Abstract
Transcriptomics approaches have been advancing quickly in the past 20 years. Technologies including various arrays and sequencing technologies are used to determine expression of RNA transcripts in cells and tissues. While the earlier approaches focus on the messenger RNA (mRNA), all other types of RNA transcripts, such as micro RNAs and non-coding RNAs, can be easily studied using current approaches. For skeletal muscle research, investigators use transcriptomics approaches to study basic muscle biology; molecular responses to physiological and environmental stimuli; effects of aging on muscles; disease mechanism of muscle disorders; molecular changes in muscles of non-muscle diseases; and molecular responses to therapeutic. This chapter focuses on current approaches and platforms used in the studies. Platform selections and limitations as well as data analysis will be discussed. Emerging approaches such as single cell profiling, single nucleus profiling, modified RNA profiling, and spatial transcription profiling are described in the chapter.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
The transcriptome refers to all RNA molecules transcribed from our genome, including messenger RNA (mRNA), ribosomal RNA (rRNA), transfer RNA (tRNA), as well as other regulatory noncoding RNAs. Some well-known regulatory noncoding RNA molecules are long noncoding RNA (lncRNA), microRNA (miRNA), small nuclear RNA (snRNA), and small nucleolar RNA (snoRNA). Among the different types of RNA, the protein-coding mRNA received the most attention, particularly when the tools for RNA profiling, or gene expression profiling, were first developed in the late twentieth century. Since then, different tools and sample preparation techniques have been developed to target various groups of RNAs for study. This chapter will focus on mRNA profiling approaches primarily; however, the same or modified technologies can be used to study other RNA groups. The approach for studying miRNA, miRNA profiling, is described in a separate chapter.
Transcriptomic studies are conducted to understand how transcriptome differences and changes contribute to biological functions and diseases. For skeletal muscle research, a large number of studies have been published in the past 20 years using arrays and sequencing approaches to investigate (1) basic muscle biology [1,2,3,4,5]; (2) molecular responses to physiological and environmental stimuli [6,7,8,9,10]; (3) effects of aging on muscles [11, 12]; (4) disease mechanism of muscle disorders [13,14,15,16,17,18]; (5) molecular changes in muscles of non-muscle diseases [19,20,21]; and (6) molecular responses to therapeutic interventions [22,23,24]. Most of these transcriptome data sets have been deposited into public databases, such as the Gene Expression Omnibus (GEO) database hosted by the National Center for Biotechnology Information (NCBI), National Institutes of Health (NIH) (https://www.ncbi.nlm.nih.gov/geo/). This provides a rich source of information for researchers today. For example, one can query and download data of interest and conduct further analyses. In this chapter, we will focus on the most commonly used platforms and approaches to generate these data sets.
1 Theory and History of the Technique
Gene expression is the process wherein information from a gene is used in the production of a functional gene product. When the final product is a protein, the process involves “transcription” which produces mRNA using DNA as the template and “translation” which produces proteins using the information provided by the mRNA transcripts. The size of human genome is estimated to be 3.3 billion base pairs, and approximately 5% of it can be transcribed and produce RNA products. Of the whole transcriptome, only ~4% of the transcribed RNAs are protein-coding mRNAs [25]. The amount of mRNA changes slightly depending on the cell types. Dividing cells that are more active transcriptionally produce more mRNA transcripts compared to terminally differentiated cells, such as myofibers. In addition to mRNA quantity, gene expression profiles are different in different cell types and cell states. Controlling which genes are expressed at a given time enables the cell to control its size, shape, and functions. In other words, while all cells carry the same DNA content, only a small portion of the DNA is actively transcribed at any one point in time. Among the transcripts produced, only a small portion is protein-coding RNAs. Transcriptomic approaches allow us to investigate differences in these transcripts in muscle tissues and cells in different physiological conditions and disease states. By comparing samples of interests to the controls, one may discover important molecular changes and pathways that play critical roles in the conditions or diseases under study. Because many regulatory steps in addition to transcription are involved in regulating protein synthesis and function, one should not make conclusions solely based on mRNA profiling data. Previous studies showed that mRNA expression levels positively correlate with protein levels; however, the change of mRNA level does not fully explain changes in proteins because posttranslational modifications such as phosphorylation and methylation may have further effects on protein function [26,27,28,29,30].
Gene expression profiling measures RNA transcripts at a given moment, which provides a “snapshot” of the gene activities in cells in a specific state or condition. In general, methods for determining RNA transcript levels can be based on (1) transcript visualization, (2) transcript hybridization, or (3) transcript sequencing. Because RNA molecules are prone to degradation, they are usually reverse-transcribed into cDNA (complementary DNA) before further processing. However, there are exceptions which will be discussed later. Polymerase chain reaction (PCR) arrays use fluorescent dyes to detect and quantify the amount of PCR amplicons of each transcript in a sample. Microarrays use cDNA or oligonucleotide probes to detect the transcripts by hybridization methods. The next-generation sequencing approach directly sequences the transcripts and determines the expression level of each transcript by the number of reads. Earlier technologies for expression profiling (cDNA arrays or PCR-based arrays) measure the expression of hundreds to thousands of genes at a time. These arrays can be made in-house or custom-made by companies. Afterward, microarrays were commercially developed by several companies to increase coverage, throughput, sensitivity, and specificity. Some of the platforms are capable of surveying tens of thousands of transcripts and allow genome-wide profiling. Commonly used platforms include Affymetrix GeneChip® Microarrays, Illumina High-Density Silica Bead-Based Microarrays, and Agilent Expression Microarrays. For these microarray platforms, oligonucleotide probes at various lengths accompanied by different array designs are used to capture the target transcripts by hybridization methods. Transcripts that have not been discovered or are not included on the microarrays will not be detected or measured using these platforms. This limitation was resolved by the next-generation sequencing technologies, which first became available at the beginning of the 2000s [31, 32]. Different from the traditional Sanger sequencing, next-generation sequencing (NGS) does not target specific sequences. Instead, all transcripts in a sample are sequenced. This unbiased approach in theory can survey the whole transcriptome including both coding and noncoding RNA. However, due to the limitation of how many reads can be obtained in one run and one may only be interested in a specific type of RNA transcripts, different protocols, including primer types and enrichment methods, were developed to examine specific types of RNA transcripts.
2 Major Applications
2.1 Hybridization-Based Platforms
The principle of microarrays is nucleic acid hybridization, in which two complementary strands of DNA or RNA molecules join to form a double-stranded molecule, following the complimentary base pairing rules: adenine (A) pairs with thymine (T) and cytosine (C) pairs with guanine (G). Northern blotting uses probes that are complementary to the target RNA to detect an RNA that is immobilized on a nitrocellulose or nylon membrane. To detect many RNA transcripts at the same time, instead of immobilizing the RNA samples, many different probes are immobilized, i.e., spotted onto the membrane or glass slide and then used to detect the target RNA transcripts in samples adding to the immobilized probes. In this case, RNA transcripts are reverse-transcribed into cDNA and then hybridized to the probes on the slides. The cDNA fragments are fluorescently labeled for visualization. The prototype of the cDNA microarray made by spotting known cDNAs into a 96-well microplate was reported in 1995, on which cDNAs were printed by robot [33]. Differential expression of 45 Arabidopsis genes were measured simultaneously using two-color fluorescence approach. In1996, 870 cDNAs were spotted on glass slides to determine gene expression for cancer classification [34]. Two different colors are used for the platform, the sample of interest is labeled with one color (e.g., Cy3), and a reference sample is labeled with a different color (e.g., Cy5). Both samples are hybridized to the same array and the ratio between the experiment and the reference samples are used for further data analyses. This design allows proper normalization and comparisons among different arrays (Fig. 5.1). One of the earliest expression profiling studies done in skeletal muscles was reported in 1996, in which membranes with cDNA clones spotted on them were used [5]. The early versions of cDNA arrays provide platforms for examining hundreds of transcripts simultaneously. There are several issues associated with the cDNA arrays, such as sensitivity, specificity, normalization, reproducibility, throughput, and coverage which were addressed in later generations of microarrays. Next, we describe three commercial platforms that are commonly used for expression profiling studies. A comparison of the three platforms is in Table 5.1 at the end of this section.
2.1.1 Affymetrix
Affymetrix makes GeneChip arrays for transcriptome analysis using millions of 25-base DNA probes that have been synthesized directly onto a glass chip using light-directed oligonucleotide synthesis method [35]. The probes are used to “probe” the sample for target RNA segments. Hybridization is the basis for the detections. Because the 25-mer is relatively short and can potentially bind to more than one transcript, instead of one probe, a set of 11 probes are designed for each transcript. The intensity data from all 11 probes are analyzed to determine whether the transcript is detected (called “present”) or not detected (called “absent”) and give a measure of the level of expression. In an experiment, the mRNA is reverse-transcribed into double-stranded cDNA. The cDNA is then used as the template for in vitro transcription. The RNA produced is known as cRNA, in which the uracil bases are labeled with biotin. The fragments of biotin-labeled cRNA are then loaded into the array so that the sample can hybridize with the probes on the glass chip of the array. Afterward the array is washed to remove RNA that has not hybridized to the probes. The hybridization is then visualized using streptavidin-linked fluorescent dyes that bind to the biotin-labeled cRNA. The array is scanned with a laser scanner and the image is analyzed to determine which transcripts are detected and how much of each transcript is present.
