICE for MNAR data - this is always the case for higher missing rates (15, 20 and 40%), regardless of the used metric.
We also performed a statistical test (Wilcoxon rank-sum) in order to verify if there were significant differences between the results obtained by the SDAE and the best method for the classification and imputation (MICE for RMSE and SVMimp for F-measure). In terms of RMSE and for most of the studied scenarios, the p-value reveals strong evidence against the null hypothesis, so we reject it - meaning that there are significant differences between the two methods, MICE and SDAE. For some scenarios where SDAE seems to be superior – MNAR unifo under 15 and 20% of MR – the p-value reveals weak evidence against the null hypothesis and therefore we can not ensure that there are significant differences between SDAE and MICE. Regarding F-measure and for almost all of the studied scenarios, we obtained a p-value that indicates strong evidence against the null hypothesis, so we reject it, meaning that there are significant differences between the two methods, SVMimp and SDAE. Since SVMimp has a higher performance, it does not seem that using SDAE brings any advantage in terms of classification performance.
5 Conclusions and Future Work
This work investigates the influence of different missing mechanisms on imputation methods (including a deep learning-based approach) under several missing rates. This influence is evaluated in terms of imputation quality (RMSE) and classification performance (F-measure). Our experiments show that MICE performs well in terms of imputation quality while SVMimp seems to be the method that guarantees the best classification results.
We also compare the behaviour of SDAE with well-established imputation techniques included in related work: for standard datasets, such as those we have used, SDAE does not seem to be superior to the remaining approaches, since the obtained results do not outperform all of the state-of-the-art methods. Furthermore, the simulations become more complex with the use of deep networks due to both computational time and space/memory required.
As future work, we will investigate the usefulness of SDAE when handling more complex datasets (higher number of samples and dimensionality). Also, as the advantage of SDAE seems to be more clear for higher missing rates (40%), a smoother step of missing rates (between 20% and 40%) could bring new insights.
References
Abreu, P.H., Santos, M.S., Abreu, M.H., Andrade, B., Silva, D.C.: Predicting breast cancer recurrence using machine learning techniques: a systematic review. ACM Comput. Surv. (CSUR) 49(3), 52 (2016)
Amorim, J.P., Domingues, I., Abreu, P.H., Santos, J.: Interpreting deep learning models for ordinal problems. In: 26th European Symposium on Artificial Neural Networks, Computational Intelligence and Machine learning (ESANN), pp. 373–378 (2018)
Azur, M.J., Stuart, E.A., Frangakis, C., Leaf, P.J.: Multiple imputation by chained equations: what is it and how does it work? Int. J. Methods Psychiatr. Res. 20, 40–49 (2011)
Beaulieu-Jones, B.K., Moore, J.H.: Missing data imputation in the electronic health record using deeply learned autoencoders. In: Altman, R.B., Dunker, A.K., Hunter, L., Ritchie, M.D., Klein, T.E. (eds.) PSB, pp. 207–218 (2017)
Charte, D., Charte, F., García, S., del Jesus, M.J., Herrera, F.: A Practical Tutorial on Autoencoders for Nonlinear Feature Fusion: Taxonomy, Models, Software and Guidelines, vol. 44, pp. 78–96. Elsevier (2018)
Dempster, A.P., Laird, N.M., Rubin, D.B.: Maximum likelihood from incomplete data via the EM algorithm. 39, 1–22 (1977)
Duan, Y., Lv, Y., Kang, W., Zhao, Y.: A deep learning based approach for traffic data imputation. In: ITSC, pp. 912–917. IEEE (2014)
Duan, Y., Lv, Y., Liu, Y.L., Wang, F.Y.: An efficient realization of deep learning for traffic data imputation. Transp. Res. Part C: Emerg. Technol. 72, 168–181 (2016)
García-Laencina, P.J., Sancho-Gómez, J.L., Figueiras-Vidal, A.R.: Classifying patterns with missing values using multi-task learning perceptrons. Expert Syst. Appl. 40, 1333–1341 (2013)
