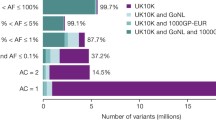
Abstract
From 2010 to 2013, UK10K was Britain’s largest genomic sequencing consortium, awarded £10.5 million by the Wellcome Trust to investigate how low-frequency and rare genetic variants contribute to human disease (www.uk10k.org). This collaborative project brought together researchers working on obesity, autism, schizophrenia, and a number of rare conditions (familial hypercholesterolemia, thyroid disorders, learning disabilities, ciliopathies, congenital heart disease, coloboma, neuromuscular disorders, and severe insulin resistance) to generate whole genome and exome sequence data for almost 10,000 highly phenotyped individuals. The data generated by UK10K not only enabled the discovery of novel disease-causing genes by the consortium, but was also made available to the research community during the life of the project as a managed access data resource; providing access to data an order of magnitude deeper than was previously possible, and empowering future research into human genetics.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- UK10K Project
- Severe Insulin Resistance (SIR)
- Ciliopathies
- Coloboma
- Avon Longitudinal Study Of Parents And Children (ALSPAC)
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
What Is UK10K?
From 2010 to 2013, UK10K was Britain’s largest genomic sequencing consortium, awarded £10.5 million by the Wellcome Trust to investigate how low-frequency and rare genetic variants contribute to human disease (www.uk10k.org). This collaborative project brought together researchers working on obesity, autism, schizophrenia, and a number of rare conditions (familial hypercholesterolemia, thyroid disorders, learning disabilities, ciliopathies, congenital heart disease, coloboma, neuromuscular disorders, and severe insulin resistance) to generate whole genome and exome sequence data for almost 10,000 highly phenotyped individuals. The data generated by UK10K not only enabled the discovery of novel disease-causing genes by the consortium, but was also made available to the research community during the life of the project as a managed access data resource; providing access to data an order of magnitude deeper than was previously possible, and empowering future research into human genetics.
Motivation Behind the Project
Although many hundreds of genes involved in disease processes have already been discovered, the picture is far from complete. For most traits, only a small fraction of the genetic contribution has been explained, suggesting that many more disease loci remain unknown. Whilst highly valuable, linkage analyses and genome-wide association studies (GWAS) are restricted to the identification of those genes whose variants either have strong and distinctive effects, or those that have weaker effects but are more common (minor allele frequency [MAF] ≥ 5 %). Candidate gene re-sequencing studies have demonstrated that some mutations can, however, have an effect on disease phenotypes whilst existing at a rare or low allele frequency (MAF < 5 %). Taking advantage of new technology-sequencing platforms and falling sequencing costs, the UK10K project set out to detect variants with allele frequencies as low as 0.1 %.
It was anticipated that the project’s outcomes would have a far-reaching impact across the scientific, research, and medical community. The unprecedented scale and quality of the data generated has already been recognized as an excellent resource for further research into human genetics, whilst the data processing pipeline and the statistical analyses developed during the project provided examples of current best recommended practice. It is hoped that the discovery of novel, rare disease-causing variants identified by UK10K will lead to further insight into disease processes, and improvements in disease diagnoses and the development of new therapies.
UK10K Project Design
The project consisted of five key stages:
Genome-Wide Sequencing of 4,000 Cohort Samples
To maximize the amount of variation detected, whole genome sequencing at 6× depth was performed on the DNA of 4,000 highly phenotyped individuals of UK origin. It was anticipated that this coverage would provide enough power to detect all accessible SNVs, indels, and structural variants down to a 0.1 % allele frequency, and improve the accuracy of genotype calls on sequenced individuals. This ‘Cohort’ group as it was referred to within the project was composed equally of subjects recruited from two well-established studies: the Avon Longitudinal Study of Parents and Children (ALSPAC, www.bristol.ac.uk/alspac) and the TwinsUK study (www.twinsuk.ac.uk).