In 2000, Affymetrix GeneChip® HuGeneFL Arrays were used to identify transcriptomic changes underlying disease mechanisms of muscular dystrophies [18]. In this study, approximately 30–40% of the known human transcripts that were on the array were called “present”. Based on the data collected from the array, the group developed a custom-made muscle array which contains genes that are expressed in skeletal muscles [36, 37]. The Affymetrix GeneChip® HuGeneFL Array is the first version of human whole genome array and contained approximately 5000 full-length human sequences. Several different versions containing more sequences to cover the whole genome were subsequently developed, and the final/latest human version, Clariom™ D Assay, consists of more than 5,40,000 transcripts, including alternative splicing isoforms of both coding and noncoding RNAs. Microarrays for other species, including mouse, rat, nonhuman primates, insects, livestock, bird and fish, and small mammals, are also currently available for expression profiling studies. In addition to the arrays for mRNA and long noncoding RNA, GeneChip™ miRNA Arrays are available for studying small noncoding RNA, including miRNA, snoRNA, and scaRNA.
2.1.2 Illumina
Illumina Whole-Genome Gene Expression BeadChip consists of oligonucleotides immobilized to beads which are held in microwells on the array [38]. Up to 30 beads are available for each probe to improve data quality and reproducibility. The beads are randomly distributed across the array, one bead per well, and a 29-mer address sequence present on each bead is used for mapping the location of the beads on the array. In addition to the unique bead design, the BeadChip microarray is deployed on multi-sample array formats. Four to 24 uniform pits can be on each array, and multiple samples can be loaded to the Illumina Expression BeadChip arrays, which increases throughput and reduced sample-to-sample variations. The length of the probe is 50 bases which is synthesized in solution and then cross-linked to the beads. Labeled cRNA segments are hybridized to the probes on the BeadChip. After hybridization, washing, and staining, the image data are acquired by a scanner. The HumanHT-12 Expression BeadChip simultaneously profiles more than 47,000 transcripts representing 28,688 well-annotated genes. The transcription level is calculated using average of the signals from all the beads. In addition, some genes can be detected by more than one probe. For formalin-fixed paraffin-embedded (FFPE) samples, the Illumina developed the DASL Assay for handling degraded RNA, including muscle samples [39, 40].
2.1.3 Agilent Technologies
Agilent SurePrint G3 Gene Expression Microarrays are made with probes of 60 bases long [41]. The probes are synthesized onto the glass slides directly by printing A, T, C, G using an inkjet-like printer at hundreds of thousands of spots on the slide. After each nucleotide is added, a chemical de-blocking step is used to allow the next nucleotide in the chain to be added. The probes grow to the full length at the end. Agilent SurePrint used to be a two-color system Cy3 and Cy5). Now a one-color only (Cy3) system is available for the users. The advantage of this approach is that it makes updating of stock microarrays possible/easier as new gene information becomes available. In addition, custom-designed arrays targeting transcripts of specific interest can be created using this platform. The probes used here are relatively longer compared to the probes on the Affymetrix arrays but shorter than the traditional cDNA array. The length of the oligonucleotides is at a balance point for better sensitivity and specificity. Currently, human, mouse, and rat microarrays that cover coding and noncoding transcripts from the NCBI Reference Sequence (RefSeq) database are available. The coding and noncoding transcripts from RefSeq database are curated and nonredundant sequences.
2.2 Sequencing-Based Platforms
Next-generation sequencing (NGS)-based RNA sequencing (short for RNA-seq) is a highly sensitive and accurate method for gene expression profiling analysis that provides insight to previously undetectable changes in gene expression, as well as enabling the characterization of multiple forms of noncoding RNA. With RNA-seq, researchers can detect the various structures of the transcriptome, such as transcript isoforms, gene fusions, single nucleotide variants, and other features, without the limitation of prior knowledge. RNA-seq (1) provides sensitive, accurate measurement of gene expression at the transcript level; (2) generates both qualitative and quantitative data; (3) detects and sequences small RNAs and multiple forms of noncoding RNA, such as small interfering RNA (siRNA), microRNA (miRNA), small nucleolar (snoRNA), and transfer RNA (tRNA); (4) identifies alternatively spliced isoforms, splice sites, and allele-specific expression in a single experiment; (5) provides data sets that are not biased or restrained by existing knowledge; (6) obtains allele-specific information in the data; and (7) scales for large studies and high sample numbers. As researchers seek to understand how the transcriptome shapes biology, RNA-seq is becoming one of the most significant and powerful tools in modern science. In addition, these sequencing-based methods are more cost-effective in comparison to microarrays and real-time RT-PCR. A comparison of the major platforms is in Table 5.2 at the end of this section.
2.2.1 Illumina
The Illumina platforms are the most commonly used sequencers for next-generation sequencing. The technology has been used extensively for diagnosis of muscle diseases [42, 43]. It has also been used for studying muscle transcriptome to answer various biological questions, such as annotations of muscle transcripts, muscle disease mechanisms, and basic muscle biology [44,45,46]. Sample preparation for mRNA-seq using the Illumina platform involves isolation of RNA and chemical fragmentation of RNA, followed by reverse transcription to generate double-stranded cDNA fragments that are then sequenced. Figure 5.2 illustrates a simplified workflow of the process. Sequencing adaptors which contain barcodes are ligated to the cDNA fragments before size selection by gel electrophoresis. The fragments at desired size are then excised for sequencing. For example, fragments ranging from 250 to 400 bases are collected for regular RNA-seq. To sequence shorter fragments, e.g., to target shorter miRNA, fragments of 150 bases or less are collected. A relatively low number of cycles (9–12 cycles) of PCR amplification are performed to increase the template. Alternatively, PCR-free kits are now available to reduce biases and contamination that may be introduced during PCR amplification.
Workflow of Illumina RNA-seq. Total RNA (or mRNA) is isolated, followed by RNA fragmentation, cDNA generation, linker ligation, fragment enrichment, and size selection to prepare library for sequencing. Next-generation sequencing is a massive parallel sequencing which generates tens of billions of bases of sequences. Downstream data analyses followed
DNA libraries are hybridized to the primer lawn on the flow cell by an automated Cluster Station (cBot). Single-stranded cDNA fragments are washed across the flow cell and bind to primers on the surface of the flow cell. DNA that doesn’t attach is washed away. The DNA attached to the flow cell is then replicated to form small clusters of DNA with the same sequence. This design (a cluster of the same molecule instead of only one molecule) allows florescent signals emitted during the sequencing process strong enough to be detected by a camera. During the sequencing process, the primers are first added. The DNA polymerase then adds the first fluorescently labeled terminator bases (A, C, G, and T) to the new DNA strand. Laser lights are used to activate the fluorescent label on the nucleotide base. This fluorescence is detected by a camera and recorded on a computer. Each of the terminator bases (A, C, G, and T) give off a different color. The fluorescently labeled terminator group is then removed from the first base, and the next fluorescently labeled terminator base can be added, and the process continues until the fragments are fully sequenced. Illumina sequencer sequences short reads (50 or 250 bp depending on the version) and can sequence both ends of the molecules (paired ends).
2.2.2 Ion Torrent
The major advantage of the Ion Torrent platform is lower cost and rapid sequencing speed [47]. After reverse transcription, single-stranded DNA templates are loaded to a semiconductor chip. Unmodified A, C, G, or T dNTP are then added individually along with DNA polymerase enzyme. If an introduced dNTP is complementary to the next unpaired nucleotide of the DNA template, it will be incorporated into the complementary strand by DNA polymerase. During the incorporation process, hydrogen and pyrophosphate are released. The releasing of the hydrogen is detected by a sensor and used to determine the base at that position in the DNA template. With a portfolio of chips of varying outputs, the Ion GeneStudio™ S5 Plus and Ion PGM™ Sequencers scale to a variety of RNA-seq applications for a broad range of transcriptome sizes. For example, the Ion 550 Chip generates 100–130 million sequencing reads on the Ion GeneStudio™ S5 Plus and Prime Systems using the automated workflow of the Ion Chef System. The sequencing run time is as little as 2.5 h, with only 15 min of sample preparation time. Torrent Suite Software processes and exports the sequence reads in FASTQ or BAM formats, which can be easily imported into third-party software, such as Partek Flow software packages, for further analyses. The read length has increased from 50 bases in the original model to 400 bases in the latest model. Few examples of using this technology include studies to dissect molecular pathways involved in myogenic differentiation and de novo assembly of a muscle transcriptome [48,49,50].
2.2.3 Pacific Biosciences
PacBio uses single-molecule real-time (SMRT) sequencing technology for long-read sequencing, which allows sequencing full-length cDNA without read assembly [51]. The strengths of the system are that it is able to easily identify and quantify new transcripts and alternative splicing isoforms. The SMRT sequencing is built upon zero-mode waveguides (ZMWs) and phospholinked nucleotides. Zero-mode waveguide is an optical waveguide that guides light energy into a volume that is small in all dimensions compared to the wavelength of the light. Tens of thousands of tiny wells with ZMWs are in the SMRT cell, in which one cDNA template molecule is immobilized. Phospholinked nucleotides labeled with four different fluorophores allow observation of the addition of the nucleotides as the DNA polymerase producing the complementary strand. PacBio sequencer has advantages of long read lengths, simultaneous epigenetic characterization, and single-molecule resolution. The disadvantage is that the error rate is higher compared to the traditional short-read sequencing. Currently, muscle transcriptomic data generated using this technology were frequently used to improve annotation of an incomplete genome [52, 53].
