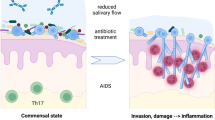
Abstract
Interactions of C. albicans with co-colonizing bacteria at mucosal sites can be synergistic or antagonistic in disease development, depending on the bacterial species and mucosal site. Mitis group streptococci and C. albicans colonize the oral mucosa of the majority of healthy individuals. These streptococci have been termed “accessory pathogens,” defined by their ability to initiate multispecies biofilm assembly and promote the virulence of the mixed bacterial biofilm community in which they participate. To demonstrate whether interactions with Mitis group streptococci limit or promote the potential of C. albicans to become an opportunistic pathogen, in vitro and in vivo co-infection models are needed. Here, we describe two C. albicans-streptococcal co-infection models: an organotypic oral mucosal tissue model that incorporates salivary flow and a mouse model of oral co-infection that requires reduced levels of immunosuppression compared to single fungal infection.
Access provided by CONRICYT – Journals CONACYT. Download protocol PDF
Similar content being viewed by others
Key words
1 Introduction
Oropharyngeal candidiasis (OPC) is an opportunistic infection afflicting humans in a variety of immunosuppressed states, which may also predispose them to invasive infection [1]. Although Candida albicans is the primary etiologic pathogen in OPC, the microbial ecology of this infection is complex since it contains members of the endogenous mucosal bacterial flora [2–4]. Interactions of C. albicans with co-colonizing bacteria at mucosal sites can be synergistic or antagonistic in disease development, depending on the bacterial species and mucosal site [5]. Synergistic interactions in biofilm development between C. albicans and oral streptococci have been demonstrated by our group and others in vitro [6–10]. More recently we examined the outcome of these interactions in a mouse oral co-infection model with C. albicans and S. oralis [11]. In this model S. oralis promoted the virulence of C. albicans in the oral mucosa by enhancing the inflammatory response to infection, without significantly affecting fungal burdens.
Here we describe our in vitro mucosal flow cell system and mouse oral co-infection models which can be used to further characterize the role of the interaction between C. albicans and oral streptococci in the pathogenesis of mucosal infection.
2 Materials
2.1 In Vitro Salivary Flow Three-Dimensional Mucosal Model
2.1.1 Plates/Inserts
-
1.
Plates: six-well Transwell Carrier (Organogenesis, TS01-001, Canton, MA).
-
2.
Corning Costar Transwell 3414 (24 mm diameter inserts, 3.0 μm pore size, Tewksbury, MA).
2.1.2 Fibroblast Line
-
1.
3T3 cells (ATCC).
2.1.3 Fibroblast Culture Media: DMEM-10 % FBS-1 % P/S
-
1.
DMEM: Dulbecco’s Modified Eagle Medium (DMEM) (1×) liquid (high glucose) without pyruvate: DMEM (−) (Gibco #11965-084).
-
2.
FBS: fetal bovine serum (HyClone).
2.1.4 Epithelial Cells
OKF6/TERT2 cells (floor of the mouth epithelium, obtained from J. Rheinwald, Harvard University) (see Note 1 ).
2.1.5 Epithelial Cell Culture Media
KSFM (Invitrogen, #17005-042) with Bovine Pituitary Extract (BPE) (1 tube/each bottle), hEGF (0.2 ng/ml), CaCl2 (0.4 mM), and Pen/Strep (1:100).
2.1.6 Matrix and Cell Culture Media Components
-
1.
DMEM (−) (Gibco #11965-084).
-
2.
Ham’s F12 (Gibco #11765-054).
-
3.
10× EMEM (BioWhittaker #12-684F).
-
4.
7.5 % Na-Bicarbonate (BioWhittaker #17-613E).
-
5.
FBS (HyClone).
-
6.
l-Glutamine (Cellgro #25-005CI).
-
7.
Type I Collagen (Organogenesis #200/50).
-
8.
Matrigel (BD Biosciences #354234).
-
9.