The Cohorts
TwinsUK is Britain’s largest adult twin registry. Composed of more than 12,000 identical and non-identical twins, TwinsUK is an invaluable resource for studying the genetic and environmental aetiology of age-related complex traits and diseases. The 2,000 samples selected for sequencing (one per twin pair) were taken from unrelated females from all over the UK, approximately three-fifths of which were dizygotic and two-fifths monozygotic. Where possible twins who were already part of the MuTHER (multiple tissue human expression resource), and/or HATS study (healthy ageing twin study) were preferentially selected for inclusion in Cohorts.
A core set of 63 UK10K phenotypes were selected to ensure as much overlap as possible between TwinsUK and ALSPAC phenotypic data (derived from physical examinations and questionnaires for both groups), and was made available alongside sequence data in the European genome-phenome archive (EGA). The Cohorts phenotypes included measurements for liver, kidney and lung function, cardiovascular function and hypertension, and anthropometric data such as waist and hip size, leg length, and head circumference.
ALSPAC is a longitudinal, population-based birth cohort study that recruited over 13,000 pregnant women in the Avon area, collecting data from the eighth gestational week onwards. DNA was collected from approximately 9,000 children who continued to supply phenotypic data up until the age of 18 years after which many participants re-consented their participation in the study, providing data into adulthood. In contrast to the TwinsUK samples, the 2,000 samples supplied by ALSPAC were from teenage individuals, based in and around a single region of the UK (Avon).
By collaborating with established longitudinal studies such as TwinsUK and ALSPAC, UK10K was able to investigate the contribution of genetic variants to phenotypic variation over time. Modelling correlated and longitudinal phenotypic measurements in association tests reduced phenotypic variance and increased the power of analyses. Further gains in power were achieved by imputing low-frequency variants into non-sequenced individuals with existing genome-wide association scan (GWAS) data. By preferentially including samples that overlapped with existing studies for which DNA methylation, gene expression, and metabolic profiling data were available, it was possible to explore genetic associations in functionally relevant variation, and to develop new analytical methods for incorporating functional annotation into association testing. It was anticipated that by including individuals from a continuum of trait, age, and geographical distribution, the resulting data would be widely applicable and used internationally.
Whole Genome Sequencing
A ‘production pipeline’ was developed specifically for the project, managing the flow of samples from the point of arrival through to DNA quality control (which included Picogreen quantification and Sequenom Genotyping), multiplexed sequencing and data generation, and lane QC (confirming sample identity by genotype matching, checking base quality, even-GC representation, and library insert size), prior to read pair mapping (BAM format) and variant calling (VCF format).
For whole genome sequencing, 1–3 μg DNA was sheared to 100–1,000 bp then subjected to Illumina paired-end DNA library preparation. Following size selection (300–500 bp insert size, sufficient to span Alu repeats), DNA libraries were multiplexed in a single pull-down experiment (with indexing barcodes attached prior to pull-down, enabling the sample of origin to be determined for each read) and sequenced using the Illumina HiSeq platform as paired-end 100 base reads (according to the manufacturer’s protocol).
Realignment was made around known indels from the 1000 Genomes Project Pilot (1000 Genomes Project Consortium 2010) to improve raw BAM alignment for SNP calling, and then base quality scores were recalibrated using GATK (DePristo et al. 2011). BAQ tags were added using SAMtools, the BAMs were merged and then any duplicates removed. SNP and indel variants were called on the data using both SAMtools (Li 2011) mpileup and GATK UnifiedGenotyper, then merged and annotated with allele frequencies from 1000 Genomes, dbSNP entry date, and rsIDs. Functional annotation was added using the Ensembl Variant Effect Predictor against Ensembl 64, and finally BAM and VCF files were deposited in the EGA. Cumulative single-sample and multiple-sample releases were made throughout the duration of the project, enabling the scientific community to benefit from access to the data long before the end of UK10K.