2.2.4 Oxford Nanopore
Oxford Nanopore sequencing can directly sequence single molecule of DNA or RNA without the need for PCR amplification or chemical labeling of the sample. The flow cell contains more than a thousand nanopores which are nanoscale holes on a membrane. In the device, ionic currents pass through the nanoscale holes, and changes in current that occur as biological molecules pass through the nanopore are recorded by a sensor [54, 55]. Computational algorithms are used to analyze the data to reduce which of the four nucleotides passed through the pore. The approach can be used to sequence DNA, RNA, and protein. When the RNA transcripts are sequenced, the RNA samples can be reverse-transcribed into cDNA for sequencing or directly sequenced. When the RNA is sequenced directly using the technology, modifications of the bases (e.g., inosine, N6-methyladenosine, and N5-methylcytosine) can be detected based on the subtle differences in the current changes [56]. However, the call heavily relies and depends on the algorithms used for data analyses. While quickly improving, this platform still has the higher error rates in comparison to other major platforms. One advantage of the platform is the transportability of the smaller device, such as MinION. The pocket-size device has been brought into space and used in Antarctica as well as in rural areas for experiments and fieldwork [57,58,59,60,61]. This provides new opportunities for clinical diagnostic and research use. Nanopore sequencing has the potential to offer relatively low-cost sequencing, high mobility for testing, and rapid processing of samples with the ability to display results in real time. The device is the only sequencer to date that is able to sequence full-length RNAs.
2.3 Reverse Transcriptase Polymerase Chain Reaction (RT-PCR) Assay
Microarrays and next-generation sequencing are high-throughput approaches, which are great screening tools for identifying transcriptional changes in samples. The RT-PCR assay is the most commonly used method to validate the changes identified. Real-time RT-PCR allows accurate quantification because the amount of PCR amplicons in a sample is measured real time after each PCR cycle, which allows proper selection of data points for RNA quantification based on the rate of amplification. In the method, RNA is reverse-transcribed into cDNA, followed by detecting and real-time monitoring of the presence of PCR products using florescent dyes. The dyes can be either SYBR™ Green, which can be incorporated into the DNA amplicons directly, or a fluorescently labeled probe that can bind to the target sequence. In addition to individual assay, multiple assays can be performed to examine gene networks and pathways. One example is the TaqMan® Gene Expression Assay developed by Applied Biosystems. The assay utilizes the 5′′ nuclease activity of the Taq DNA polymerase to cleave the fluorescently labeled probe. Each assay includes a single FAM™ dye-labeled TaqMan® probe with a minor groove binder (MGB) moiety and two unlabeled oligonucleotide primers. The assays were designed based on transcripts obtained from the NCBI Reference Sequence Project database (RefSeq). QuantStudio™ 12K Flex OpenArray® Plate allows users to specify the TaqMan® Gene Expression Assays to be included on the plate. Each plate generates 2600 data points. The advantage is the flexibility of the design which allows easy customization. The TaqMan® Array Human MicroRNA Card Set v3.0 is a two-card set containing a total of 384 TaqMan® MicroRNA Assays per card, which can be used to assay miRNAs that are differentially expressed. It was used to identify miRNAs that are differentially expressed between disease and healthy muscle cells in order to understand disease mechanisms [62].
The PCR-based technique is highly sensitive, and no pre-amplification is needed. In addition to being performed in an arrayed format for large-scale analysis, real-time RT-PCR is the gold standard technique for validating differential expressed genes identified by other high-throughput methods. Either absolute or relative quantification can be performed. Standard curve can be included to allow comparison among different plates. For validating expression changes less than twofold, digital PCR (dPCR) can be considered for their better resolution. The dPCR is a quantitative PCR method that the initial sample mix is partitioned into many individual wells prior to the PCR amplification step, resulting in either 1 or 0 targets being present in each well. Following PCR amplification, the number of positive and negative reactions is determined, and the absolute quantification of target transcript is calculated using Poisson statistics.
2.4 Single-Cell RNA Sequencing
Traditional gene expression profile analyses analyze the expression of RNAs from tissues or large populations of cells. In such mixed-cell populations, these measurements may obscure critical differences that exist between individual cells, e.g., diseased vs viable, replicating vs senescent. Single-cell RNA sequencing (scRNA-seq) allows expression profiling in individual cells. This can reveal the existence of rare cell types within a cell population that have not previously been known. In addition, scRNA-seq can be used to examine expression variations among the same type of cells. Briefly a scRNA-seq experiment involves several steps: (1) single-cell capture; (2) single-cell lysis; (3) reverse transcription of the RNA; (4) library preparation; and (5) sequencing and data analysis (Fig. 5.3). Currently, common single-cell sequencing platforms include 10X Genomics Chromium, Drop seq, and Fluidigm C1. These platforms are primarily used for single-cell capture, processing, and library preparation. A few potential applications of scRNA-seq include characterization of cell types, elucidation of gene regulatory networks, drug resistance clone identification, noninvasive biopsy diagnosis, stem cell lineage regulatory networks, assessing tumor heterogeneity, and CRISPR screening.
Workflow of single-cell sequencing. Single-cell RNA sequencing starts with single-cell dissociation from the tissue sample, followed by single-cell capture. The captured cells then undergo cell lysis, reverse transcription, and fragment enrichment in preparation for sequencing and downstream data analysis
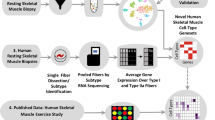
Several studies utilizing the scRNA-seq technology to study skeletal muscles suggested that muscle stem cells are a heterogeneous cell population with substantial biochemical and functional diversity [63,64,65]. scRNA-seq was also used to provide critical insights to disease mechanisms of the muscle disorder, such as facioscapulohumeral muscular dystrophy (FSHD). The majority of the FSHD (FSHD1) cases are caused by the contraction of a macrosatellite array called D4Z4 at the chromosome 4q35. This mutation in combination with a permissive genomic feature in the region allows the aberrant expression of double homeobox protein 4 (DUX4) protein. The aberrant expression of the DUX4 leads to downstream molecular changes that cause the disease [15, 16, 66, 67]. It has been reported that the DUX4 is not expressed in all cells and is present in only approximately 1 in 1000 proliferating myoblasts in culture [68]. Additional studies suggested that the expression of DUX4 is stochastic due to the epigenomic changes at the D4Z4 region [69, 70]. The scRNA-seq studies allowed the investigators to determine expression patterns in individual cells and conclude that a DUX4 expression induced a series of downstream expression changes [71].
3 Data Analyses
3.1 Microarrays
There are many bioinformatics tools/algorithms available to analyze microarray data. Some are command line based (mostly in R) and some are user interface (UI) based. GeneSpring from Agilent and Transcriptome Analysis Console (TAC) from Affymetrix are UI based, while limma and affy are R command line based [72, 73]. The basic analysis of microarray data can be broadly divided into steps. The first step is data extraction, preprocessing, and normalization and the second is identifying differentially expressed genes between samples, across different conditions. The affy package from the Bioconductor suite of tools in R extracts intensity data from .CEL files. Similarly, GeneSpring from Agilent Technologies helps in intensity extraction from raw microarray data sets from Agilent. The beadArray package from the R/Bioconductor suite of tools can be used to extract intensity values from raw images from beadArray experiments [74].
For any experiment related to the evaluation of expression differences between two groups, it is important to carry out the same experiment multiple times. This increases the sample size to perform statistical significance tests for differential expression, as well as reducing the bias in the experiment. This is typically done using one of the following methods depending on the purpose of the repeated samples. The biological replicates are samples extracted from multiple biological entities, under the same biological condition. For example, to study the effect of a drug on a muscle disease, 3 affected mice are given a drug (experimental condition) and 3 mice are not administered the drug (control). RNA is extracted from these 6 mice individually, to yield 3 biological replicates for the experiment and the control conditions. The purpose of biological replicates is to draw conclusions on the larger population of samples/controls. The technical replicates are samples extracted multiple times from the same biological entity. In this case, one mouse affected is given the drug (experiment) and the other mouse receives no drug (control). RNA is extracted once from each of the mice and RNA is separated into 3 portions each, to run 3 controls and 3 experimental samples in the microarray experiment. The purpose of technical replicates is to measure how much variation in the quantification can be expected due to technical conditions. Note technical replicates are not, and should not be, considered biological replicates; they are not used to make conclusions about the population of the mice.
In any experiment, not all replicates can be run at the same time on the same machine under the exact same conditions. Replicates may have to be run on consecutive days or on two different instruments. These conditions introduce systemic variations which, in turn, can cause variation in the expression of genes that should be identical. To quantify and account for these run-to-run variations, normalization methods are applied. There are two types of normalization, between and within arrays. Between array methods normalize the expression of genes across the multiple arrays (i.e., replicates), while within array methods normalize expression across genes within an array (within the same replicate). One of the most commonly used methods of between array normalization is quantile normalization [75]. The RMA function in the affy package, part of the Bioconductor suite of tools and GeneSpring from Agilent technology, not only performs quantile normalization but also performs background correction, probe-level intensity calculation, and probe set summarization. For within array normalization, Loess normalization is typically used and implemented by the limma package, part of the Bioconductor suite of tools. Other platforms like nimblegen and illumine have the packages oligo [76] and beadArray, respectively, which extract intensity and normalize data using the RMA function.