Hydrocortisone (Sigma #H0888): MW = 362.46.
-
Dissolve 0.0269 g in 2.5 ml EtOH.
-
Add into 97.5 ml DMEM (−): 0.74 mM.
-
Filter-sterilize, dispense into aliquots, and store at −20 °C.
-
-
10.
ITES—Insulin, Transferrin, Ethanolamine, and Selenium (BioWhittaker #17839Z).
-
11.
O-phosphorylethanolamine (Sigma #P0503).
-
Dissolve 0.705 g in 100 ml DMEM (−): 50 mM.
-
Filter-sterilize, dispense into aliquots, and store at −20 °C.
-
-
12.
Adenine (Sigma #A9795).
-
Dissolve 1.55 g in 100 ml warm (37 °C) ddH2O: 0.09 M.
-
Filter-sterilize, dispense into aliquots, and store at −20 °C.
-
-
13.
Progesterone (Sigma #P8783).
-
Dissolve 1 mg in 1 ml ETOH.
-
Add 14.7 ml ddH2O.
-
Dilute 1 ml of that in 100 ml DMEM (−): 2.0 μM.
-
Filter-sterilize, dispense into aliquots, and store at −20 °C.
-
-
14.
Triiodothyronine (Sigma #T5516).
-
Dissolve 1 mg in 1 ml 1N NaOH.
-
Add 19 ml of DMEM (−).
-
Dilute 4 μl of that in 31 ml plain DMEM (−): 1 nM.
-
Filter-sterilize, dispense into aliquots, and store at −20 °C.
-
-
15.
Newborn Calf Serum (HyClone #SH 3011802).
-
16.
Gentamicin, 50 mg/ml (Cellgro #MT30-0005-CR).
-
17.
Saliva, collected from systemically healthy volunteers (IRB approval is required). Whole stimulated saliva is collected in polypropylene tubes on ice, pooled, and treated with 2.5 mM dithiothreitol (Sigma-Aldrich, St. Louis, MO) for 10 min to reduce salivary protein aggregation. The saliva is then centrifuged at 7500 × g, at 4 °C, for 20 min, and supernatants are diluted with Dulbecco’s phosphate-buffered saline (D-PBS) to obtain a 25 % (vol/vol) saliva/D-PBS solution. Diluted saliva is then filtered through a 0.22-μm-pore-size polyethersulfone low-protein-binding filter (Nalgene; Thermo Fisher Scientific, Rochester, NY), divided into aliquots, and frozen at −80 °C until further use in culture media.
2.2 In Vivo Oral Infection Mouse Model
Mice: 6−8 weeks old female C57BL/6 mice are purchased from the Jackson Laboratory (Maine, US). Animals are maintained in BSL-2 biocontainment during the infection process.
2.3 Microorganisms Used and Microbiological Media
Organisms that can form mixed mucosal biofilms in both in vitro and in vivo models are Candida albicans SC5314 [12], Streptococcus oralis 34 (kindly provided by P. E. Kolenbrander), and Streptococcus gordonii Challis CH1 (kindly provided by J. M. Tanzer) [8, 11]. Streptococcus sanguinis SK36 (ATCC BAA-1455) can also form a biofilm with C. albicans under flow in vitro [8]. C. albicans is routinely maintained in yeast extract-peptone-dextrose (YPD) agar and grown in YPD medium, aerobically, at room temperature, in a rotor shaker. YPD medium consists of 5 g of yeast extract (Fisher Scientific, Pittsburgh, PA) liter−1, 10 g of peptone (Fisher Scientific) liter−1, and 20 g dextrose (Fisher Scientific) liter−1. Streptococci are routinely grown in brain heart infusion (BHI) medium (Oxoid, Ltd., Cambridge, UK) under static conditions at 37 °C, 5 % CO2.
3 Methods
3.1 In Vitro Flow Three-Dimensional Mucosal Model
3.1.1 Preparation of Oral Mucosa Tissue Analogue [13–15]
Day 0
-
Thaw Matrigel overnight (one vial for 2.5 OTC plates) at 4 °C.