Direct Association of Traits in the Sequenced Individuals to the Variants Found in Section ‘Genome-Wide Sequencing of 4,000 Cohort Samples’
The next stage of the project involved directly associating newly discovered variants with quantitative traits, and identifying those variations linked with disease.
Primary association analyses focused specifically on testing the associations of intermediate and rare sequence variants with selected quantitative traits (including cardiometabolic traits, blood pressure, and body mass index), using single variant association tests and also collapsing together multiple low-frequency/rare variants in order to detect association to a gene or region. With almost 4,000 whole genome samples sequenced in total, imputed into a further 10,000 samples (approximately) using 1000 Genomes panels and IMPUTE2, power was sufficient to detect single variant associations contributing 0.1 % variance.
Secondary analyses were directed at determining the impact of novel loci on longitudinal analyses and age effects (using linear and logistic regression methods), maternal and parent of origin effects (for ALSPAC; exploiting maternal genome-wide SNP data and phenotypic records, and matching of the surrounding haplotype), and the analysis of correlated traits and pleiotropy (analysing correlated intermediate traits to assess potential pleiotropic effects at novel QTLs).
Sequencing and Association Analysis of 6,000 Exomes from Samples with Extreme Phenotypes
The UK10K project was constrained in terms of having a fixed duration and budget, and as a result limited itself to investigating three key areas of disease for which there were already some recognized rare causal variants: obesity, neurodevelopmental disorders (autism and schizophrenia), and a selection of rare conditions (including familial hypercholesterolemia, thyroid disorders, learning disabilities, ciliopathies, congenital heart disease, coloboma, neuromuscular disorders, and severe insulin resistance). To identify novel and rare variants associated with these diseases, close to 6,000 DNA samples from subjects with extreme disease phenotypes were whole exome sequenced to an average depth of 72×. High depth sequencing was necessary to enable the precise calling of rare variants, and the increased costs associated with higher depth sequencing were met by compromising on exome, rather than whole genome sequencing. Sequencing exomes (protein-coding exons and flanking conserved sequence) greatly reduced the overall sequencing costs for this stage of the project, though it was acknowledged that exome sequencing would not capture variants outside of these regions.
The objectives of this stage were to identify novel variants and genes involved in these conditions, first by association and then by determining as far as possible causal variants and mode of action. Selecting for extreme traits of interest substantially increased the power of analyses (using the Cohorts data as a common control set), and exome sequencing enabled more than 90 % of the target region to be sequenced to a sufficient enough depth to accurately call heterozygous sites (and singletons), from 4Gb total sequence per sample. The resulting publications, describing the contribution of novel variants to the genetic variation underlying these disease phenotypes, can be found on the UK10K website (All UK10K publications are available at: www.uk10k.org/publications_and_posters.html).
Whole Exome Sequencing
For whole exome sequencing 1–3 μg DNA was sheared to 100–400 bp, then subjected to Illumina paired-end DNA library preparation and enriched for target sequences according to the manufacturer’s recommendations (Agilent Technologies; SureSelectXT Automated Target Enrichment for Illumina paired-end Multiplexed Sequencing). Enriched libraries were multiplexed (see section ‘Whole Genome Sequencing’) and sequenced using the Illumina HiSeq platform as paired-end 75 base reads (according to the manufacturer’s protocol). Algorithms were developed to call base substitutions, indels, and CNVs from the exome data, using a read-depth approach.
Statistical Methods
The statistical methods applied in this stage of the project were similar to those employed in earlier stages, and broadly included:
-
Imputation (both internally and into other GWAS cohorts).
-
Family-based method development (for TwinsUK, Neurodevelopmental and Rare data).
-
Meta-analysis of rare variants. Multivariate methods (such as Fisher’s combined probability test, Stouffer’s z-score method, SKATmeta (http://cran.r-project.org/web/packages/skatMeta/vignettes/skatMeta.pdf) and metaSKAT (Lee et al. 2013)) were used as an alternative to single-point analyses, which would have be underpowered given the size of sample sets used.