To evaluate possible changes in expression pattern across replicates, it is important to use visualization in combination with statistical methods. A useful visualization method is the heatmap.2 function in the gplots R package [77], which combines heatmap functionality with hierarchical clustering to visualize the expression patterns across replicates. Ideally, a normalized data set should show a similar expression pattern for a given gene across multiple replicates. If a significant difference in expression patterns is observed, one needs to first verify that the correct file was used for a given replicate, an easy mistake to make given the similarity in file names. Next, different normalization methods should be used to verify that they show the same general pattern. Principal component analysis (PCA) [77] and box plots are used by Genespring to visualize the expression patterns across samples. This normalization allows one to be confident that the quantification of gene expression from different samples can be directly compared when assessing differences in gene expression affected by an introduced factor (i.e., treatment, disease, etc.).
When comparing two or more groups, one can identify genes that are differentially expressed using statistical analyses. Difference in expression is expressed as fold change (experimental gene expression/control gene expression) or logarithmic fold change [log2 (experimental gene expression/control gene expression)]. Statistical significance is measured using hypothesis testing based on the design of the experiment. See the chapter of bioinformatics and statistics for omics data for details on the appropriate statistical method to use. Often, for designs where there is a single experimental condition and a single control condition, Student’s t-test is used [78], whereas ANOVA is commonly used for complex designs where there are multiple experimental and control conditions [79]. Be advised however that both Student’s t-test and ANOVA have assumptions that the data must meet for the tests to be valid. Multiple testing correction methods such as the Benjamini-Hochberg [80] or Bonferroni [81] are used to limit false positives. Base R packages are available to perform t-tests (t.test()), ANOVAs (aov()), and multiple testing p-value correction (p.adjust). GeneSpring uses ANOVA in their differential expression analysis. Another program, limma, is a Bioconductor package which is based on linear regression models and is one of the most popular differential expression analysis tools. Before beginning the analysis of differential expression, a clear understanding of the appropriate statistical method to use is essential, as using the incorrect method can lead to invalid conclusions. Once genes are found to be differentially expressed between conditions, they can be further analyzed by various means including clustering analyses and functional annotation and gene ontology methods. In addition, Ingenuity Pathway Analyses, a web-based commercial tool, can be used to identify networks, pathways, and regulatory relationships among the identified transcripts.
3.2 RNA-Sequencing (RNA-Seq)
High-throughput sequencing techniques produce either short (50–75 bp) reads like those generally produced by Illumina sequencers or longer reads produced by newer methods such as PacBio (30–50kbp) [82] and Oxford Nanopore (can be longer than 100K) [55, 83] systems. Read length is an important parameter, as longer reads are better able to estimate the number of counts of larger genes. Another important parameter is the coverage of reads across a gene. To get a greater coverage, the reads are sequenced from both ends (also known as paired end reads) when the Illumina platform is used. Although the read lengths are different, major workflow is similar and described below.
The major workflow, from raw sequences obtained from the sequencers (here we specifically discuss short-read sequencers) to a list of differentially expressed genes, can be divided into four major steps (workflow is depicted visually in Fig. 5.4):
Step 1 is preprocessing of raw sequences. This step uses the same tools as used in Quality Control in Genomics analysis section.
Step 2 is aligning the sequence. Multiple tools/algorithms are available for the alignment/mapping of the transcriptome sequence to the reference genome. The important aspects in choosing an alignment algorithm/software are the accuracy of alignment to the reference genome, computational memory used, and the time taken for the alignment. In addition, for transcriptome alignments, it is important that the aligners are splice junction aware. This is because the RNA-seq is performed on mature messenger RNAs (mRNAs), which are devoid of introns, but the reference genome contains the intron sequences. If the aligner is not able to accurately identify the region where splicing or removal of introns (splicing junction) happens, then it would treat it as a deletion, and not estimate the transcript count accurately. Tophat2 [84], part of the Tuxedo [85] suite of tools, is a read aligner that is splice junction aware. The Tuxedo suite of tools takes a raw FASTQ file as an input, aligns it to the reference genome (Tophat2), and assembles it to accurately calculate expression. Cufflinks [86] calculates the differential expression between samples (cuffdiff) and finally visualizes expression plots (Cummerbund). Tophat2 uses Bowtie2 [87] (a fast aligner that uses Burrows-Wheeler transform (BWT) [88] for compression and storage of the reference genome) and FM index [89] (a compressed indexing method for Burrows-Wheeler transformation), so that it can be accessed rapidly. The regions that do not align to the reference genome using bowtie are divided into smaller segments using Tophat2. Tophat2 determines splice junction, when it observes read segments aligning to the reference genome with a gap of 100–1000 bases between them.
STAR (spliced transcripts alignment to a reference) [90] is a fast aligner which uses the concept of Maximal Mappable Prefix (MMP), on uncompressed suffix arrays. Suffix arrays are a type of data structure, which enables faster searching of sequences for alignment, by breaking the larger sequence into smaller subsections (suffix), sorting and storing the address in the form of arrays. The MMP algorithm searches these arrays to find the longest subsection of sequence reads that map to the reference genome sequence. After the alignment, the software performs clustering and stitching of all reads aligned to the genome to form a complete realigned sequence. The disadvantage of this algorithm is that it is memory intensive as the MMP search algorithm is performed on an uncompressed suffix array instead of a compressed one. HiSat [91], a newer alignment algorithm, is part of the new Tuxedo 2 [92] pipeline [HiSat (alignment), StringTie (assembly) [93], and Ballgown (differential expression calculation) [94]]. Like Tophat2, HiSat also uses bowtie for alignment utilizing the FM index functionality but also implements two different FM indexes that increase the accuracy of alignment. Of the FM indices, one is a global FM index and comprises the whole reference genome. The other index contains several small FM indices, each made up of a 64,00-bp subsection, which together covers the whole genomic region. By using memory optimization algorithms, it can reduce memory usage and can align a whole human genome using only 4GB of memory.
Step 3 is assembly and quantification. The output from the alignment processes produces aligned reads in the Sequence Alignment Map (SAM) format [95]. The SAM file is text-based and includes mandatory fields of chromosome number, location on the chromosome, and quality annotated, i.e., no genes are associated to the reads. The assembly and quantification tools are responsible for annotating the reads and estimating read counts associated with each transcript in the genome. SAM files are converted into BAM files (binary SAM files), sorted, indexed, and then given as an input to the assembler or read count estimation tools. Quality check of the BAM files can be done using RSeQC [96]. Preprocessing is performed by samtools software. There are multiple processes that perform assembly and quantification. The choice of read count quantification tool depends on whether one wants to have normalized read counts or raw read counts that can be normalized using custom scripts.
Normalized read counts can be divided into two basic types, reads per kilo million bases (RPKM) and transcripts per kilobase millions (TPM). RPKM is a normalized read count for single-read RNA sequences [97]. It is measured as the ratio of the number of reads depicting a region and the product of total reads divided by 1 million and region length divided by 1000. Fragments per kilo million (FPKM) bases is the paired end interpretation of RPKM. One of the tools that produce FPKM values is Cufflinks. Cufflinks, part of the Tuxedo pipeline, assembles the reads by first creating an overlap graph which represents all the reads that map to a region. The algorithm then traverses through the graph to assemble the isoforms, by identifying the minimum number of transcripts that can signify a particular intron junction. The assembled transcript fragments (transfrags) are counted using a statistical model for RNA-seq experiments [86]. The output from this tool is a Gene transfer format (GTF) file, which, along with information like chromosome name and location of the gene, also contains the gene abundance or read counts in FPKM. TPM is another normalization method [98] that is measured as the ratio of reads per kilobase (RPK) per million scaling factors. The RPK value is the read counts of each gene divided by the length of each gene in kilobases. The per million scaling factor is the sum of all RPK in a sample divided by 10,00,000. As the denominator remains constant, TPM for a given gene remains constant across replicate conditions if the same number of reads is aligned to the reference genome. This is not the case for RPKM (or FPKM) as its denominator is the length of the genes, which is a variable factor.
RSEM tool is responsible for the calculation of TPM for reads [99]. In the workflow, rsem-prepare-reference function takes in whole genome reference sequence in fasta format and GTF files and converts it to a transcript reference sequence. RSEM utilizes the transcript reference to get a clear count of isoforms. Next, rsem-calculate-expression calculates the TPM read counts. The input to this tool can be either FASTQ files or aligned BAM files. If fastq is provided as an input, the function first aligns the reads to a reference genome (default aligner is bowtie, STAR is the other aligner available in RSEM). Then using the expectation-maximization algorithm [100], maximum likelihood abundance (expected counts) is calculated and converted into TPM values. In case of aligned BAM files, special caution must be taken, if they are produced by aligners other than BWA and STAR. Because RSEM uses an enhanced read generation model for estimating read abundance compared to other aligners, aligner parameters must be changed to report all aligned reads. Outputs from this tool are 2 text files reporting gene TPM and isoform TPM values, respectively.
There are few other tools that do not perform any normalization in the read estimation step. These provide users with raw read counts which can then be normalized either by using available R/Python scripts or by functions available in downstream processes. One of the tools, Htseq a python package [101], not only quantifies read counts but can also be used as a parser for different genomic data files like FASTQ, SAM/BAM, a quality assessment tool (htseq-qa) for aligned reads, and GenomicArray class that stores the information from genomic data. The htseq-count function counts the reads based on the idea that the reads should cover an exon either completely or partially. The read counts are raw read counts, which can be normalized into FPKM, by custom scripts in python or R. Other useful tools are featureCounts [102] and summarizeOverlaps of the Genomic Alignment package in R [103].
