Abstract
GlobeImmune’s Tarmogen® immunotherapy platform utilizes recombinant Saccharomyces cerevisiae yeast as a vaccine vector to deliver heterologous antigens for activation of disease-specific, targeted cellular immunity. The vaccines elicit immune-mediated killing of target cells expressing viral and cancer antigens in vivo via a CD8+ CTL-mediated mechanism. Tarmogens are not neutralized by host immune responses and can be administered repeatedly to boost antigen-specific immunity. Production of the vaccines yields stable off-the-shelf products that avoid the need for patient-specific manufacturing found with other immunotherapeutic approaches. Tarmogens for the treatment of chronic hepatitis B and C and various cancers were well tolerated and immunogenic in phase 1 and 2 clinical trials encompassing >600 subjects. The platform is being widely utilized in basic vaccine research and the most rapid path to success in these endeavors follows from optimal immunoassay selection and execution. This chapter provides detailed methods for the construction and preclinical immunogenicity testing of yeast-based immunotherapeutic products to support the rapid and efficient use of this versatile technology.
Access provided by CONRICYT – Journals CONACYT. Download protocol PDF
Similar content being viewed by others
Keywords
- Tarmogen®
- Recombinant yeast
- Cellular immunity
- CD8 T cells
- CD4 T cells
- Th17 cells
- Immunotherapy
- Therapeutic vaccine
1 Introduction
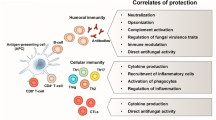
Tarmogens are comprised of heat-inactivated, whole recombinant Saccharomyces cerevisiae yeast cells expressing disease-related target antigens, most typically intracellularly. These vaccines possess unique cell surface molecular signatures (pathogen-associated molecular patterns) that are key agonists for phagocytic and toll-like receptors expressed by antigen-presenting cells (APCs). Following vaccination with Tarmogen, receptor binding and activation of APCs trigger yeast uptake and cross-presentation of heterologous yeast-expressed antigens with class I and class II MHC molecules, in turn stimulating CD4+ and CD8+ T-cell responses in vivo [1]. The yeast also induces the Th17 pathway resulting in reduced regulatory T-cell activity [2, 3]. With help from the CD4+ T-cell population, the CD8+ T cells specifically kill and/or clear virus-infected cells and tumor cells expressing the target antigen.
Tarmogens are being developed clinically for the treatment of chronic human viral infections and a variety of cancers [4]. The broad applicability of the platform is also being widely exploited in basic vaccine research.
Functional evaluation of Tarmogens is accomplished by any of a wide array of in vivo and in vitro assays that can illuminate the activity and mechanism in different host immunological backgrounds. For Tarmogens targeting tumor-associated antigens, many clinically relevant and tractable rodent models exist that require only lower biosafety level (e.g., ABSL1) animal facilities for execution and that inherently evaluate activity in the context of immunological tolerance and suppression [5–7]. Achieving a similar level of immunological relevance for infectious diseases often involves challenge of immunized animals with a target microorganism. As selection of a lead candidate Tarmogen would ideally include testing of multiple candidates and immunization regimen optimization, such infectious animal models can be prohibitively long and expensive.
These latter obstacles can be ameliorated with a project plan that includes (1) infectious ex vivo assays with T cells and monocyte-derived dendritic cells from a patient’s blood [8] and (2) noninfectious in vivo murine experiments emphasizing the cellular mechanisms of activity in the context of Tarmogen immunization. Following on this theme, we describe four methods that in our hands provide high-magnitude antigen-specific T-cell responses in a relatively short time frame with moderate resources.
2 Materials
Supplier details are provided where reagent source or quality is particularly important.
-
1.
S.cerevisiaehaploid yeast (e.g., genotype ade2-1; ura3-1; his3-11,15; trp1-1; leu2-3,112; can1-100; or a closely related strain).
-
2.
Two μm circle-based shuttle vector with yeast and E.coliorigins of replication, an auxotrophic selectable marker (e.g., URA3 or HIS3), a bacterial antibiotic resistance gene (β-lactamase), and a multiple cloning restriction cluster adjacent to the copper-inducible CUP1 promoter.
-
3.
General DNA cloning reagents (restriction enzymes, T4 DNA ligase, DH5-alpha competent E.coli, Luria broth liquid and solid plates each containing 50 μg/mL of ampicillin, and standard materials for agarose gel electrophoresis).
-
4.
Frozen-EZ yeast transformation kit II (Zymo Research, cat # T2001).
-
5.
Synthetic complete medium agar plates lacking uracil for selection of transformed yeast containing a URA3 selectable marker.
-
6.
Liquid synthetic complete medium lacking uracil (“U2”): 15 g/L glucose, 6.7 g/L yeast nitrogen base containing ammonium sulfate but lacking amino acids, 0.04 g/L adenine, 0.04 g/L histidine, 0.04 g/L tryptophan, and 0.06 g/L leucine. Sterilized by 0.2 μM filtration.
-
7.
Sterile 1 M copper sulfate solution.
-
8.
Calcium- and magnesium-free phosphate-buffered saline (PBS).
-
9.
Sterile disposable Erlenmeyer flasks with vented caps (2 L, 1 L, 250 mL sizes).
-
10.
2× complete yeast cell lysis buffer, per 100 mL: 4 g SDS, 2 mL β-me, 1 mg bromophenol blue, 1× protease inhibitor cocktail, and 1 mM EDTA.