-
Identification and evaluation of appropriate controls to alleviate bias in case–control analyses. Controls were selected from the Cohorts data, as well as from other exomes of non-related phenotypes both from within UK10K and from other sources such as dbGAP, the NHLBI exome-sequencing project, and the 1000 Genomes Project.
-
Development of robust pipelines for quantitative trait and case–control analyses.
-
Correcting for population stratification.
-
Defining a genome-wide significance threshold for testing.
Great care was taken when matching cases to controls to consider the effects of population structure.
The Exome Collections
The full lists of studies sequenced as part of UK10K are described on the project website (http://www.uk10k.org/studies/) as well as in the EGA; however, a summary is presented below (and in more detail in section ‘UK10K Sample Sets’):
Neurodevelopmental Disorders Group
It is estimated that neurodevelopmental traits such as autism spectrum disorders (ASDs) and schizophrenia affect up to 2 % of the world’s population. Autism and schizophrenia are complex conditions involving multiple susceptibility genes and environmental factors, and often overlap in terms of characteristic clinical features. It has been proposed that these traits are part of a continuum of genetic and molecular events in the nervous system, and that rare variants in multiple genes may account for much of the unexplained susceptibility observed for these particular neurodevelopmental traits. Thus, further characterization of underlying genes and pathways as part of UK10K could significantly improve diagnostic classification for these conditions.
As both autism and early onset schizophrenia are uncommon and evidence suggests that rare, relatively penetrant alleles might be involved—it was decided at the outset that including families and individuals from isolated populations would be beneficial to enrich for genetic effects. The project selected 3,000 well-characterized cases of autism or schizoaffective disorders from founder populations and from collections of families demonstrating a clustering of cases to enrich for genetic effects and allow validation by segregation. Subjects were predominantly of UK origin (four Finnish studies were also used), and all represented genetically enriched cases—coming from families with multiple affected members (ASD and schizophrenia), representing early onset cases (schizophrenia), or being part of special interest populations (such as the Kuusamo schizophrenia study). Some cases presenting with intellectual disability as well as schizophrenia were also included, as variants associated with this more severe phenotype might have been more penetrant. Analyses to identify variants for these conditions fell into three categories:
-
Family sample analyses—for families with a high loading of autism or schizophrenia, where one or a few highly penetrant variants were likely to contribute to the observed phenotype.
-
Singleton analyses—where inheritance patterns could be dominant, recessive, or oligogenic.
-
De novo analysis—where trios of two unaffected parents and one affected child were sequenced to identify de novo variants present in the child, but neither parents.
-
Population analyses—performing single point tests and tests for aggregation of variants in genes and pathways, separately for autism and schizophrenia data.
Variants identified in these ‘extreme’ phenotype populations were assessed for relationships with ‘normal’ cognitive and behavioural traits as observed in controls.
Rare Diseases Group
Although linkage and homozygosity mapping have identified many of the causal variants underlying many mendelian diseases, the basis for many rare genetic diseases (where significant locus heterogeneity can attenuate the power of linkage studies) is much less clear. To further our understanding of such conditions, exomes were sequenced from 125 cases of each of the following conditions spanning a broad range of extreme phenotypes, some of which have the potential to respond well to therapeutic intervention:
-
Severe insulin resistance
-
Thyroid disorder
-
Learning disabilities
-
Ciliopathies
-
Familial hypercholesterolaemia
-
Neuromuscular disease
-
Coloboma
-
Congenital heart disease
Limiting the study to eight rare diseases maximized power in the presence of probable locus heterogeneity. In total 1,000 ‘rare disease’ samples were submitted by collaborating PIs from existing collections, and sequenced. This number of samples was sufficiently powered to detect genes with causal mutations in 10 % of patients with any false ‘discoveries’ removed during segregation analyses and follow-up re-sequencing of candidate genes. Power was further increased as the number of causal variants per exome reduced, due to improved specificity of variant detection algorithms and better variant sampling in control datasets. Wherever possible samples from families with multiple affected members were used to enrich for genetic aetiology and enable segregation analyses. As for the neurodevelopmental disorders, there were three tiers of analyses used to discover candidate genes in this group:
-
Within-family analyses—identifying candidate variants shared by affected individuals within the same family, examining trios to identifying candidate de novo variants, and examining single affecteds.