Day 1
-
15–30 min prior to matrix preparation:
-
Place FBS, l-Glutamine, 10× EMEM and 7.5 % Na-bicarbonate, Matrigel, and type I collagen on ice.
-
Place transwell inserts into each well in six-well Transwell Carrier plate.
3.1.2 Making Acellular Layer
-
1.
Add 10× EMEM, FBS, l-Glutamine, Na-bicarbonate, and type I collagen in this order in 50 ml tube on ice (Table 1).
Table 1 Acellular layer media -
2.
Mix gently using a 25 ml pipette in a 50 ml tube on ice (see Note 2 ).
-
3.
Pour 1 ml per insert using a 10 ml pipette.
-
4.
Leave inside the tissue culture hood while preparing fibroblasts and cellular layer (Table 1).
3.1.3 Making Cellular Layer
-
1.
Prepare 6 × 105/ml 3T3 cell suspension.
-
2.
Add 10× EMEM, FBS, l-Glutamine, Na-bicarbonate, type I collagen, Matrigel, and fibroblasts in this order into a new 50 ml tube on ice (Table 2).
Table 2 Cellular layer media -
3.
Mix gently using a 25 ml pipette in 50 ml tube on ice (see Note 2 ).
-
4.
Pour 3 ml per insert using a 10 ml pipette.
-
5.
Incubate for 30–45 min in the tissue culture incubator (37 °C, 5 % CO2).
-
6.
Add DMEM-10 % FBS-1 % P/S: 10 ml into the bottom of the wells, 2 ml into the insert.
Day 2—Dislodge matrix and add 2 ml of DMEM-10 % FBS-1 % P/S into the insert.
-
1.
Use a sterile glass Pasteur pipette to go around (two to three times) the matrix along the inner wall of the insert. Feel the friction at the tip on the transwell membrane, but try not to pierce it (see Note 3 ).
-
2.
No need to change medium. Wait at least 4 days before seeding epithelial cells.
Day 5 or later—Seeding epithelial cells.
-
1.
Make DMEM (−)/F12 (Table 3) to pre-saturate the OTC matrix.
Table 3 DMEM/F12 media -
2.
Remove old medium from the OTC plates/inserts.
-
3.
Add DMEM+F-12. Add 10 ml to the bottom well and 2 ml into the insert.
-
4.
Incubate for 1 h to equilibrate the matrix.
-
5.
Prepare 1 × 107/ml epithelial cell suspension in the regular medium used to grow epithelial cells (e.g., KSFM for OKF6/TERT2) (see Note 4 ).
-
6.
Remove DMEM+F-12.
-
7.
Seed epithelial cells by adding 50 μl (5 × 105) of cell suspension per well onto the center of the surface of contracted matrix.
-
8.
Incubate for 2 h in the tissue culture incubator without medium.
-
9.
Make EP2 (Table 4). Store at 4 °C.
Table 4 EP2 media -
10.
Add EP2 to plate—10 ml into the bottom well and 2 ml into the insert.
Day 7 (or 2 days after seeding epithelial cells)—Medium change with EP2.
Day 9 (or 4 days after seeding epithelial cells, Air Lifting)—Medium change with EP3.
-
1.
Make EP3 (Table 5). Store at 4 °C.
Table 5 EP3 media -
2.
Remove old medium from both inserts and the bottom well.
-
3.
Add 7.5 ml of EP3 into the bottom wells only.
Day 11 (or 6 days after seeding epithelial cells)—Medium change with EP3, prepared on day 9.
-
1.
Remove old medium from both inserts and the bottom well (see Note 5 ).
-
2.
Add 7.5 ml of EP3 into the bottom wells only.
Day 13 (or 8 days after seeding epithelial cells)—Medium change with EP4 (infection medium).
-
1.
Make EP4 (Table 6). Store at 4 °C. This will be used for the rest of the experiment as well. Remove old medium from both inserts and the bottom well.