-
11.
Acid-washed glass beads, 0.5 mm.
-
12.
User selected materials for SDS-PAGE and Western blot analyses.
-
13.
Hexahistidine-tagged NS3-his standard protein or equivalent purified his-tagged antigen for Western blot standard curves.
-
14.
Mouse monoclonal antibody recognizing a hexahistidine epitope.
-
15.
Horseradish peroxidase-coupled goat anti-mouse antibody.
-
16.
Enhanced chemiluminescence substrate (seeNote1).
-
17.
Bio-Rad digital imager with Chemidoc software or equivalent system for light emission quantification.
-
18.
Aim-V base medium.
-
19.
100× penicillin-streptomycin solution.
-
20.
Human type AB serum (Invitrogen/Life Technologies cat # 34005100).
-
21.
Complete Aim-V medium: Aim-V containing 10 % human type AB serum plus 1× penicillin-streptomycin.
-
22.
A nonenzymatic cell dissociation buffer.
-
23.
Recombinant human GM-CSF.
-
24.
Recombinant human IL-4.
-
25.
Recombinant human IL-2.
-
26.
PE-coupled antihuman interferon gamma (IFNγ) clone 4SB3.
-
27.
APC-Cy7-coupled antihuman CD8, clone SK1.
-
28.
PerCP/Cy5.5-coupled antihuman CD4, clone SK3.
-
29.
Peptides of 98 % purity.
-
30.
Intracellular cytokine staining kit.
-
31.
Human IFNγ ELISpot kit.
-
32.
Murine IFNγ ELISpot kit.
-
33.
Cesium-137 cell irradiator.
-
34.
Multichannel flow cytometer (recommended 6-color or higher capability).
-
35.
ViraPower Lentiviral Expression System (Invitrogen/Life Technologies cat # K4990-00; Note2).
-
36.
Thermal cycler.
-
37.
Animal research facility/vivarium with biological safety cabinet.
-
38.
Controlled temperature CO2 tissue culture incubator.
-
39.
Microcentrifuge and clinical swinging bucket centrifuges.
-
40.
Heat-inactivated fetal bovine serum.
-
41.
Lipofectamine 2000 (Invitrogen/Life Technologies cat # 11668027).
-
42.
25 g × 5/8 1 mL TB syringe (for subcutaneous (SC) injections).
-
43.
28 g insulin syringe (for intradermal (ID) injections).
-
44.
Electric razor for shaving murine injection sites (for ID vaccination and tumor implantation).
-
45.
Digital calipers for tumor measurements.
-
46.
Autoclavable stainless steel dissection tools (for lymph node and spleen removal).
-
47.
6-well flat bottom and 96-well round bottom tissue culture plates.
-
48.
Sterile 70 μm cell strainers.
-
49.
Sterile 50 mL polypropylene Falcon tubes.
-
50.
Anti-CD40 antibody (Bio-X-Cell cat. # BE0016-2).
-
51.
PKH26 labeling kit (Sigma cat # PKH26GL-1Kt).
-
52.
CFSE (carboxyfluorescein succinimidyl ester) labeling kit.
-
53.
Opti-MEM® reduced serum medium.
-
54.
Tissue culture grade DMSO.
-
55.
Gas pressure regulator and veterinary manifold.
-
56.
Isoflurane controlled release unit (seeNote3).
-
57.
Mouse cage with Plexiglass cover connected to gas supply hose.
-
58.
Compressed oxygen; 24 ft3-sized tank.
-
59.
Leucosep™ tubes for PBMC preparation from human whole blood (VWR cat # 89048-936).
3 Methods
3.1 Recombinant Yeast Vaccine Construction and Antigen Quantification
Design of heterologous antigen to be expressed in yeast
The heterologous disease antigen should ideally possess the following attributes: (1) high intracellular expression (preferably > 5 % of total yeast cellular protein), (2) the presence of key epitopes and antigenic regions that are relevant to the stage/cycle of the target disease, and (3) high solubility to avoid inducing the unfolded protein response or other deleterious pathways in the yeast [7, 9, 10]. We have generally included a C-terminal epitope tag (e.g., hexahistidine) to facilitate quantification of the antigen in yeast lysates (see below).
3.1.1 Expression Vector and Promoter Sequences
To obtain stable and reliable antigen expression in S.cerevisiae, select a shuttle vector plasmid with a 2 μm circle high-copy origin of DNA replication, a yeast auxotrophic “selectable” marker such as wild-type URA3, a β-lactamase (AmpR) for antibiotic selection, and a standard bacterial origin of replication. There are many choices of promoters that may be used to achieve high antigen expression in yeast, as reviewed in [11]. We have had good success with the copper-inducible CUP1 promoter [8, 9].
3.1.2 Cloning
-
1.
Standard procedures are deployed to insert a PCR-amplified or commercially synthesized DNA insert proximal to the promoter. The insert may be codon optimized for expression in S.cerevisiaebut this is usually not essential (seeNote4).
-
2.
Kozak sequence. Yeast mRNA translation is not as sensitive to the identity of the start codon-proximal nucleotides as are mammalian cells. Nevertheless, effects on translation have been observed and good guides on this topic are published [12].
3.1.3 Tarmogen Construction
This protocol is for use with CUP1 promoter-driven heterologous antigens. For strains with constitutive promoters, perform the same steps but omit the addition of copper sulfate (step 8).