Step 5 is differential expression calculation to identify genes or noncoding RNAs that are statistically significantly different between two or more conditions. In these steps raw or normalized reads are taken as an input, and using either parametric or nonparametric methods, differentially expressed genes are evaluated. The reads are given as an input directly as text files (in the case of Cuffdiff) or as R data objects extracted by tools like tximport [104], for downstream processing with R Bioconductor packages. Sample size is an important criterion to identify truly statistically significant differentially expressed genes. For RNA-seq tools, the efficiency of detection increases with sample size, and most of the statistical tools work reasonably well with a sample size of 3–5 per condition. Many of these tools can identify statistically significant targets with no replicates; however, extreme caution should be taken with interpretation. Please see the chapter of bioinformatics and statistics for omics data to fully understand the appropriate sample size and statistical methods to use. In general, differential expression tools can be divided into two types.
The first one is parametric analysis. The read counts generated from the quantification tools are either raw read counts or normalized read counts (FPKM/TPM). Most of the downstream differential expression tools have methods of normalization built into the packages. EdgeR [105], a R Bioconductor package, takes as input the read counts and stores them in a list-based data object called DGEList. This contains the read count matrix and the sample information, including the library information, in the form of a R type data frame and an optional data frame containing annotated gene information. EdgeR has methods to filter data based on count per million (CPM; which is a normalization method similar to TPM) read counts, to normalize data (calcNormFactors), and to estimate dispersion in read counts across samples using a quantile-adjusted conditional maximum likelihood negative binomial model (estimateDisp). For the differential expression calculation, a design matrix containing the condition and sample information (i.e., which samples are controls and which are experimental) must be provided as an input. Differential expression is calculated using an exact negative binomial test (exactTest). Differentially expressed genes are listed with parameters showing statistical significance (p-value and adjusted p-value) and fold change by the topTags function. Deseq2 is another Bioconductor package that uses similar statistics as EdgeR [106]. It takes as input raw, non-normalized read counts and stores them as a DESeqDataSet, similar to DGEList for EdgeR. Instead of three different functions to calculate differential expression, Deseq2 has the function DESeq which contains the functions for normalization, dispersion estimation, and negative binomial generalized linear model (GLM) [107] fitting followed by a Wald test [108] to test for differential expression. This tool provides an output similar to EdgeR. Cuffdiff, part of the Tuxedo suite of tools, uses a merged GTF file produced by Cufflink along with aligned SAM files from the conditions (output from Tophat) as the input for finding differential expression between conditions. The GTF files for each of the samples produced by Cufflinks are merged using the Cuffmerge function. The Cuffdiff function works twofold: First, it quantifies the reads to FPKM values. Second, it performs a statistical significance test based on negative binomial distribution as the previous two functions. The output is quite similar to the previous two methods.
The second is nonparametric analysis. Although parametric methods are quite powerful, it is completely dependent on the assumption that the distribution of the expression values fit the distribution of the statistical test being used. Methods like GFold [109] and Noiseq [110] use nonparametric methods of differential expression calculation. GFold uses a posterior distribution of logarithmic value of the raw fold changes and then ranks them based on their values. Those with higher ranks are upregulated, whereas low-ranked ones are downregulated. GFold can be used for both single replicates and multiple replicates. For Noiseq, several distributions are created; a noise distribution is for exhibiting change in counts, a contrasting fold change distribution (M), and an absolute expression difference (D), for each gene in a given condition. These are then used to compare whether M and D values between conditions fall within the noise or are actual differences. NoiSeq has 2 functions, NOISeq-real for replicates and NOISeq-sim for non-replicated samples. The output from these steps is a list of genes exhibiting statistically significantly different expressions between conditions. This list is used in further downstream analysis as described in the chapter of bioinformatics and statistics for omics data.
3.3 Single-Cell Sequencing
Different cells in an organism exhibit different phenotypic functionalities. These functionalities are governed by their genetic architecture; thus, it is important to understand the expression of genes in individual cells. The single-cell sequencing technique uses NGS methods on RNA isolated from single cells [111]. A review of library preparation and sequencing technique for single cell can be found in [112]. The bioinformatics pipeline is similar to RNA-seq, wherein quality check is done by FastQC, alignment performed by STAR, followed by read count estimation by HTSEq, and differential expression analysis performed by Deseq2. Custom-made codes to differentiate cells based on barcodes are present in pipelines developed by companies that develop the libraries such as 10Xgenomics. Identification of different cell types is done using PCA or t-distributed stochastic neighbor embedding (t-SNE) [113]. These assist in differentiation and visualization of gene clusters based on expression similarities. For example, genes having similar differential expression patterns among conditions would be clustered together and by identifying markers we can identify known cell types or unknown cell types. T-SNE is implemented in Seurat [114].
4 Platform Selection and Limitations
Gene expression profiles of different tissues, cells, conditions, disease states, or even single cells are now routinely generated. This explosion in gene expression profiling has been deeply affected by the rapid development of new technologies with an improved sensitivity and cost effect. Which platform to be used for a study depends on the goal of the study. The advantages of next-generation sequencing are that the method provides direct counts of molecules; it can be used to study all types of RNA transcripts; it provides better resolution for transcripts that share similar sequences; alternative splice isoforms can be directly detected; and it can be used to examine allele-specific expression. The disadvantages of the NGS approach are that it may not be able to detect low-abundance transcripts; data analyses and statistical analyses are more challenging, and the final results heavily rely on how the raw data were processed; computational demands are high; data storage and sharing are challenging; and there are concerns of privacy when sequence information is examined. In addition to the above, the cost and availability of the platform and bioinformatics expertise are something to be considered.
To select a proper platform and plan a profiling study, which RNA population will be studied is an important factor. NGS will be the choice if whole transcriptome, including those that are not on the microarrays, is studied. If one is only interested in a specific group of RNA transcripts or few specific pathways, it may be more cost-effective to use arrays and PCR-based assays. In addition to commercially available stock arrays and PCR panels, researchers can custom-design arrays and PCR-based assays to include specific transcripts of interests. The transcripts are well annotated, and data analyses process is straightforward; therefore, the turnaround time is shorter. Another factor to be considered is that NGS is limited by its total read counts for each run. For example, the highly abundant rRNAs need to be removed from the samples to increase the reads of the rest of the transcripts. The transcripts that are low in abundance are often missed by direct sequencing. These include low-abundant mRNA transcripts, alternatively spliced variants, and lncRNA transcripts. In general, 20–50 million reads are sufficient for detecting ~20,000 transcripts. 100 million reads will increase coverage and allow proper quantification and identify differentially expressed transcripts. Three hundred million reads give enough depth for studying alternative splicing and higher number of reads may be needed for studying lncRNA. Since commercial microarrays that cover the whole transcriptome, noncoding RNA, and splice variants are available, one can decide which platform to use based on the experimental aims, turnaround time, and costs. While the microarrays are generally good at detecting low-abundant transcripts, depending on the dynamic range of different microarray platforms, sensitivities to changes of highly abundant transcripts may reduce due to saturation of the intensity signals at the high end.
One of the important considerations when conducting the profiling studies is normalization of the data. The method used for normalization and for hybridization-based assays is usually less an issue when a large number of genes are examined at the same time. For example, Affymetrix normalizes the intensity of individual data point to the average intensity of the whole array. This strategy is based on the assumption that most of the genes in a sample do not change significantly. Any conditions that change the expression of a large number of genes toward the same direction will not be suitable to use this normalization method. Instead, one can identify genes that express at consistent level in all samples to be normalized to. The same strategies can be used when the platform does not examine a large number of genes, such as cDNA arrays or PCR arrays. For muscle research, commonly used internal controls such as glyceraldehyde-3-phosphate dehydrogenase (GAPDH), β-actin, and 18S rRNA may have inconsistent expression levels in samples that are severely affected by a condition or disease. An important question is whether the condition or disease you are studying will cause genome-wide transcriptional changes toward one direction or changes of the gene level of the internal controls.
Increased heterogeneity of pathological specimen needs to be kept in mind when interpreting transcriptomic data. In addition to degeneration/regeneration, a muscle sample may contain different degrees of pathological changes such as inflammation, fibrosis, and fatty tissue replacement. One can imagine that severe fat infiltration or inflammation can completely change the profiles. When the pathological changes are prominent, we are comparing different tissues in such situations. This will cause a challenge when analyzing data as well as interpreting results. To answer the questions regarding the origin of the changes, additional studies are needed, such as immunostaining or immunohistochemistry to visualize and locate the protein products of the genes that were shown to change in the profiling data [6, 7, 18]. Now one can also use single-cell sequencing to examine gene profiles in each cell individually and determine which cells contribute to the changes [115]. However, it is still a good idea to validate the protein changes in the tissue directly.
While transcriptomic approaches can potentially generate large amount of data, usually only a limited number of genes and pathway are reported in a publication. This can be due to (1) insufficient statistical power due to small sample sizes; (2) lack of bioinformatics expertise to fully analyze the data; and (3) the researcher selecting and focusing on only part of the genes and pathways to follow up. Placing expression profiling results in a publicly accessible microarray database makes it possible for other researchers to access the data and have new discoveries beyond the scope of published results [116].