Table 6 EP4 media -
2.
Transfer inserts to six-wells dish (Corning 3516) with tweezers.
-
3.
Add 1.5 ml of EP4 into both inserts and the bottom well.
Day 14 (or 9 days after seeding epithelial cells)—Infection.
Three-dimensional mucosal organotypic culture is now ready to use in flow cell assembly and infection process.
3.1.4 Fabrication and Assembly of Tissue Harboring Flow Cells (Day 13)
In order to grow mucosal biofilms under an environment that resembles the upper GI tract, a flow cell system was designed to harbor the mucosal tissue analogue, pre-grown in a porous membrane, with saliva flowing over the apical epithelial surface and cell culture medium flowing basally under the tissue [8]. The flow cell apparatus was custom-built by Sirois Tool Company (Berlin, CT) and consists of two discrete pieces that can be assembled to form two flow chambers separated by a membrane. The body of the upper and lower flow cell components was milled from a block of polytetrafluoroethylene (MSC Industrial Direct, Inc.). The connectors used between the components and the tubing were 0.0625-in. hose barb connectors with a #10–32 threaded port made of Kynar polyvinylidene fluoride (Small Parts, Inc.). The flow cell apparatus is assembled as illustrated in Fig. 1 by connecting two medium reservoirs to the upper and lower components of the flow cell via silicone manifold tubing (Watson-Marlow, Inc., Wilmington, MA). For convenient visualization, a window was built into the upper flow cell chamber and is sealed by attaching a 22-by-22-mm glass coverslip to the outside flow cell surface. The flow cell is assembled with an O-ring placed between the membrane and the upper component in order to seal the system. The upper and lower components are held in place by screws. One medium reservoir is filled with saliva-supplemented medium, which flows through the upper chamber. The second medium reservoir is filled with a 3:1 (vol/vol) mixture of Dulbecco’s modified Eagle’s medium and Ham’s F-12 medium, and EPES buffer to a final concentration of 15 mM. This medium flows through the lower chamber. A peristaltic pump model 205S/CA (Watson-Marlow Inc., Wilmington, MA) is connected downstream of the flow cells to establish flow (Fig. 1).
Mucosal flow cell system. Panels (a) and (b) depict the flow cell apparatus and setup. Panel (c) is a schematic representation of the flow cell system with organisms inoculated on the mucosa to promote biofilm formation. In panel (d) the flow cell was inoculated on the glass coverslip to promote biofilm formation on an abiotic surface in close proximity to the mucosa
Day 13
-
1.
Assemble flow cells.
-
2.
Glue glass slides to flow cells with silicone.
-
3.
Autoclave medium reservoirs with appropriate tubing, grease, twisters, distilled water, and beakers.
-
4.
Inoculate bacteria or yeast for overnight growth in BIH or YPD broth, respectively.
-
5.
Place mucosal tissues to be used in infection medium and incubate overnight at 37 °C with 5 % CO2 (Fig. 1).
3.1.5 Mucosal Biofilm Growth in Tissue Harboring Flow Cells
Day 14
-
1.
Clean flow cell with 0.1 M HCl and 10 % hypochlorite 1 and 2 h, respectively.
-
2.
Cut membranes out of PET inserts with sterile twisters and blade. Place on a petri dish. Assemble flow cell with PET membrane in the middle. Make sure this is done under sterile conditions.
-
3.
Check flow rate at this point, 4.5 rpm gives around 95 μl/min, which is around 6 ml per track.
-
4.
Calculate volume of saliva/10 % BHI and DMEM: F12 (3:1) needed (see Note 6 ).
-
5.
Condition flow cell surface with saliva/10 % BHI in upper track and DMEM: F12 (3:1) in the lower chamber for 15 min (see Note 7 ).
-
6.
Cut out tissue-harboring inserts under the hood and connect to flow cell chambers.
-
7.