-
1.
Obtain a Saccharomyces cerevisiaehaploid yeast strain harboring multiple auxotrophic markers for selection and maintenance of plasmid expression vectors (see Subheading 2). Revive the cells from a frozen vial by aseptic streaking on CM glucose agar plates followed by incubation at 30 °C for 3 days.
-
2.
Inoculate a 25 mL volume of synthetic complete liquid medium (“CLM”) with a single colony. Shake the culture at 250 rpm at 30 °C until a density of 3 × 107 cells/mL ± 0.5 × 107 cells/mL has been reached (seeNote5).
-
3.
Harvest cells and render them competent for DNA uptake using the Frozen-EZ yeast transformation kit II and conduct transfection of the yeast cells with plasmid DNA according to the kit protocol (Zymo Research).
-
4.
Plate 150 μL of the transfectant mixture on CM glucose minus uracil plates, and return to a 30 °C incubator for 3 days.
-
5.
Re-streak three individual colonies onto a fresh CM glucose minus uracil plates and return to 30 °C incubator for three additional days.
-
6.
Inoculate a small portion of each re-streaked transfectant into 20 mL of liquid CM glucose minus uracil medium and incubate at 30 °C (250 rpm) for 16 h (starter culture).
-
7.
Obtain a cell count of the three starters and use these to inoculate final cultures to 0.3 YU/mL (one YU = 107 yeast cells).
-
8.
Incubate the cultures to a density of 3 YU/mL ± 1 YU/mL and then add copper sulfate from a sterile 1 M stock to a final concentration of 400 μM. Return to shaker and incubate for a further 3 h at 30 °C (250 rpm).
-
9.
Harvest the cells by centrifugation at 2600 × g for 7 min at 20 °C. Resuspend the cell pellet by hand vortexing and then add PBS to 500 mL per liter of original culture, invert the container, and harvest again by centrifugation. Decant the supernatant and resuspend the yeast pellet in the residual liquid as above.
-
10.
Heat inactivate the yeast by adding the cell slurry to pre-warmed 56 °C PBS for 1 h (final yeast concentration: 10–50 YU/mL).
-
11.
Wash the cells three times in PBS as in step 9. Resuspend the Tarmogen at an estimated 50 YU/mL, sonicate an aliquot for 10 s to disperse any aggregates, and then obtain an exact cell density by hemocytometer count.
3.1.4 Preparation of Yeast Cell Lysate (See Note 6)
-
1.
Pellet 20 YU of Tarmogen by centrifugation at 5900 × g in a tabletop microcentrifuge for 4 min. Keep the cell pellet on ice.
-
2.
Prepare 500 μL of 2× complete cell lysis buffer per strain to be evaluated (materials).
-
3.
Remove the supernatant (step 1) and resuspend the cell pellet in 200 μL of 2× complete cell lysis buffer and add the slurry to a 3 mL polystyrene snap-cap tube containing 300 μL of acid-washed glass beads. Vortex by hand at maximum speed for 90 s and then heat at 95 °C for 3 min. Place the tube on ice for 1 min.
-
4.
Using a p200 pipet attached to a tip from which the very end has been cut off, stick the tip into the beads and withdraw all of the liquid and transfer it to a fresh 1.5 mL microcentrifuge tube.
-
5.
Add 200 μL of 2× SDS buffer to the beads and vortex for 15 s. Remove the liquid with a fresh cut tip and pool it with the previous aliquot of cell lysate.
-
6.
Spin the lysate at 9300 × g for 4 min to pellet debris, transfer the supernatant to aliquots, and freeze at −80 °C (seeNote7).
3.1.5 Measurement of Heterologous Antigen Content
-
1.
Conduct measurement of total protein in the lysate using a nitrocellulose-binding/TCA protein precipitation method as described previously [13] (seeNote8).
-
2.
Expression of the heterologous antigen is measured by standard Western blot. Estimation of antigen content is typically determined by interpolation of heterologous antigen against a standard curve comprised of known quantities of his-tagged HCV NS3 protein. Lanes with the following amounts of antigen generally produce a linear response: 200, 100, 50, and 25 ng (seeNote9).
3.2 Immunogenicity Assays
A wide array of in vivo and ex vivo assays were developed by our laboratory. The four assays described here are examples that provide high-amplitude antigen-specific Th1 T-cell responses. The use of human dendritic cell/peripheral blood mononuclear cell (PBMC) assays enables the study of cellular immune responses in the context of varied exposure to pathogens or in a variety of tumor patient backgrounds. The murine assays are relevant because T-cell responses are mounted in vivo by Tarmogen immunization. The in vivo cytotoxic T-lymphocyte (CTL) assay is an especially attractive test in cases where a dominant target epitope is known, and it attests to the primary mechanism by which Tarmogens function.
3.2.1 Ex Vivo Stimulation of PBMCs with Tarmogen-Pulsed, Autologous Dendritic Cells (DCs; ELISpot Readout)
In this assay, human donor PBMCs are cultured with Tarmogen-treated autologous DCs. Two or three 1-week periods (rounds) of stimulation trigger activation and expansion of disease antigen-specific T cells, resulting in a polyclonal population of lymphocytes whose specificity and activity can be evaluated with ELISpot, intracellular cytokine staining, pentamer staining, or CTL-mediated killing assays [8, 14].
-
1.