-
Across-family analyses—identifying candidate genes shared by affected individuals in different families.
-
Association analyses—looking at single gene and gene-set enrichment of functional variants.
Whole genome amplification and re-sequencing of candidate genes in additional patients (provided by each of the collaborating disease groups), and functional analyses in model systems were also used to determine causality for candidate genes.
Obesity Group
Obesity (defined in Caucasians as having a body mass index (BMI) > 30 kg/m2) is a widely recognized and growing public health problem associated with type 2 diabetes, cardiovascular disease, and some cancers. Once considered a problem exclusive to high-income countries, obesity is becoming more prevalent in middle- and low-income countries.
Over the last decade, much progress has been made in the detection of monogenic causes of obesity; however, the variants found to be associated with high BMI in cohort studies are estimated to account for less than 1 % of the variance of BMI in European adults, and little is known about the causal genes underlying early onset obesity. By including both clinically extreme (obese children) and population extreme obesity (BMI > 40 kg/m2) as a phenotype in UK10K, it was hoped to gain a better understanding of rare variants associated with this condition. A total of 1,500 samples from obese individuals were submitted from three separate studies: the Severe Childhood Onset Obesity Project (or ‘SCOOP’ study), the Generation Scotland: Scottish Family Health Study (http://www.genetics.med.ed.ac.uk/generation-scotland), and obese individuals from the TwinsUK cohort. Generation Scotland is a multi-institution population-based resource, aiming to identify the genetic basis of common complex diseases. The SCOOP cohort is a subset of Caucasian patients with severe early onset obesity in whom all monogenic causes of obesity have been excluded, derived from the larger ‘Genetics of Obesity (GOOS) Study’ (http://www.goos.org.uk/) that consists of children with an age-adjusted BMI greater than 3 standard deviations above the mean, and obesity onset at less than 10 years old. Prior to inclusion in the study, SCOOP subjects were sequenced for MC4R, which contains the highest proportion of variants that cause obesity, and their leptin levels were measured.
The majority of samples were supplied by SCOOP (1,000 samples in total), with just over 400 samples from Generation Scotland, and 69 provided by the TwinsUK registry.
To uncover variants underlying both monogenic and polygenic forms of obesity, tiered filtering analyses were directed towards identifying 33 known human obesity genes and 88 functional candidates using single affected and cross-family analyses (monogenic disease-causing candidates) and case–control analyses employing regression and collapsing methods (complex obesity-associated variants). Exome-wide single variant and gene-region-based association tests were used to identify associations with obesity, and trio and family-based analyses were used (within the Generation Scotland dataset) to search for causal de novo mutations or segregating variants.
Imputation into Additional GWAS Samples
Association analyses were extended by imputing low-frequency variants into non-sequenced individuals with existing GWAS data. Genotype imputation makes predictions at un-genotyped markers in GWAS samples based upon the correlation between markers in reference panels with known sequence/dense genotypes, and less dense genotypes in GWAS data. Imputing into additional TwinsUK and ALSPAC samples, and other case–control and cohort studies with GWAS data in this way increased the power of UK10K analyses, and the potential for discovering novel variant candidates. Using a reference panel of 4,000 whole genome Cohort samples sequenced at 6×, it was possible to impute down to below 0.1 % allele frequency.