Prepare microorganism inoculum with 105 yeast and 106 bacteria in 500 μl total volume. Inoculate microorganisms on the mucosal surface or glass slide using a sterile syringe. Incubate under static conditions for 30 min. After 30 min adhesion period, start flow with 4.5 rpm.
-
8.
Incubate flow cell overnight at 37 °C with 5 % CO2 (see Note 6 ).
-
9.
After overnight culture, stop flow and remove tissue samples from flow cell chambers. For histology, place samples in 4 % paraformaldehyde for fixation at least 2 h at 4 °C (see Note 8 ). After fixation, process for paraffin embedding.
3.1.6 In Vivo Murine Oral Co-infection Model
After arrival of mice at the animal facility, allow them to adapt to the new housing environment for 10 days (see Note 9 ).
Day 1—Animal setup and immunosuppression.
Organize animal cages by infection group, and set up individual mouse identification system by ear punch. Immunosuppress by subcutaneous injection with cortisone acetate (225 mg/kg) dissolved in 200 μl PBS containing 0.5 % Tween-80.
Days 1 to 4—Overnight culture of microorganisms.
Prepare fresh overnight cultures of C. albicans SC5314 in YPD broth (aerobically, at room temperature, on a rotor shaker) and S. oralis 34 or S. gordonii Challis CH1 culture in BHI medium (aerobic, static conditions, at 37 °C) for each day of infection (days 2–4).
Day 2—Oral infection under anesthesia.
Anesthetize mice with intraperitoneal (IP) injection of ketamine: xylazine (90–100 and 10 mg/kg of body weight, respectively). Prepare small cotton pads soaked with 100 μl of a C. albicans cell suspension (6 × 108 yeast/ml), or 100 μl of streptococcal cell suspension (2.5 × 109 bacteria/ml), or 50 μl of C. albicans cell suspension (1.2 × 109 yeast/ml) combined with 50 μl of streptococcal cell suspension (5 × 109 bacteria/ml). Place two cotton pads into oral cavity of anesthetized mice and swab the entire oral cavity (see Note 10 ). The cotton pads are left for 2 h under the tongue and are removed before the animals awake. Add fresh microorganism cultures daily in drinking water (C. albicans 6 × 108 yeast organisms/ml, or streptococci 2.5 × 109 bacteria/ml) or the combination of the two, to maintain high oral carriage loads throughout the experimental period [16].
Day 3—Repeat cortisone injection and oral infection through drinking water.
Day 4—Continue oral infection through drinking water.
Day 5—Sacrifice animals.
Sacrifice animals after CO2 inhalation by cervical translocation and excise tongues. Immediately photograph dorsal surface digitally (1:1 lens) for Image J analysis of the surface area covered with lesions, as we previously described [16] (Fig. 2a). For histology cut ½ tongue and place in 4 % paraformaldehyde for fixation at least 2 h at 4 °C (see Notes 8 and 11 ). Process for paraffin embedding.
Pathogenic synergy between C. albicans and S. oralis in a mouse oral co-infection model. (a) Tongues of mice were excised after 5 days of infection and the dorsal aspect was digitally photographed. Representative pictures are shown from one mouse in each group. Note the extensive biofilm lesion forming on the tongues of co-infected animals. (b) Overlay images of tongue tissue sections stained with a FITC‐labeled anti‐ Candida antibody (green), followed by FISH with an Alexa 546‐labeled S. oralis‐specific probe (red), and counterstained with the nucleic acid stain Hoechst 33258 (blue). The FISH signal was completely absent in biofilms formed by C. albicans only, and the FITC signal was completely absent in animals infected with S. oralis only, showing staining specificity. Bars = 50 μm
3.2 Quantification of Fungi and Bacteria in Vitro and In Vivo
3.2.1 Molecular Method of Streptococcal Quantification in Murine Fecal Samples
-
1.
Bacterial genomic DNA from mouse fecal samples collected daily is isolated and purified by QIAGEN DNA Stool mini kit according to the manual. DNA quantity and quality is assessed using the NanoDrop device (ND-1000 spectrophotometer, NanoDrop Technologies).