Prepare PBMCs from fresh whole blood of donors using Leucosep tubes.
-
2.
Incubate ten million viable PBMCs in a single well of a 6-well plate for 2 h in 5 mL of complete AIM-V medium (cAIM-V).
-
3.
Discard the nonadherent cells and gently rinse once with cAIM-V to remove loosely adherent cells.
-
4.
Add five mL of cAIM-V containing 100 ng/mL recombinant human GM-CSF plus 20 ng/mL recombinant human IL-4 to the adherent cells and incubate for 5 days at 37 °C.
-
5.
Estimate the number of DCs by trypan blue dye exclusion of a (destructive) sister well and then add Tarmogen at a ratio of 1 yeast cell to 1 DC. Return the plate to the incubator for an additional 40 h (seeNote10).
-
6.
Rinse the adherent yeast-fed cells twice with calcium- and magnesium-free PBS and then incubate for 15 min with 2–3 mL nonenzymatic cell dissociation buffer at 37 °C. Gently and repeatedly pipet the Tarmogen-pulsed DCs (TPDCs) to detach them from the plastic, irradiate the cells (30 Gy), and store on ice.
-
7.
Thaw 10–20 million frozen autologous PBMCs, wash them in pre-warmed cAIM-V, count, and combine the PBMCs with irradiated yeast-pulsed DCs at a ratio of 1:10 (DC:PBMC) in a T-75 tissue culture flask (seeNote11).
-
8.
Incubate the DC/PBMC co-culture for 3 days at 37 °C and then add recombinant human IL-2 at a final concentration of 20 U/mL for 4 additional days. This process comprises one round of stimulation (1 week).
-
9.
Transfer the suspension cells from the round 1 stimulation to a new flask and conduct a round 2 stimulation with freshly prepared TPDCs produced from frozen/banked autologous PBMCs.
-
10.
Repeat the process of DC co-culture once more (steps 7–9), using the suspension cells from round 2. This is the third round stimulation.
-
11.
On day 6 of the third round, thaw a vial of fresh autologous PBMCs, wash them in cAIM-V, and incubate with 3 μg/mL of purified recombinant target antigen for 24 h to allow antigen uptake and presentation by APCs.
-
12.
Wash the pulsed APCs (step 11) 1× in cAIM-V, count, and add them to the third round DC-stimulated effector population at a T cell/APC ratio of 10:1 for 36 h directly in an IFNγ ELISpot plate (200,000 cells/well). Develop the ELISpot plate per manufacturer’s protocols. ELISpot counting can be conducted by Cellular Technology, Ltd.
3.2.2 Ex Vivo Stimulation of PBMCs with Tarmogen-Pulsed, Autologous DCs (Intracellular Cytokine Staining Readout)
As an alternative or in addition to the ELISpot assay of step 12, an intracellular cytokine staining assay can be conducted that is useful for establishing the T-cell subsets that have been activated as well as their epitope sequence specificity (seeNote12). In the present example, protocols are listed for determining the frequency of CD4+ or CD8+ IFNγ+ T cells. For antibody concentrations used for staining steps, follow manufacturer’s recommendations.
-
1.
Generate effectors by stimulation with TPDCs as in steps 1–11, Subheading 3.2.1.
-
2.
Collect an aliquot of cells following complete three-round stimulation, by centrifugation at 300 × g for 6 min.
-
3.
Treat the cells with 1× GolgiStop from the BD Cytofix-Cytoperm kit in the presence of disease antigen-specific peptides for 5 h at 37 °C. Stain the cells with antihuman CD8/APC-Cy7 antibody and antihuman CD4/PerCP-Cy5.5 antibody and then wash the cells 3 times with cold PBS + 1 % BSA.
-
4.
Fix the stained cells with 4 % paraformaldehyde for 10 min at 25 °C, permeabilize with the Cytofix-Cytoperm kit, and stain the cells with a PE-coupled antihuman IFNγ antibody. Analyze the cells by flow cytometry to determine the percentages of each T-cell subset that produced IFNγ in response to peptide stimulation.
3.2.3 Tumor Challenge in Tarmogen-Vaccinated Mice
Tumor challenge assays are a time-proven gold standard measure of immunogenicity, and yet establishing the optimal conditions for use with Tarmogens involves knowledge of tumor target construction, careful selection of immunization dose/regimen, tumor challenge dose and timing, and, in the case of adoptive transfer studies, procedural details to maximize the expansion of transferred T cells prior to challenge. Following these protocols will accelerate the completion of conclusive tumor challenge studies with Tarmogens.
3.2.4 Target Construction
It is recommended to use a system for antigen expression that results in stable chromosomal integration of the target gene such that cells can ultimately be propagated in the absence of drug selection in vivo (seeNote13).
-
1.
EL4 thymoma cells (ATCC) are grown in cRPMI-10 to mid-exponential phase and then transduced by lentivirus expressing the target gene (seeNote14). Cloning, transduction, and drug selection procedures recommended by Invitrogen (Life Technologies) work well for most genes.
3.2.5 Adoptive Transfer-Based Tumor Protection
-
1.
Immunize C57BL/6 mice subcutaneously once per week for 3 weeks (this regimen has not been fully optimized). Immunize with 2.5 YU of Tarmogen in the outer flank and 2.5 YU in the scruff of the neck. Rotate sides of flank injection each week to minimize irritation.
-
2.