5 Vital Future Directions
Technologies for studying transcriptome have been evolving quickly. New methods and approaches to address common issues and concerns associated with the current approach are becoming available. Companies have been improving reagents for better sample preparation and producing new instruments for higher throughput and lower cost per base. In addition, new utilization and approaches are developed by both the companies and users to answer specific questions. Here we discuss few examples. One of the questions is issues associated with tissue heterogeneity in muscle samples. scRNA-seq allows single-cell resolution of transcriptomes; however, the information on the location of the cells and the relationship among the cells are lost during the sample processing. To allow researchers to learn not only what is in a cell but how the cells interact with other cells provides an invaluable insight into understanding muscle biology and disease mechanisms. Recently 10X Genomics acquired Spatial Transcriptomics which provides such technology. The technology for the spatial gene expression profiling was originally developed at Science for Life Laboratory in Stockholm, Sweden, which allows RNA sequencing to be performed from tissue sections [117]. The process includes first attaching a frozen-sectioned tissue section to a specialized chip. The tissue section is imaged before the RNA transcripts in the tissue section are reverse-transcribed. The image with coordinate information will later be used to be matched to the gene expression data generated from each specific location. The chip contains an array of probes which have poly-T tails at the end. After the tissue section is fixated and permeabilized, the RNA with poly-A sequence will be captured by the probes and reverse-transcribed. The cDNA-RNA hybrids are cleaved off the chip, followed by library preparation for RNA-seq. The RNA-seq data are integrated with the histology data for visualizing the transcriptomes of cells at different locations in the section.
One challenge in performing single-cell profiling on skeletal muscle samples is that the myofibers are multinucleated cells. It is not feasible to capture individual myofiber for profiling using current platforms. On the other hand, single-nucleus profiling will provide transcriptome information of individual nucleus in a sample. A recent study showed that comparable results were obtained by using scRNA-seq and snRNA-seq approaches although the snRNA-seq is better for sequencing nucleus-enriched lncRNAs and miRNA precursors [64].
In addition to sequences, new technologies allow researchers to study RNA base modifications [118]. More than a hundred RNA base modifications are known but the role of the modifications in mRNA is mostly unclear. The Oxford Nanopore platform allows direct sequencing of RNA molecules and identifying RNA modifications. The epitranscriptome will be one future direction that can take advantage of the new technologies.
Transcriptomic approaches allow the researcher to determine the transcription activities of all active genes. Temporal profiling approaches use a series of profiles obtained at different time points to construct temporal changes of gene activities over a period of time. Meanwhile, researches combine more than one omics approaches to understand how the other processes, such as epigenomic, posttranscriptional, and posttranslational regulations, affect the cell functions and to gain a more integrated picture. Using multiple omics approaches to obtain temporal data makes construction of comprehensive molecular pathways involved in specific conditions possible. Data from single-cell profiling and spatial gene profiling increases resolution to changes in individual cell.
References
Li, Z., et al. (2018). Systematic transcriptome-wide analysis of mRNA-miRNA interactions reveals the involvement of miR-142-5p and its target (FOXO3) in skeletal muscle growth in chickens. Molecular Genetics and Genomics: MGG, 293, 69–80. https://doi.org/10.1007/s00438-017-1364-7.
Li, R., et al. (2019). Characterization and expression profiles of muscle transcriptome in Schizothoracine fish, Schizothorax prenanti. Gene, 685, 156–163. https://doi.org/10.1016/j.gene.2018.10.070.
Cote, L. E., Simental, E., & Reddien, P. W. (2019). Muscle functions as a connective tissue and source of extracellular matrix in planarians. Nature Communications, 10, 1592. https://doi.org/10.1038/s41467-019-09539-6.
Burniston, J. G., et al. (2013). Gene expression profiling of gastrocnemius of “minimuscle” mice. Physiological Genomics, 45, 228–236. https://doi.org/10.1152/physiolgenomics.00149.2012. physiolgenomics.00149.2012 [pii].
Pietu, G., et al. (1996). Novel gene transcripts preferentially expressed in human muscles revealed by quantitative hybridization of a high density cDNA array. Genome Research, 6, 492–503.
Chen, Y. W., Hubal, M. J., Hoffman, E. P., Thompson, P. D., & Clarkson, P. M. (2003). Molecular responses of human muscle to eccentric exercise. Journal of Applied Physiology, 95, 2485–2494.
Chen, Y. W., et al. (2002). Response of rat muscle to acute resistance exercise defined by transcriptional and translational profiling. The Journal of Physiology, 545, 27–41.
Bonafiglia, J. T., Menzies, K. J., & Gurd, B. J. (2019). Gene expression variability in human skeletal muscle transcriptome responses to acute resistance exercise. Experimental Physiology, 104, 625–629. https://doi.org/10.1113/EP087436.
Turner, D. C., Seaborne, R. A., & Sharples, A. P. (2019). Comparative transcriptome and methylome analysis in human skeletal muscle anabolism, hypertrophy and epigenetic memory. Scientific Reports, 9, 4251. https://doi.org/10.1038/s41598-019-40787-0.
Dickinson, J. M., et al. (2018). Transcriptome response of human skeletal muscle to divergent exercise stimuli. Journal of Applied Physiology (1985), 124, 1529–1540. https://doi.org/10.1152/japplphysiol.00014.2018.
Mahmassani, Z. S., et al. (2019). Age-dependent skeletal muscle transcriptome response to bed rest-induced atrophy. Journal of Applied Physiology (1985), 126, 894–902. https://doi.org/10.1152/japplphysiol.00811.2018.
Vechin, F. C., et al. (2019). Low-intensity resistance training with partial blood flow restriction and high-intensity resistance training induce similar changes in skeletal muscle transcriptome in elderly humans. Applied Physiology, Nutrition, and Metabolism = Physiologie appliquee, nutrition et metabolisme, 44, 216–220. https://doi.org/10.1139/apnm-2018-0146.
Chen, Y. W., et al. (2005). Early onset of inflammation and later involvement of TGFbeta in Duchenne muscular dystrophy. Neurology, 65, 826–834.
Dadgar, S., et al. (2014). Asynchronous remodeling is a driver of failed regeneration in Duchenne muscular dystrophy. The Journal of Cell Biology, 207, 139–158. https://doi.org/10.1083/jcb.201402079. jcb.201402079 [pii].
Sharma, V., Harafuji, N., Belayew, A., & Chen, Y. W. (2013). DUX4 differentially regulates transcriptomes of human rhabdomyosarcoma and mouse C2C12 cells. PLoS One, 8, e64691. https://doi.org/10.1371/journal.pone.0064691. PONE-D-13-08552 [pii].
Dixit, M., et al. (2007). DUX4, a candidate gene of facioscapulohumeral muscular dystrophy, encodes a transcriptional activator of PITX1. Proceedings of the National Academy of Sciences of the United States of America, 104, 18157–18162.
Wang, E. T., et al. (2019). Transcriptome alterations in myotonic dystrophy skeletal muscle and heart. Human Molecular Genetics, 28, 1312–1321. https://doi.org/10.1093/hmg/ddy432.
Chen, Y. W., Zhao, P., Borup, R., & Hoffman, E. P. (2000). Expression profiling in the muscular dystrophies: Identification of novel aspects of molecular pathophysiology. The Journal of Cell Biology, 151, 1321–1336.
Zhang, N., et al. (2018). Dynamic transcriptome profile in db/db skeletal muscle reveal critical roles for long noncoding RNA regulator. The International Journal of Biochemistry and Cell Biology, 104, 14–24. https://doi.org/10.1016/j.biocel.2018.08.013.
Scott, L. J., et al. (2016). The genetic regulatory signature of type 2 diabetes in human skeletal muscle. Nature Communications, 7, 11764. https://doi.org/10.1038/ncomms11764.
Gallagher, I. J., et al. (2012). Suppression of skeletal muscle turnover in cancer cachexia: Evidence from the transcriptome in sequential human muscle biopsies. Clinical Cancer Research: An Official Journal of the American Association for Cancer Research, 18, 2817–2827. https://doi.org/10.1158/1078-0432.CCR-11-2133.
Chen, Y. W., et al. (2017). Molecular signatures of differential responses to exercise trainings during rehabilitation. Biomedical Genetics and Genomics, 2. https://doi.org/10.15761/BGG.1000127.
Boehler, J. F., et al. (2017). Effect of endurance exercise on microRNAs in myositis skeletal muscle—A randomized controlled study. PLoS One, 12, e0183292. https://doi.org/10.1371/journal.pone.0183292.
Benoit, B., et al. (2017). Fibroblast growth factor 19 regulates skeletal muscle mass and ameliorates muscle wasting in mice. Nature Medicine, 23, 990–996. https://doi.org/10.1038/nm.4363.
Wu, J., et al. (2014). Ribogenomics: The science and knowledge of RNA. Genomics, Proteomics and Bioinformatics, 12, 57–63. https://doi.org/10.1016/j.gpb.2014.04.002.
Varemo, L., et al. (2016). Proteome- and transcriptome-driven reconstruction of the human myocyte metabolic network and its use for identification of markers for diabetes. Cell Reports, 14, 1567. https://doi.org/10.1016/j.celrep.2016.01.054.
Lundberg, E., et al. (2010). Defining the transcriptome and proteome in three functionally different human cell lines. Molecular Systems Biology, 6, 450. https://doi.org/10.1038/msb.2010.106.
Vogel, C., & Marcotte, E. M. (2012). Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nature Reviews. Genetics, 13, 227–232. https://doi.org/10.1038/nrg3185.
Nie, L., Wu, G., Brockman, F. J., & Zhang, W. (2006). Integrated analysis of transcriptomic and proteomic data of Desulfovibrio vulgaris: Zero-inflated Poisson regression models to predict abundance of undetected proteins. Bioinformatics, 22, 1641–1647. https://doi.org/10.1093/bioinformatics/btl134.
Greenbaum, D., Colangelo, C., Williams, K., & Gerstein, M. (2003). Comparing protein abundance and mRNA expression levels on a genomic scale. Genome Biology, 4, 117. https://doi.org/10.1186/gb-2003-4-9-117.