-
2.
To calculate the S. oralis 34 cell numbers in stools, S. oralis 34 cultures are used to generate pure genomic DNA. To create a standard curve, pure gDNA is prepared for a tenfold serial dilution of known amounts of gDNA: 0.32 ng, 0.032 ng, 0.0032 ng, and 0.00032 ng. According to genomic size and mass of S. oralis, the corresponding genomic copy numbers of the single copy glucosyltransferase R (GtfR) gene, or cell numbers (for this single copy gene) are: 1.45 × 105, 1.45 × 104, 1.45 × 103, and 1.45 × 102, respectively.
-
3.
Primers for the GtfR gene (gtf-F: TCCCGGTCAGCAACTCCAGCC, gtf-R: GCAACCTTTGGATTTGCAAC) are used to target S. oralis 34 DNA in the samples and the cell numbers of S. oralis 34 are calculated according to the standard curve [15] (see Note 12 ).
-
4.
Real-time PCR is performed with BIO-RAD CFX96 cycler real-time PCR detection system. All PCR reaction mixtures contain the following: 10 μl 2 × iQTM SYBR® Green Supermix (for cDNA) or Soso Advanced SYBR green Mix (DNA) (Bio-Rad, Hercules, CA), 1 μl of first-strand cDNA or DNA, 0.1 μM of primers and H2O to bring the final volume to 20 μl (see Note 13 ).
-
5.
The program for cDNA amplification is 95 °C incubation for 5 min, followed by 40 cycles of 95 °C for 10 s and 60 °C for 30 s.
-
6.
The program for gDNA amplification is 98 °C incubation for 5 min, followed by 40 cycles of 98 °C for 10 s and 67 °C for 50 s.
-
7.
Data are analyzed by the CFX96 cycler system software (Bio-Rad) (Fig. 3) [11].
Fig. 3 Following infection stool samples were collected daily for five days, pooled from each group and analyzed by qPCR in triplicate. qPCR quantification of S. oralis in fecal samples. S. oralis 34 genomic DNA in stool samples of mice infected S. oralis alone (So), or co-infected with C. albicans and S. oralis (CaSo) were analyzed by qPCR using primers specific for the S. oralis-species-specific gtfR gene and the cell numbers were calculated by a standard curve. * P < 0.05. Linear regression software in Excel® is used to calculate cell numbers from standard curve
3.2.2 Confocal Microscopy/Immunohistochemistry: Immuno-FISH Staining for Mucosal Analogues and Tongue Tissue
-
1.
After sectioning, deparaffinize sections by immersion in xylene and dehydration in a series of ethanol washes.
-
2.
Stain with anti- C. albicans -FITC antibody (300 μl/slide, dilute 1:20 in PBS, Meridian B65411F). Incubate at room temperature for 2 h (see Note 11 ).
-
3.
Wash three times with PBS (200–300 μl/slide).
-
4.
Permeabilize bacterial cells with lysozyme (200–300 μl/slide, 70,000 U/ml in 20 mM Tris–HCl, 5 mM EDTA pH 7.5. Lysozyme Sigma-Aldrich L6876) for 9 min at 37 °C in a humid atmosphere (see Note 12 ).
-
5.
Rinse with PBS and dehydrate with a series of ethanol washes (50, 80, and 100 %) 2–3 min each wash.
-
6.
Expose cells to 200 μl hybridization buffer, 10 ng/ml Streptococcus -specific oligonucleotide probe STR405, labeled with Alexa 546 in 0.9 M NaCl, 20 mM Tris–HCl (pH 7.5), 0.01 % SDS, 25 % formamide. Incubate slides at 46 °C for 90 min in a humid atmosphere (see Note 13 ).
-
7.
Wash slides with pre-warmed washing buffer (20 mM Tris–HCl (pH 7.5), 5 mM EDTA, 0.01 % SDS, and 159 mM NaCl) for 15 min at 46 °C.
-
8.