One week after the third immunization, harvest splenocytes by gentle dispersal over a 70 μm cell strainer and conduct one wash with cRPMI-10. Do not perform ammonium chloride-potassium lysis of red blood cells.
-
3.
Adjust the preparation to 2 × 107 viable cells/mL and inject 1 mL into the intraperitoneal cavity of 5–7 week old scid mice.
-
4.
Twenty four hours later (to allow for in vivo T-cell expansion), challenge the scid mice subcutaneously in the (shaved) ribcage with 30,000 EL4 tumor cells expressing a relevant (i.e., target) or irrelevant control antigen (seeNote15).
-
5.
Monitor tumor growth daily starting 48 h after challenge. Tumors will likely become visible in control (PBS or irrelevant yeast) groups ~day 6 or 7 post-challenge (seeNote16).
-
6.
When tumors are large enough to be measured by digital caliper, collect tumor diameter measurements daily, in two perpendicular dimensions for each mouse. Plot average tumor diameter or tumor volume as a function of time to estimate growth rate.
3.2.6 Interferon Gamma ELISpot in Tarmogen-Immunized Mice
Ex vivo T-cell activation assays are valuable protocols for assessing the antigen specificity of therapeutic vaccines. For Tarmogens, we have found that several differences from traditional methods result in enhanced antigen-specific responses. Heterologous antigen expression level, immunization regimen/dose, period of rest prior to dissection, and the type and purity of antigen are all factors that can affect assay quality.
-
1.
Immunize mice with 5 YU of Tarmogen by the intradermal or subcutaneous routes (2.5 YU each outer flank) at days 0 and 7. To achieve representative results, it is recommended to immunize at least seven mice per treatment group (seeNote17).
-
2.
Nine to 21 days post-immunization, remove the draining inguinal lymph nodes from the 7 mice/group, pool the organs for each group, and immediately disperse the cells into cold cRPMI-10 over a 70 μM cell strainer using the flat end of a 3 mL syringe plunger.
-
3.
Wash the cells once in 50 mL cRPMI-10, resuspend in 3 mL medium, and obtain viable cell counts by hemocytometer counts of trypan blue-stained cells.
-
4.
Adjust the preparation to 2.5 million viable cells per mL and aliquot 100 μL per well into 96-well round bottom tissue culture plates.
-
5.
Add 100 μL of target or irrelevant antigen in cRPMI-10 to each well, mix by gentle pipetting, and place the covered plate in a humidified 37 °C/5 % CO2 tissue culture incubator for 4 days (seeNote18).
-
6.
Pipet the cells up and down 3 times with a multichannel pipet and transfer 150 μL of the mixture to a pre-blocked 96-well IFNγ ELISpot plate. Return the plate to the CO2 incubator for 24 h.
-
7.
Wash the cells off of the ELISpot plate and develop the assay exactly per R&D systems kit directions. Dry plates at 37 °C for 30–60 min prior to spot counting.
3.2.7 In Vivo CTL
The ability of Tarmogens to trigger cytotoxic T-cell-mediated killing is central to the mechanism of action, and here we present a rapid (8-day), simple, and highly relevant in vivo assay to assess this function. A key requirement is that a class I MHC-restricted dominant epitope is known. The assay can be run in most any strain of mouse including HLA transgenic mice to allow the possibility of testing for CTL against epitopes of known importance in human disease.
The key steps are (1) immunization of mice (day 0), (2) injection of immunized and naive mice with dye-labeled and peptide-pulsed and unpulsed splenocytes (day 7), and (3) removal of spleens followed by analysis of percent killing of the dye-labeled population by flow cytometry.
3.2.7.1 Immunization of Mice with Tarmogen
-
1.
Immunize mice retro-orbitally or by tail vein injection with 2 YU of Tarmogen mixed with 25 μg of α-CD40 antibody (seeNote19).
3.2.7.2 Target Preparation
Except for step 6, all centrifugation steps are performed at 400 × g for 10 min.
-
1.
On day 7 post-immunization, harvest spleens from naive syngeneic mice and prepare them as targets for killing as follows. The following is written for a single spleen which contains sufficient cells to inject four recipient mice. A total of 4 × 106 cells/recipient are required (2 × 106 peptide-pulsed and 2 × 106 unpulsed cells).
-
2.
Macerate spleen in PBS + 1 % BSA (wash medium; W1) and filter cells into a 50 ml Falcon tube using a 70 μM mesh filter and centrifuge. Discard supernatant and continue with step 3. Perform all subsequent steps at room temperature.
-
3.
Lyse red blood cells for 5 min by resuspending pellet in 2 mL ACK (ammonium chloride-potassium) lysing solution per spleen.
-
4.
Add 15 mL complete RPMI-10 % FBS (cRPMI-10) to stop the reaction. Centrifuge, resuspend the cells in 10 mL W1, and obtain a viable cell count.
-
5.
Reserve 4 × 106 unstained cells as a control (2 × 106 to be used as no staining control and 2 × 106 for CFSE (carboxyfluorescein succinimidyl ester)-only stained control).
-
6.
Centrifuge the cells from step 5 at 400 × g for 5 min into a loose pellet.
-
7.
Carefully aspirate the supernatant leaving no more than 25 μL of supernatant on the pellet.
-
8.
Based on the estimated number of cells from the count in step 5, resuspend the cells at 2 × 107/ml in Diluent C from the PKH26 staining kit (this is a 2× cell suspension). Ensure the cell suspension is uniform but do not vortex.