Margulies, M., et al. (2005). Genome sequencing in microfabricated high-density picolitre reactors. Nature, 437, 376–380. https://doi.org/10.1038/nature03959.
Heather, J. M., & Chain, B. (2016). The sequence of sequencers: The history of sequencing DNA. Genomics, 107, 1–8. https://doi.org/10.1016/j.ygeno.2015.11.003.
Schena, M., Shalon, D., Davis, R. W., & Brown, P. O. (1995). Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science, 270, 467–470.
DeRisi, J., et al. (1996). Use of a cDNA microarray to analyse gene expression patterns in human cancer. Nature Genetics, 14, 457–460. https://doi.org/10.1038/ng1296-457.
Dalma-Weiszhausz, D. D., Warrington, J., Tanimoto, E. Y., & Miyada, C. G. (2006). The affymetrix GeneChip platform: An overview. Methods in Enzymology, 410, 3–28. https://doi.org/10.1016/S0076-6879(06)10001-4.
Kostek, M. C., et al. (2007). Gene expression responses over 24 h to lengthening and shortening contractions in human muscle: Major changes in CSRP3, MUSTN1, SIX1, and FBXO32. Physiological Genomics, 31, 42–52.
Borup, R. H., et al. (2002). Development and production of an oligonucleotide MuscleChip: Use for validation of ambiguous ESTs. BMC Bioinformatics, 3, 33.
Fan, J. B., et al. (2006). Illumina universal bead arrays. Methods in Enzymology, 410, 57–73. https://doi.org/10.1016/S0076-6879(06)10003-8.
Kotorashvili, A., et al. (2012). Effective DNA/RNA co-extraction for analysis of microRNAs, mRNAs, and genomic DNA from formalin-fixed paraffin-embedded specimens. PLoS One, 7, e34683. https://doi.org/10.1371/journal.pone.0034683.
Kibriya, M. G., et al. (2010). Analyses and interpretation of whole-genome gene expression from formalin-fixed paraffin-embedded tissue: An illustration with breast cancer tissues. BMC Genomics, 11, 622. https://doi.org/10.1186/1471-2164-11-622.
Wolber, P. K., Collins, P. J., Lucas, A. B., De Witte, A., & Shannon, K. W. (2006). The agilent in situ-synthesized microarray platform. Methods in Enzymology, 410, 28–57. https://doi.org/10.1016/S0076-6879(06)10002-6.
Wu, L., Brady, L., Shoffner, J., & Tarnopolsky, M. A. (2018). Next-generation sequencing to diagnose muscular dystrophy, rhabdomyolysis, and HyperCKemia. The Canadian Journal of Neurological Sciences. Le journal canadien des sciences neurologiques, 45, 262–268. https://doi.org/10.1017/cjn.2017.286.
Nigro, V., & Piluso, G. (2012). Next generation sequencing (NGS) strategies for the genetic testing of myopathies. Acta myologica: Myopathies and Cardiomyopathies: Official Journal of the Mediterranean Society of Myology, 31, 196–200.
Hestand, M. S., et al. (2010). Tissue-specific transcript annotation and expression profiling with complementary next-generation sequencing technologies. Nucleic Acids Research, 38, e165. https://doi.org/10.1093/nar/gkq602.
Colangelo, V., et al. (2014). Next-generation sequencing analysis of miRNA expression in control and FSHD myogenesis. PLoS One, 9, e108411. https://doi.org/10.1371/journal.pone.0108411.
Cardoso, T. F., et al. (2017). RNA-seq based detection of differentially expressed genes in the skeletal muscle of Duroc pigs with distinct lipid profiles. Scientific Reports, 7, 40005. https://doi.org/10.1038/srep40005.
Pennisi, E. (2010). Genomics. Semiconductors inspire new sequencing technologies. Science, 327, 1190. https://doi.org/10.1126/science.327.5970.1190.
Tripathi, A. K., et al. (2014). Transcriptomic dissection of myogenic differentiation signature in caprine by RNA-Seq. Mechanisms of Development, 132, 79–92. https://doi.org/10.1016/j.mod.2014.01.001.
Parmakelis, A., Kotsakiozi, P., Kontos, C. K., Adamopoulos, P. G., & Scorilas, A. (2017). The transcriptome of a “sleeping” invader: De novo assembly and annotation of the transcriptome of aestivating Cornu aspersum. BMC Genomics, 18, 491. https://doi.org/10.1186/s12864-017-3885-1.
Possidonio, A. C., et al. (2014). Cholesterol depletion induces transcriptional changes during skeletal muscle differentiation. BMC Genomics, 15, 544. https://doi.org/10.1186/1471-2164-15-544.
Cartolano, M., Huettel, B., Hartwig, B., Reinhardt, R., & Schneeberger, K. (2016). cDNA Library Enrichment of Full Length Transcripts for SMRT Long Read Sequencing. PLoS One, 11, e0157779. https://doi.org/10.1371/journal.pone.0157779.
Chen, S. Y., Deng, F., Jia, X., Li, C., & Lai, S. J. (2017). A transcriptome atlas of rabbit revealed by PacBio single-molecule long-read sequencing. Scientific Reports, 7, 7648. https://doi.org/10.1038/s41598-017-08138-z.
Masonbrink, R. E., et al. (2019). An annotated genome for Haliotis rufescens (Red Abalone) and resequenced green, pink, pinto, black, and white abalone species. Genome Biology and Evolution, 11, 431–438. https://doi.org/10.1093/gbe/evz006.
Loman, N. J., & Watson, M. (2015). Successful test launch for nanopore sequencing. Nature Methods, 12, 303–304. https://doi.org/10.1038/nmeth.3327.
Mikheyev, A. S., & Tin, M. M. (2014). A first look at the Oxford nanopore MinION sequencer. Molecular Ecology Resources, 14, 1097–1102. https://doi.org/10.1111/1755-0998.12324.
Ayub, M., & Bayley, H. (2012). Individual RNA base recognition in immobilized oligonucleotides using a protein nanopore. Nano Letters, 12, 5637–5643. https://doi.org/10.1021/nl3027873.
Johnson, S. S., Zaikova, E., Goerlitz, D. S., Bai, Y., & Tighe, S. W. (2017). Real-time DNA sequencing in the Antarctic dry valleys using the Oxford nanopore sequencer. Journal of Biomolecular Techniques: JBT, 28, 2–7. https://doi.org/10.7171/jbt.17-2801-009.
McIntyre, A. B. R., et al. (2016). Nanopore sequencing in microgravity. NPJ Microgravity, 2, 16035. https://doi.org/10.1038/npjmgrav.2016.35.
Runtuwene, L. R., Tuda, J. S. B., Mongan, A. E., & Suzuki, Y. (2019). On-site MinION sequencing. Advances in Experimental Medicine and Biology, 1129, 143–150. https://doi.org/10.1007/978-981-13-6037-4_10.
Walter, M. C., et al. (2017). MinION as part of a biomedical rapidly deployable laboratory. Journal of Biotechnology, 250, 16–22. https://doi.org/10.1016/j.jbiotec.2016.12.006.
Jain, M., Olsen, H. E., Paten, B., & Akeson, M. (2016). The Oxford nanopore MinION: Delivery of nanopore sequencing to the genomics community. Genome Biology, 17, 239. https://doi.org/10.1186/s13059-016-1103-0.
Narola, J., Pandey, S. N., Glick, A., & Chen, Y. W. (2013). Conditional expression of TGF-beta1 in skeletal muscles causes endomysial fibrosis and myofibers atrophy. PLoS One, 8, e79356. https://doi.org/10.1371/journal.pone.0079356. PONE-D-13-27811 [pii].
Cho, D. S., & Doles, J. D. (2017). Single cell transcriptome analysis of muscle satellite cells reveals widespread transcriptional heterogeneity. Gene, 636, 54–63. https://doi.org/10.1016/j.gene.2017.09.014.
Zeng, W., et al. (2016). Single-nucleus RNA-seq of differentiating human myoblasts reveals the extent of fate heterogeneity. Nucleic Acids Research, 44, e158. https://doi.org/10.1093/nar/gkw739.
Dell’Orso, S., et al. (2019). Single cell analysis of adult mouse skeletal muscle stem cells in homeostatic and regenerative conditions. Development, 146. https://doi.org/10.1242/dev.174177.
Winokur, S. T., et al. (2003). Expression profiling of FSHD muscle supports a defect in specific stages of myogenic differentiation. Human Molecular Genetics, 12, 2895–2907.
Lemmers, R. J., et al. (2010). A unifying genetic model for facioscapulohumeral muscular dystrophy. Science, 329, 1650–1653. https://doi.org/10.1126/science.1189044.
Snider, L., et al. (2010). Facioscapulohumeral dystrophy: Incomplete suppression of a retrotransposed gene. PLoS Genetics, 6, e1001181. https://doi.org/10.1371/journal.pgen.1001181.
Jones, T. I., et al. (2015). Individual epigenetic status of the pathogenic D4Z4 macrosatellite correlates with disease in facioscapulohumeral muscular dystrophy. Clinical Epigenetics, 7, 37. https://doi.org/10.1186/s13148-015-0072-6.
Himeda, C. L., et al. (2014). Myogenic enhancers regulate expression of the facioscapulohumeral muscular dystrophy-associated DUX4 gene. Molecular and Cellular Biology, 34, 1942–1955. https://doi.org/10.1128/MCB.00149-14.
van den Heuvel, A., et al. (2019). Single-cell RNA sequencing in facioscapulohumeral muscular dystrophy disease etiology and development. Human Molecular Genetics, 28, 1064–1075. https://doi.org/10.1093/hmg/ddy400.