Rinse with ice-cold water.
-
9.
Counterstain with Hoechst 33258 (2 μg/ml in PBS, Invitrogen H3569) for 30 min at room temperature.
-
10.
Rinse with water, air dry, and mount with coverslip. Observe under epifluorescence microscope (Fig. 2b, 4a, and 5a).
Fig. 4 Fig. 5 C. albicans and S. oralis biofilms forming on an oral mucosa analogue. (a) Confocal microscopy images used for 3D reconstructions and biovolume estimates of monospecies and mixed-species mucosal biofilms of C. albicans and S. oralis after 16 h of biofilm growth. C. albicans was stained with a FITC-labeled anti- Candida antibody (green), and S. oralis was visualized with an Alexa 546-labeled Streptococcus -specific probe (red). (b) Biovolume quantifications from CLSM image stacks of C. albicans and S. oralis in monospecies or mixed-species mucosal biofilms. * P < 0.05
3.2.3 Biovolume Quantification Using Confocal Microscopy
-
1.
After immuno-FISH staining as described above, confocal images of biofilms are analyzed for biovolume quantification using the IMARIS 7.0 software package (Bitplane AG).
-
2.
3D reconstructions of biofilm must be generated using the “Surpass” volume rendering option and a new “Surface” must be created for each channel used on confocal microscopy (Fig. 4) (see Note 14 ).
-
3.
To start the “Surface” creation, click on the Surface button (blue icon at the top left) and it will automatically appear at your object list.
-
4.
Four steps are necessary to create a “Surface” for biovolume measurement: selection of a “Region of Interest,” “Source Channel,” “Threshold” adjustment, and “Surface Classification,” when working with more than one channel/organism (see Notes 15 and 16 ). Once “Surface” creation is finished, it now has the same dimensions of the original confocal image, displayed as a “Solid Surface” (Fig. 4a, b).
-
5.
At the Object properties area, several tabs are now available. Go to “Statistics” tab, select “Detailed”—“Specific Values”—“Volume” to measure the volumes of the created “Surfaces” and save as an excel file for further analysis (Fig. 5a, b).
4 Notes
-
1.
Use freshly thawed vial of OKF6/TERT2 cells from Liquid N2. Subculture when cells are 70–80 % confluent. Subculture 1 week after starting culture.
-
2.
When making acellular and cellular layers, mix components gently on ice to avoid bubbles.
-
3.
The matrix will contract over next few days.
-
4.
5 × 105 epithelial cells are seeded per insert/well. One ~80 % confluent 75-cm2 flask yields about 2–3 × 106 OKF6/TERT2 cells.
-
5.
After air lifting, further significant contraction will be seen. Contraction can be better assessed during medium change.
-
6.
Prepare extra media, to avoid drying the tissues during overnight culture after inoculation.
-
7.
Check for any media leakage. If leakage is found, tighten all screws. It may be necessary to disassemble flow cell and add grease between upper and lower chambers.
-
8.
Do not over-fix. It may mask certain antigens. 2–3 h is sufficient for fixation with 4 % paraformaldehyde.
-
9.
If animals are bred in animal facility, the 10-day adjustment period is not necessary.
-
10.
Some animals may awake during this period; it may be necessary to inject additional anesthetic solution to maintain cotton pads in the mouth.
-
11.
Some animals may have visible thrush on their tongues at day 4. Tongues may be checked daily with ring forceps (Fine Science Tools 11101-09).
-
12.
gtfR primers are specific for S. oralis species and target all S. oralis strains. If more specificity is needed, new primers need to be designed. For S. oralis 34, we designed strain-specific wefA-wefH primers to confirm the data with gtfR [11].
-
13.
Both iQTM SYBR® Green Supermix and Soso Advanced SYBR green Mix can be used to amplify gDNA, but Soso Advanced SYBR green Mix works better.
-
14.
Histology slides should be kept in the dark in plastic boxes with some water inside to avoid evaporation.
-
15.