-
9.
Immediately prior to staining, prepare 8 × 10−6 M PKH26 dye (2× mix) in a polypropylene tube using Diluent C. If an intermediate dilution of dye is necessary (unlikely), do so in 100 % ethanol. Hold this preparation at 25 °C.
-
10.
Add 8 μL of 1 × 10−3 M PKH dye stock per 1 ml Diluent C. Rapidly add the 2× cells to the 2× dye and immediately mix the sample by pipetting. Rapid and homogenous mixing is critical for uniform labeling because staining is nearly instantaneous.
-
11.
Incubate the cells at 25 °C for 3 min. Periodically, invert the tube gently to ensure mixing during this staining period at 25 °C.
-
12.
Stop the staining reaction by adding an equal volume of 100 % fetal bovine serum (2 mL cell/dye mix + 2 mL serum). Incubate for 1 min at 25 °C.
-
13.
Dilute the serum-stopped sample with an equal volume of cRPMI-10.
-
14.
Centrifuge the cells at 400 × g for 10 min at room temperature to remove cells from staining solution (seeNote20).
-
15.
Remove the supernatant and wash cells with 15 ml cRPMI-10 (a minimum of three washes is recommended). Perform the last wash using 15 mL PBS. Count an aliquot of the cells while in the last centrifugation step and resuspend the cells to 4–5 × 107 cells/mL in PBS. Retain 5–10 μL of the PKH26 stained preparation as a single-stained control.
-
16.
Divide the PKH26-stained cells equally into two polypropylene tubes. One sample will be stained with 1 μM CFSE (CFSE high) and the second sample with 0.1 μM CFSE (CFSE low).
-
17.
Prepare a 2× CFSE pre-mixture (2 μM and 0.2 μM, respectively) in PBS and add an equal volume to the PKH26-labeled cells (1 mL cells + 1 mL 2× CFSE). Stain at room temperature for 10 min.
Also in parallel, treat 2 × 106 unstained splenocytes from above with 1 μM CFSE in a volume of 2 mL for 10 min (1 mL cells + 1 mL 1 μM CFSE).
-
18.
Stop the CFSE labeling by adding equal volume of 100 % FBS for 1 min and then add one volume of cRPMI-10 and centrifuge as above.
-
19.
Wash the cells once more with cRPMI-10 and once with W1. Resuspend the cells stained with 1 μM CFSE (CFSE-high cells) in serum-free medium (SFM) for peptide pulsing. Use 1 mL SFM for cells from up to five donor spleens and increase volume accordingly dependent on number of spleens used. Resuspend the cells stained with 0.1 μM CFSE (CFSE-low cells) in 5 mL PBS and hold at 4 °C until further use.
-
20.
Pulse the CFSE high population with 100 μM peptide (seeNote21) in polypropylene tubes at 37 °C 5 % CO2 for 60 min.
-
21.
Dilute the peptide-pulsed splenocytes to 20 mL with SFM and centrifuge for 10 min. Aspirate the supernatant carefully to remove as much medium as possible without losing cells.
-
22.
Wash the CFSE-high cells with 20 mL PBS, centrifuge, and resuspend the pellet in 5 mL PBS.
-
23.
Count both CFSE-high and CFSE-low preparations and mix 2 × 106 pulsed (CFSE-high) cells with 2 × 106 unpulsed (CFSE-low) cells in 100 μL total volume per mouse to be injected. Inject the suspension retro-orbitally into naïve and into Tarmogen-immunized mice using a U-100 insulin syringe, 28G1/2 (0.36 mm × 13 mm).
3.2.7.3 Harvest and Processing of Spleens for Flow Cytometry
-
1.
20-h post-challenge, remove spleens and process organs as described in steps 2–4.
-
2.
Transfer 10–15 × 106 cells to a 15 ml Falcon tube and adjust the cell concentration to 5 × 106/ml using W1.
3.2.7.4 Flow Cytometric Analysis
-
1.
Transfer 2 ml of each sample prepared in step 2 above to a 5 mL polystyrene tube. Filter over a 70 μM mesh cell strainer if cell clumps are visible. Resuspend the cells used as single stain controls from steps 5, 15, and 17 in 1 mL W1 and also transfer to a 5 mL polystyrene tube. Keep samples on ice.
-
2.
On the flow cytometer, create the experiment layout for analysis. Include a dot plot for forward (x-axis) and side scatter (y-axis), as well as a dot plot displaying PKH26 (x-axis) against side scatter (y-axis). Add a histogram plot for displaying CFSE (x-axis) against counts (y-axis).
-
3.
Use the unstained control to adjust forward and side scatter as well as photomultipliers for the detectors used (seeNote22).
-
4.
Compensate the spillover between CFSE and PKH26 using the single stain controls (seeNote23).
-
5.
Run the sample corresponding to the “naïve” mouse (seestep 23) to set and adjust the gates to display the different populations. Begin with adding a lymphocyte gate to the forward-side scatter plot (gate 1). In the “PKH26 vs. side-scatter” plot, display events from gate 1 and set a 2nd gate around the PKH26-positive population (seeNote24). In the CFSE histogram plot, display the PKH26-positive cells (gate 2). The CFSE-labeled cells should be visible as two separate peaks, CFSE-high and CFSE-low cells, respectively. Set “region” gates for each peak to determine the frequency of CFSE-high cells versus CFSE-low cells (seeNote25).