Ritchie, M. E., et al. (2015). limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Research, 43, e47–e47. https://doi.org/10.1093/nar/gkv007.
Gautier, L., Cope, L., Bolstad, B. M., & Irizarry, R. A. (2004). affy – Analysis of Affymetrix GeneChip data at the probe level. Bioinformatics, 20, 307–315. https://doi.org/10.1093/bioinformatics/btg405.
Dunning, M. J., Smith, M. L., Ritchie, M. E., & Tavare, S. (2007). beadarray: R classes and methods for Illumina bead-based data. Bioinformatics, 23, 2183–2184. https://doi.org/10.1093/bioinformatics/btm311.
Bolstad, B. M., Irizarry, R. A., Astrand, M., & Speed, T. P. (2003). A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics, 19, 185–193. https://doi.org/10.1093/bioinformatics/19.2.185.
Carvalho, B. S., & Irizarry, R. A. (2010). A framework for oligonucleotide microarray preprocessing. Bioinformatics, 26, 2363–2367. https://doi.org/10.1093/bioinformatics/btq431.
Warnes, G. R., Bolker, B., Bonebakker, L., Gentleman, R., Huber, W., Liaw, A., Lumley, T., et al. (2009). gplots: Various R programming tools for plotting data. R package version 2.
Student. (1908). The probable error of a mean. Biometrika.
Fisher, R. (1919). A. XV.—The correlation between relatives on the supposition of Mendelian inheritance. Transactions of the Royal Society of Edinburgh, 52, 399–433. https://doi.org/10.1017/S0080456800012163.
Benjamini, Y., & Hochberg, Y. (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. Journal of the Royal Statistical Society: Series B (Methodological), 57, 289–300. https://doi.org/10.2307/2346101.
Bonferroni, C. (1936). Teoria statistica delle classi e calcolo delle probabilità. Pubblicazioni del R Istituto Superiore di Scienze Economiche e Commerciali di Firenze, 8, 3–62.
Schadt, E. E., Turner, S., & Kasarskis, A. (2010). A window into third-generation sequencing. Human Molecular Genetics, 19, R227–R240. https://doi.org/10.1093/hmg/ddq416.
Eisenstein, M. (2012). Oxford nanopore announcement sets sequencing sector abuzz. Nature Biotechnology, 30, 295–296. https://doi.org/10.1038/nbt0412-295.
Kim, D., et al. (2013). TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biology, 14, R36. https://doi.org/10.1186/gb-2013-14-4-r36.
Trapnell, C., et al. (2012). Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nature Protocols, 7, 562–578. https://doi.org/10.1038/nprot.2012.016.
Trapnell, C., et al. (2010). Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nature Biotechnology, 28, 511–515. https://doi.org/10.1038/nbt.1621.
Langmead, B., & Salzberg, S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nature Methods, 9, 357–359. https://doi.org/10.1038/nmeth.1923.
Adjeroh, D., Bell, T., & Mukherjee, A. (2008). The Burrows-Wheeler Transform: Data compression, suffix arrays, and pattern matching. New York: Springer.
Ferragina, P., & Manzini, G. (2001). An experimental study of a compressed index. Information Sciences, 135, 13–28. https://doi.org/10.1016/S0020-0255(01)00098-6.
Dobin, A., et al. (2013). STAR: Ultrafast universal RNA-seq aligner. Bioinformatics (Oxford, England), 29, 15–21. https://doi.org/10.1093/bioinformatics/bts635.
Kim, D., Langmead, B., & Salzberg, S. L. (2015). HISAT: A fast spliced aligner with low memory requirements. Nature Methods, 12, 357–360. https://doi.org/10.1038/nmeth.3317.
Pertea, M., Kim, D., Pertea, G. M., Leek, J. T., & Salzberg, S. L. (2016). Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nature Protocols, 11, 1650–1667. https://doi.org/10.1038/nprot.2016.095.
Pertea, M., et al. (2015). StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nature Biotechnology, 33, 290–295. https://doi.org/10.1038/nbt.3122.
Frazee, A. C., et al. (2015). Ballgown bridges the gap between transcriptome assembly and expression analysis. Nature Biotechnology, 33, 243–246. https://doi.org/10.1038/nbt.3172.
Li, H., et al. (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics (Oxford, England), 25, 2078–2079. https://doi.org/10.1093/bioinformatics/btp352.
Wang, L., Wang, S., & Li, W. (2012). RSeQC: Quality control of RNA-seq experiments. Bioinformatics, 28, 2184–2185. https://doi.org/10.1093/bioinformatics/bts356.
Mortazavi, A., Williams, B. A., McCue, K., Schaeffer, L., & Wold, B. (2008). Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nature Methods, 5, 621–628. https://doi.org/10.1038/nmeth.1226.
Li, B., Ruotti, V., Stewart, R. M., Thomson, J. A., & Dewey, C. N. (2010). RNA-Seq gene expression estimation with read mapping uncertainty. Bioinformatics, 26, 493–500. https://doi.org/10.1093/bioinformatics/btp692.
Li, B., & Dewey, C. N. (2011). RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics, 12, 323. https://doi.org/10.1186/1471-2105-12-323.
Dempster, A. P., Laird, N. M., & Rubin, D. B. (1976). Maximum likelihood from incomplete data via the EM algorithm. Journal of the Royal Statistical Society. Series B (Methodological), 39(1), 1–38.
Anders, S., Pyl, P. T., & Huber, W. (2015). HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics (Oxford, England), 31, 166–169. https://doi.org/10.1093/bioinformatics/btu638.
Liao, Y., Smyth, G. K., & Shi, W. (2014). featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics, 30, 923–930. https://doi.org/10.1093/bioinformatics/btt656.
Lawrence, M., et al. (2013). Software for computing and annotating genomic ranges. PLoS Computational Biology, 9, e1003118. https://doi.org/10.1371/journal.pcbi.1003118.
Soneson, C., Love, M. I., & Robinson, M. D. (2015). Differential analyses for RNA-seq: Transcript-level estimates improve gene-level inferences. F1000Research, 4, 1521. https://doi.org/10.12688/f1000research.7563.1.
Robinson, M. D., McCarthy, D. J., & Smyth, G. K. (2010). edgeR: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics, 26, 139–140. https://doi.org/10.1093/bioinformatics/btp616.
Love, M. I., Huber, W., & Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology, 15, 550. https://doi.org/10.1186/s13059-014-0550-8.
Nelder, J. A., & Wedderburn, R. W. M. (1972). Generalized linear models. Journal of the Royal Statistical Society. Series A (General), 135, 370. https://doi.org/10.2307/2344614.
Wald, A. (1945). Sequential tests of statistical hypotheses. The Annals of Mathematical Statistics, 16, 117–186. https://doi.org/10.1214/aoms/1177731118.
Feng, J., et al. (2012). GFOLD: A generalized fold change for ranking differentially expressed genes from RNA-seq data. Bioinformatics, 28, 2782–2788. https://doi.org/10.1093/bioinformatics/bts515.
Tarazona, S., et al. (2015). Data quality aware analysis of differential expression in RNA-seq with NOISeq R/Bioc package. Nucleic Acids Research, 43, e140. https://doi.org/10.1093/nar/gkv711.
Eberwine, J., et al. (1992). Analysis of gene expression in single live neurons. Proceedings of the National Academy of Sciences of the United States of America, 89, 3010–3014. https://doi.org/10.1073/pnas.89.7.3010.
Hwang, B., Lee, J. H., & Bang, D. (2018). Single-cell RNA sequencing technologies and bioinformatics pipelines. Experimental and Molecular Medicine, 50, 96. https://doi.org/10.1038/s12276-018-0071-8.
Van Der Maaten, L., & Hinton, G. (2008). Visualizing data using t-SNE. Journal of Machine Learning Research, 9, 2579–2605.
Butler, A., Hoffman, P., Smibert, P., Papalexi, E., & Satija, R. (2018). Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nature Biotechnology, 36, 411–420. https://doi.org/10.1038/nbt.4096.
Gatto, S., Puri, P. L., & Malecova, B. (2017). Single cell gene expression profiling of skeletal muscle-derived cells. Methods in Molecular Biology, 1556, 191–219. https://doi.org/10.1007/978-1-4939-6771-1_10.
Banerji, C. R. S., et al. (2017). PAX7 target genes are globally repressed in facioscapulohumeral muscular dystrophy skeletal muscle. Nature Communications, 8, 2152. https://doi.org/10.1038/s41467-017-01200-4.
Stahl, P. L., et al. (2016). Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science, 353, 78–82. https://doi.org/10.1126/science.aaf2403.
Saletore, Y., et al. (2012). The birth of the Epitranscriptome: Deciphering the function of RNA modifications. Genome Biology, 13, 175. https://doi.org/10.1186/gb-2012-13-10-175.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 The American Physiological Society
About this chapter
Cite this chapter
Liu, P., Bhattacharya, S., Chen, YW. (2019). Transcriptomic Approaches for Muscle Biology and Disorders. In: Burniston, J., Chen, YW. (eds) Omics Approaches to Understanding Muscle Biology. Methods in Physiology. Springer, New York, NY. https://doi.org/10.1007/978-1-4939-9802-9_5
Download citation
DOI: https://doi.org/10.1007/978-1-4939-9802-9_5
Published:
Publisher Name: Springer, New York, NY
Print ISBN: 978-1-4939-9801-2
Online ISBN: 978-1-4939-9802-9
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)