García-Laencina, P.J., Abreu, P.H., Abreu, M.H., Afonso, N.: Missing data imputation on the 5-year survival prediction of breast cancer patients with unknown discrete values. Comput. Biol. Med. 59, 125–133 (2015)
García-Laencina, P.J., Sancho-Gómez, J.L., Figueiras-Vidal, A.R.: Pattern classification with missing data: a review. Neural Comput. Appl. 19, 263–282 (2009)
Garciarena, U., Santana, R.: An extensive analysis of the interaction between missing data types, imputation methods, and supervised classifiers. Expert Syst. Appl. 89, 52–65 (2017)
Gondara, L., Wang, K.: Multiple imputation using deep denoising autoencoders. Department of Computer Science, Simon Fraser University (2017)
Gondara, L., Wang, K.: Recovering loss to followup information using denoising autoencoders. Simon Fraser University (2017)
Little, R.J., Rubin, D.B.: Statistical Analysis with Missing Data. Wiley, New York (1987)
Man-long, Z.: MkNNI: new missing value imputation method using mutual nearest neighbor. Mod. Comput. 31, 001 (2012)
Ning, X., Xu, Y., Gao, X., Li, Y.: Missing data of quality inspection imputation algorithm base on stacked denoising auto-encoder. In: 2017 IEEE 2nd International Conference on Big Data Analysis (ICBDA), pp. 84–88. IEEE (2017)
Rieger, A., Hothorn, T., Strobl, C.: Random forests with missing values in the covariates. Department of Statistics, University of Munich (2010)
Sang, G., Shi, K., Liu, Z., Gao, L.: Missing data imputation based on grey system theory. Int. J. Hybrid Inf. Technol. 27(2), 347–355 (2014)
Santos, M.S., Abreu, P.H., García-Laencina, P.J., Simão, A., Carvalho, A.: A new cluster-based oversampling method for improving survival prediction of hepatocellular carcinoma patients. J. Biomed. Inform. 58, 49–59 (2015)
Santos, M.S., Soares, J.P., Henriques Abreu, P., Araújo, H., Santos, J.: Influence of data distribution in missing data imputation. In: ten Teije, A., Popow, C., Holmes, J.H., Sacchi, L. (eds.) AIME 2017. LNCS (LNAI), vol. 10259, pp. 285–294. Springer, Cham (2017). https://doi.org/10.1007/978-3-319-59758-4_33
Sánchez-Morales, A., Sancho-Gómez, J.-L., Figueiras-Vidal, A.R.: Values deletion to improve deep imputation processes. In: Ferrández Vicente, J.M., Álvarez-Sánchez, J.R., de la Paz López, F., Toledo Moreo, J., Adeli, H. (eds.) IWINAC 2017. LNCS, vol. 10338, pp. 240–246. Springer, Cham (2017). https://doi.org/10.1007/978-3-319-59773-7_25
Twala, B.: An empirical comparison of techniques for handling incomplete data using decision trees. Appl. Artif. Intell. 23, 373–405 (2009)
Vincent, P., Larochelle, H., Bengio, Y., Manzagol, P.A.: Extracting and composing robust features with denoising autoencoders. In: International Conference on Machine Learning proceedings (2008)
Vincent, P., Larochelle, H., Lajoie, I., Bengio, Y., Manzagol, P.A.: Stacked denoising autoencoders: learning useful representations in a deep network with a local denoising criterion. J. Mach. Learn. Res. 11, 3371–3408 (2010)
Xia, J., Zhang, S., Cai, G., Li, L., Pan, Q., Yan, J., Ning, G.: Adjusted weight voting algorithm for random forests in handling missing values. Pattern Recognit. 69, 52–60 (2017)
Zeiler, M.D.: Adadelta: an adaptive learning rate method. arXiv preprint arXiv:1212.5701 (2012)
Zhu, B., He, C., Liatsis, P.: A robust missing value imputation method for noisy data. Appl. Intell. 36(1), 61–74 (2012)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer Nature Switzerland AG
About this paper
Cite this paper
Costa, A.F., Santos, M.S., Soares, J.P., Abreu, P.H. (2018). Missing Data Imputation via Denoising Autoencoders: The Untold Story. In: Duivesteijn, W., Siebes, A., Ukkonen, A. (eds) Advances in Intelligent Data Analysis XVII. IDA 2018. Lecture Notes in Computer Science(), vol 11191. Springer, Cham. https://doi.org/10.1007/978-3-030-01768-2_8
Download citation
DOI: https://doi.org/10.1007/978-3-030-01768-2_8
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-01767-5
Online ISBN: 978-3-030-01768-2
eBook Packages: Computer ScienceComputer Science (R0)