-
6.
After gates have been set, record the sample and continue to acquire the rest of the samples in the experiment. In order to obtain a satisfying analysis, it might be required to record up to 5 × 106 events in gate 1, as PKH26-/CFSE-labeled cells can be limited.
-
7.
The efficacy of cytotoxic T-cell lysis (target-cell killing) of peptide-pulsed (CFSE-high) cells is measured in percent and calculated by the following formula:
$$ \% \mathrm{killing}=\left(1-\frac{\mathrm{immunized} \mathrm{CFSE} \mathrm{h}\mathrm{i}/\mathrm{CFSE} \mathrm{low}}{\mathrm{Na}\ddot{\i}\mathrm{v}\mathrm{e} \mathrm{CFSE} \mathrm{h}\mathrm{i}/\mathrm{CFSE} \mathrm{low}}\right)\times 100 $$
4 Notes
-
1.
Optimal sensitivity is obtained with a digital imager such as Bio-Rad’s digital imaging system interfaced to ChemiDoc software.
-
2.
The polyubiquitin C or cytomegalovirus promoters are good choices for constitutive mid- to high-level expression; these may be selected upon purchase of the lentiviral kit.
-
3.
Isoflurane anesthesia is used for the intradermal and retro-orbital injection procedures described in the Methods section.
-
4.
The benefit of codon optimization for expression in yeast is controversial, and in our hands, many DNA sequences encoding viral pathogens or tumor-associated antigens express highly without optimization.
-
5.
Before counting yeast cell by hemocytometer, the culture should be sonicated on low power for 10 s to ensure dispersal of cell aggregates.
-
6.
The combination of mechanical shearing, high SDS (4 %), and heat (95 °C) is optimal to obtain high efficiency of lysis from heat-killed yeast.
-
7.
Yeast total protein lysates are optimally stored at −80 °C in aliquots.
-
8.
The use of the amido-schwartz TCA precipitation method is important for lysates produced by this method, as it is insensitive to high SDS concentrations. Unlike certain other popular methods, it is also inclusive of membrane proteins and therefore produces a more accurate estimate of total cellular protein than other commercial methods.
-
9.
Despite that the ECL product has a stabilizer, data should be collected immediately upon exposure to substrate as antigen content results can vary by as much as 100 % when longer ECL substrate incubation is conducted (e.g., 30 min) prior to digital image collection.
-
10.
The yeast/DC ratio should be titrated to achieve optimal separation of the Tarmogen-emergent T-cell effect to the background signal arising from negative control empty vector yeast cells.
-
11.
On the same day as the autologous PBMCs are thawed for co-culture with yeast-pulsed DCs, a fresh lot of DCs must be initiated as in step 2.
-
12.
As an example of evaluating other markers, antibodies that can detect degranulation may be added concomitant with CD8 marker staining to determine if the cells are cytolytic [15].
-
13.
Plasmid-based nonviral transfection methods can be used to establish antigen expression but the frequency of stable integration is low and thus antigen expression will usually be lost in the absence of drug selection in vitro and in vivo. We have isolated “breakthrough” tumors (those that continued to grow rapidly even in vaccinated mice) and found that the vast majority of cells in these tumors were devoid of heterologous antigen expression
-
14.
The operator may also choose to fuse the target gene to a ubiquitin sequence that accelerates proteasomal degradation and enhances antigen presentation with class I MHC. However, if using this proteasomal targeting approach, detection of antigen expression by Western blot can be difficult and requires the use of proteasome inhibitors prior to lysate preparation [16].
-
15.
We have found that 30,000–90,000 target cells provided optimal results in many studies, with tumors forming in 90 % of mice treated with empty vector control yeast. However, it is recommended to titrate the dose for each tumor line that expresses a unique antigen, as some heterologous proteins affect tumor growth rate. It is also noted that the well-known EL4-based tumor line E.G7-Ova grows notably more slowly than EL4 cells expressing, e.g., viral antigens. This can affect the interpretation of results when the protective effect of one Tarmogen vaccine is assessed against two different tumor targets lines. Always determine the growth rate of different EL4 lines before comparing them side by side in vaccine studies.
-
16.
The greatest separation of tumor growth rate between Tarmogen vs. control yeast-treated mice is often observed in the first few days after tumors have appeared.
-
17.
Higher numbers of Langerhans cells are found in the dermal layer than subcutaneously and these cells cross-prime CD8+ T cells with particularly high efficiency [17]. Thus, and especially if seeking to characterize CD8 T-cell responses, it is logical to immunize intradermally. C57BL/6 mice are recommended for these studies due to the Th1 bias of this strain. Nevertheless, CTL can be induced by Tarmogens in other common laboratory strains such as BALB/c and even in transgenic mice expressing high levels of self-antigens [5, 9].
-
18.
For each antigen stimulation, at least six replicate wells should be processed because the efficiency of T-cell activation and expansion can vary greatly from well to well.
-
19.
This amount of yeast and αCD40 Ab has been optimized; higher levels of αCD40 may result in sporadic death of mice.
-
20.
Do not reduce the time of these spins to 5 min, or considerable cell loss might occur.
-
21.
We recommend that peptides be of high purity (98 %) for best results and to ensure reproducibility, as crude peptide preparations can vary dramatically in quality from lot to lot.
-
22.
PKH26 can be detected in the PE channel, whereas CFSE emits in the FITC channel.
