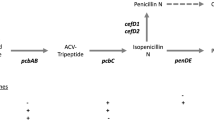
Abstract
Polymerase chain reaction amplification of conserved genes and sequence analysis provides a very powerful tool for the identification of toxigenic as well as non-toxigenic Penicillium species. Sequences are obtained by amplification of the gene fragment, sequencing via capillary electrophoresis of dideoxynucleotide-labeled fragments or NGS. The sequences are compared to a database of validated isolates. Identification of species indicates the potential of the fungus to make particular mycotoxins.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Thom C (1930) The Penicillia. Williams and Wilkins, Baltimore, p 644
Raper KB, Thom C (1949) A manual of the Penicillia. Williams and Wilkins, Baltimore, p 875
Pitt JI (1980) The genus Penicillium and its teleomorphic states Eupenicillium and Talaromyces. Academic, New York, p 634
Taylor JW, Jacobson DJ, Kroken S et al (2000) Phylogenetic species recognition and species concepts in fungi. Fungal Genet Biol 31:21–32
Mullis K, Faloona F, Scharf S et al (1986) Specific enzymatic amplification of DNA in vitro: the polymerase chain reaction. Cold Spring Harb Symp Quant Biol 51:263–273
Peterson SW (2000) Phylogenetic analysis of Penicillium species based on ITS and LSU-rDNA nucleotide sequences (pp 163–178). In: Samson RA, Pitt JI (eds) Integration of modern taxonomic methods for Penicillium and Aspergillus classification. Harwood Academic Publishers, Amsterdam, p 510
Seifert KA, Louis-Seize G (2000) Phylogeny and species concepts in the Penicillium aurantiogriseum complex as inferred from partial B-tubulin gene DNA sequences (pp 189–198). In: Samson RA, Pitt JI (eds) Integration of modern taxonomic methods for Penicillium and Aspergillus classification. Harwood Academic Publishers, Amsterdam, p 510
Samson RA, Frisvad JC (2004) Penicillium subgenus Penicillium: new taxonomic schemes, mycotoxins and other extrolites. Stud Mycol 49:1–260
Peterson SW, Jurjevic Z, Frisvad JC (2015) Expanding the species and chemical diversity of Penicillium section Cinnamopurpurea. PLoS ONE 10(4), e0121987
Seifert KA, Samson RA, Dewaard JR et al (2007) Prospects for fungus identification using CO1 DNA barcodes, with Penicillium as a test case. Proc Natl Acad Sci U S A 104:3901–3906
Schoch CL, Seifert KA, Huhndorf S et al (2012) Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc Natl Acad Sci U S A 109:6241–6246
Peterson SW (2012) Aspergillus and Penicillium identification using DNA sequences: barcode or MLST? Appl Microbiol Biotechnol 95:339–344
Osmundson TW, Robert VA, Schoch CL et al (2013) Filling gaps in biodiversity knowledge for macrofungi: contributions and assessment of an herbarium collection DNA barcode sequencing project. PLoS ONE 8(4), e62419
Seifert KA (1990) Isolation of filamentous fungi (pp 21–52). In: Labeda DP (ed) Isolation of biotechnological organisms from nature. McGraw-Hill, New York, p 322
Glass NL, Donaldson GC (1995) Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl Environ Microbiol 61:1323–1330
Hong SB, Cho HS, Shin HD et al (2006) Novel Neosartorya species isolated from soil in Korea. Int J Syst Evol Microbiol 56:477–486
White TJ, Bruns T, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics (pp 315–322). In: Innes MJ, Gelfand DH, Sninsky JJ, White TJ (eds) PCR protocols. Academic, New York, p 482
Kuczynski J, Stombaugh J, Walters WA, et al (2011) Using QIIME to analyze 16S rRNA gene sequences from microbial communities. Curr Protoc Bioinformatics, UNIT 10.7. Wiley Online Library
Zimmerman NB, Vitousek PM (2012) Fungal endophyte communities reflect environmental structuring across a Hawaiian landscape. Proc Natl Acad Sci U S A 109:13022–13027
Acknowledgments
Amy E. McGovern very carefully produced high-quality sequences to support this project. The mention of firm names or trade products does not imply that they are endorsed or recommended by the US Department of Agriculture over the firms or similar products not mentioned.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer Science+Business Media LLC
About this protocol
Cite this protocol
Peterson, S.W. (2017). Targeting Conserved Genes in Penicillium Species. In: Moretti, A., Susca, A. (eds) Mycotoxigenic Fungi. Methods in Molecular Biology, vol 1542. Humana Press, New York, NY. https://doi.org/10.1007/978-1-4939-6707-0_9
Download citation
DOI: https://doi.org/10.1007/978-1-4939-6707-0_9
Published:
Publisher Name: Humana Press, New York, NY
Print ISBN: 978-1-4939-6705-6
Online ISBN: 978-1-4939-6707-0
eBook Packages: Springer Protocols