Providing a Sequence Variation Resource for Use in Further Studies
The final aim of UK10K was to provide a genotype/phenotype resource that would support future research into human genetics, making controls available for sequence-based studies and enabling imputation into other GWAS and exome-sequencing studies. To this end, whole genome and exome sequence data from the project (including basic phenotype and quantitative trait information, as well as allele frequency summary data for the Cohorts datasets) was deposited in the EGA. Although any researcher may apply to use UK10K datasets, applicant approval and the subsequent granting of access to UK10K data is strictly managed by an independent Data Access Committee.
Managed Data Access
A major challenge for the project was creating a structure that would enable a highly diverse collection of studies to function collectively under the common goals of UK10K, without violating any of the individual studies’ terms of use. To ensure absolute clarity regarding project participation, an ethical governance framework (http://www.uk10k.org/ethics.html) was devised that clearly defined UK10K policy on ethical and regulatory approvals, informed consent, data access, and withdrawal.
Implementing a mechanism for managed data access was crucial to assuring sample providers that the terms of data use would be respected for all UK10K studies (Muddyman et al. 2013). In order to access UK10K data in the EGA all prospective data users must first complete a data access application (downloaded from the UK10K website, http://www.uk10k.org/data_access.html), outlining a research proposal for specific, named datasets. The application must then be submitted to an independent Data Access Committee for review and approval, prior to data access being granted. Broadly speaking, the access agreement requires that users will respect the confidentiality and security of the data, agree that the data will only be used for research purposes and will not be redistributed, and that no attempts will be made to identify participants. It also clearly states the specific constraints imposed by Research Ethics Committees (RECs) for each of the individual studies (for example, some exome datasets may not be used for control purposes), and how specific datasets should be acknowledged. Whilst UK10K is committed to making its data as available and widely used as possible, it is equally committed to ensuring that applicants, once approved, respect the terms of data usage. Failure to abide by these terms would result in current and future access to the data being immediately withdrawn, and journal editors being alerted to the breach in use of UK10K data.
Publications
UK10K Publications fell into three categories: a flagship consortium paper describing primary analyses using the Cohorts data (under review), papers from the consortium’s exome and statistical analysis groups on specific phenotypes and analytical methods, and manuscripts produced by researchers outside of the consortium. A one-year publication moratorium was imposed on all non-consortium Data Users, to protect first publication rights of the data generators. The moratorium expired on 2 July 2013 for the Cohorts datasets and 2 January 2014 for the exome datasets. Keen to ensure that output from the project was made publicly available as soon as possible, all consortium manuscripts were sponsored to ensure immediate open access, and uploaded to the project website (http://www.uk10k.org/publications_and_posters.html).
UK10K Sample Sets
For more information on these sample sets and constraints of use, please refer to the UK10K Data Access Agreement. The following datasets may not be used for control purposes:
-
UK10K_NEURO_ASD_SKUSE
-
UK10K_NEURO_ASD_TAMPERE
-
UK10K_RARE_FIND
-
UK10K_NEURO_ASD_BIONED
-
UK10K_NEURO_ASD_MGAS
-
UK10K_RARE_CHD
-
UK10K_NEURO_ASD_TEDS
-
UK10K_NEURO_FSZNK
-
UK10K_RARE_CILIOPATHIES
-
UK10K_NEURO_FSZ
-
UK10K_NEURO_ASD_FI
-
UK10K_NEURO_UKSCZ
-
UK10K_NEURO_IMGSAC
-
UK10K_OBESITY_SCOOP
-
UK10K_RARE_COLOBOMA
The Cohorts Group
There are no constraints attached to the use of these datasets, which may be used for control purposes.
UK10K_COHORT_ALSPAC
(EGA study ID: EGAS00001000090)
The Avon Longitudinal Study of Parents and Children (ALSPAC) is a two-generation prospective study. Pregnant women living in one of three health districts in the former county of Avon with an expected delivery date between April 1991 and December 1992 were eligible to be enrolled in the study, and this formed the initial point of contact for the development of a large, family-based resource. Information was collected on children and mothers through retrieval of biological materials (e.g. antenatal blood samples, placentas), biological sampling (e.g. collection of cord blood, umbilical cord, milk teeth, hair, toenails, blood, and urine), self-administered questionnaires, data extraction from medical notes, linkage to routine information systems and at repeat research clinics.