A new “Surface” must be created for Candida and S. oralis stains, separately, since they will be analyzed separately by selection of two different channels.
-
16.
During “Surface” creation all parameters can be set interactively at the “Object Properties Area.” “Threshold” adjustments will cover the entire confocal image.
References
Villar CC, Dongari-Bagtzoglou A (2008) Immunodefence mechanisms and immunoenhancement strategies in oropharyngeal candidiasis. Expert Rev Mol Med 10, e29
Dongari-Bagtzoglou A, Kashleva H, Dwivedi P et al (2009) Characterization of mucosal Candida albicans biofilms. PLoS One 4, e7967
Nett JE, Marchillo K, Spiegel CA et al (2010) Development and validation of an in vivo Candida albicans biofilm denture model. Infect Immun 78:3650–3659
Nobile CJ, Fox EP, Nett JE et al (2012) A recently evolved transcriptional network controls biofilm development in Candida albicans. Cell 148:126–138
Morales DK, Hogan DA (2010) Candida albicans interactions with bacteria in the context of human health and disease. PLoS Pathog 6, e1000886
Bamford CV, d’Mello A, Nobbs AH et al (2009) Streptococcus gordonii modulates Candida albicans biofilm formation through intergeneric communication. Infect Immun 77:3696–3704
Silverman RJ, Nobbs AH, Vickerman MM et al (2010) Interaction of Candida albicans cell wall Als3 protein with Streptococcus gordonii SspB adhesin promotes development of mixed-species communities. Infect Immun 78:4644–4652
Diaz PI, Xie Z, Sobue T et al (2012) Synergistic interaction between Candida albicans and commensal oral streptococci in a novel in vitro mucosal model. Infect Immun 80:620–632
Ramsey MM, Rumbaugh KP, Whiteley M (2011) Metabolite cross-feeding enhances virulence in a model polymicrobial infection. PLoS Pathog 7, e1002012
Whitmore SE, Lamont RJ (2011) The pathogenic persona of community-associated oral streptococci. Mol Microbiol 81:305–314
Xu H, Sobue T, Thompson A et al (2013) Streptococcal co-infection augments Candida pathogenicity by amplifying the mucosal inflammatory response. Cell Microbiol 16:214–231
Gillum AM, Tsay EY, Kirsch DR (1984) Isolation of the Candida albicans gene for orotidine-5′-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol Gen Genet 198:179–182
Dongari-Bagtzoglou A, Kashleva H (2006) Development of a highly reproducible three-dimensional organotypic model of the oral mucosa. Nat Protoc 1:2012–2018
Kalabis J, Wong GS, Vega ME et al (2012) Isolation and characterization of mouse and human esophageal epithelial cells in 3D organotypic culture. Nat Protoc 7:235–246
Xie Z, Thompson A, Kashleva H et al (2011) A quantitative real-time RT-PCR assay for mature C. albicans biofilms. BMC Microbiol 11:93
Dwivedi P, Thompson A, Xie Z et al (2011) Role of Bcr1-activated genes Hwp1 and Hyr1 in Candida albicans oral mucosal biofilms and neutrophil evasion. PLoS One 6, e16218
Acknowledgements
This work was supported by NIH/NIDCR grant RO1 DE013986.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2016 Springer Science+Business Media New York
About this protocol
Cite this protocol
Sobue, T., Diaz, P., Xu, H., Bertolini, M., Dongari-Bagtzoglou, A. (2016). Experimental Models of C. albicans-Streptococcal Co-infection. In: Calderone, R., Cihlar, R. (eds) Candida Species. Methods in Molecular Biology, vol 1356. Humana Press, New York, NY. https://doi.org/10.1007/978-1-4939-3052-4_10
Download citation
DOI: https://doi.org/10.1007/978-1-4939-3052-4_10
Publisher Name: Humana Press, New York, NY
Print ISBN: 978-1-4939-3051-7
Online ISBN: 978-1-4939-3052-4
eBook Packages: Springer Protocols