-
23.
Both PKH26 and CFSE have excitation and emission spectra that are different from PE and FITC, respectively, and therefore need to be compensated separately, using PKH26- and CFSE-labeled cells.
-
24.
PKH26 is used as a vital dye and only living cells will be labeled. The abundance of those cells within a whole spleen can be very limited. Thus, depending on your flow cytometer, either display at least 5000 events in the “PKH26 against side-scatter” dot plot or record approximately 1 × 106 cells in order to be able to confidently identify the PKH26-positive population.
-
25.
In the naïve mouse, the ratio of CFSE-high cells to CFSE-low cells should be approximately 1:1, as killing of the peptide-pulsed (CFSE-high) population does not occur in those animals. Ratios might be skewed toward the CFSE-high or CFSE-low population if cell counts were inaccurate and/or if the pulsed and unpulsed cells were not mixed in a 1:1 ratio in step 23. Nevertheless, the formula used to calculate the percentage of target-cell killing in the immunized animal will account for such variability.
References
Stubbs A, Martin KS, Coeshott C et al (2001) Whole recombinant yeast vaccine activates dendritic cells and elicits protective cell-mediated immunity. Nat Med 7:625–629
Cereda V, Vergati M, Huen N-Y et al (2011) Maturation of human dendritic cells with Saccharomyces cerevisiae (yeast) reduces the number and function of regulatory T cells and enhances the ratio of antigen-specific effectors to regulatory T cells. Vaccine 29:4992–4999
Cheng S-C, van de Veerdonk FL, Lenardon M et al (2011) The dectin-1/inflammasome pathway is responsible for the induction of protective T-helper 17 responses that discriminate between yeasts and hyphae of Candida albicans. J Leukoc Biol 90:357–366
Gaggar A, Coeshott C, Apelian D et al (2014) Safety, tolerability and immunogenicity of GS-4774, a hepatitis B virus-specific therapeutic vaccine, in healthy subjects: a randomized study. Vaccine 32:4925–4931
Wansley EK, Chakraborty M, Hance KW et al (2008) Vaccination with a recombinant Saccharomyces cerevisiae expressing a tumor antigen breaks immune tolerance and elicits therapeutic antitumor responses. Clin Cancer Res 14:4316–4325
Ardiani A, Farsaci B, Rogers CJ et al (2013) Combination therapy with a second-generation androgen receptor antagonist and a metastasis vaccine improves survival in a spontaneous prostate cancer model. Clin Cancer Res 19:6205–6218
Ardiani A, Gameiro SR, Palena C et al (2014) Vaccine-mediated immunotherapy directed against a transcription factor driving the metastatic process. Cancer Res 74:1945–1957
King TH, Kemmler CB, Guo Z et al (2014) A whole recombinant yeast-based therapeutic vaccine elicits HBV X, S and Core specific T cells in mice and activates human T cells recognizing epitopes linked to viral clearance. PLoS One 9(7):e101904
Haller A, Lauer GM, King TH et al (2007) Whole recombinant yeast-based immunotherapy induces potent T cell responses targeting HCV NS3 and Core proteins. Vaccine 25:1452–1463
Wu H, Ng BSH, Thibault G (2014) Endoplasmic reticulum stress response in yeast and humans. Biosci Rep 34:321–330
Shen MWY, Fang F, Sandmeyer S et al (2012) Development and characterization of a vector set with regulated promoters for systematic metabolic engineering in Saccharomyces cerevisiae. Yeast 29:495–503
Nakagawa S, Niimura Y, Gojobori T et al (2008) Diversity of preferred nucleotide sequences around the translation initiation codon in eukaryote genomes. Nucleic Acids Res 36:861–871
Schaffner W, Weissmann C (1973) A rapid, sensitive, and specific method for the determination of protein in dilute solution. Anal Biochem 56:502–514
Hamilton DH, Litzinger MT, Jales A et al (2013) Immunological targeting of tumor cells undergoing an epithelial-mesenchymal transition via a recombinant brachyury-yeast vaccine. Oncotarget 4:1777–1790
Betts MR, Brenchley JM, Price DA et al (2003) Sensitive and viable identification of antigen-specific CD8+ T cells by a flow cytometric assay for degranulation. J Immunol Methods 281:65–78
Andersson HA, Barry MA (2004) Maximizing antigen targeting to the proteasome for gene-based vaccines. Mol Ther 10:432–446
Liard C, Munier S, Joulin-Giet A et al (2011) Intradermal immunization triggers epidermal Langerhans cell mobilization required for CD8 T-cell immune responses. J Invest Dermatol 132:615–625
Acknowledgements
We thank Kirk Christoffersen and Dr. Angela Sebor for critical review of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2016 Springer Science+Business Media New York
About this protocol
Cite this protocol
King, T.H., Guo, Z., Hermreck, M., Bellgrau, D., Rodell, T.C. (2016). Construction and Immunogenicity Testing of Whole Recombinant Yeast-Based T-Cell Vaccines. In: Thomas, S. (eds) Vaccine Design. Methods in Molecular Biology, vol 1404. Humana, New York, NY. https://doi.org/10.1007/978-1-4939-3389-1_35
Download citation
DOI: https://doi.org/10.1007/978-1-4939-3389-1_35
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-4939-3388-4
Online ISBN: 978-1-4939-3389-1
eBook Packages: Springer Protocols