UK10K_COHORT_TWINSUK
(EGA study ID: EGAS00001000108)
The TwinsUK resource is the UK’s largest adult twin registry of 12,000 identical and non-identical twins, used to study the genetic and environmental aetiology of age-related complex traits and diseases. The register is predominantly female, with a mean age of mid-50. Only female twins were used to provide samples for UK10K.
The Neurodevelopmental Disorders Group
There are no constraints attached to the use of the Muir, Edinburgh, Collier, Aberdeen, Gallagher, and Gurling datasets which may be used for control purposes in analyses.
UK10K_NEURO_MUIR
(EGA study ID: EGAS00001000122)
This sample set consists of subjects with schizophrenia, autism, or other psychoses all with mental retardation (learning disability). These subjects represent the intersection of severe forms of neurodevelopmental disorders, appear to have a higher rate of familiality of schizophrenia than typical, and are likely to have more serious and penetrant forms of mutations.
UK10K_NEURO_EDINBURGH
(EGA study ID: EGAS00001000117)
This sample set comprises subjects with schizophrenia, recruited from psychiatric in- and out-patient facilities in Scotland. All diagnoses are based on standard research procedures and family histories are available. Patients have IQ > 70 and the cohort includes the following groups: 100 cases with detailed clinical, cognitive, and structural and functional neuroimaging phenotypes; 138 familial cases who are the probands of families where DNA has been collected from other affected members; 162 unrelated individuals. In most cases, patients and their families may be re-contacted to take part in further studies.
UK10K_NEURO_ASD_SKUSE
(EGA study ID: EGAS00001000114)
This sample set of UK origin consists of clinically identified subjects with Autism Spectrum Disorders, mostly without intellectual disability (i.e. verbal IQs >70). The subjects represent children and adults with autism, asperger syndrome or atypical autism, identified according to standardized research criteria (ADI-algorithm, ADOS). A minority have identified comorbid neurodevelopmental disorders (e.g. ADHD). Family histories are available, with measures of broader phenotype in first-degree relatives.
UK10K_NEURO_ASD_TAMPERE
(EGA study ID: EGAS00001000115)
This sample set consists of Finnish subjects with autism spectrum disorders (ASD) with IQs >70 recruited from a clinical centre for the diagnosis and treatment of children with ASD.
UK10K_NEURO_ASD_BIONED
(EGA study ID: EGAS00001000111)
The BioNED (Biomarkers for childhood onset neuropsychiatric disorders) study has been carrying out detailed phenotypic assessments evaluating children with an autism spectrum disorder. These assessments included ADI-R, ADOS, neuropsychology, EEG, etc.
UK10K_NEURO_ASD_MGAS
(EGA study ID: EGAS00001000113)
The MGAS (Molecular Genetics of Autism Study) samples are derived from clinical samples seen by specialists at the Maudsley hospital, and have had detailed phenotypic assessments with ADI-R and ADOS.
UK10K_NEURO_FSZ and A.2.8 UK10K_NEURO_FSZNK
(EGA study ID: EGAS00001000118 [FSZ] and EGAS00001000119 [FSZNK])
These Finnish schizophrenia samples (FSZ: Kuusamo and FSZNK: non-Kuusamo) were collected from a population cohort using national registers. The entire resource collected by the Finnish National Institute for Health and Welfare (THL) consists of 2,756 individuals from 458 families—of whom 931 were diagnosed with schizophrenia spectrum disorder, each family having at least two affected siblings.
Samples were supplied from families originating from an internal isolate (Kuusamo) with a three-fold lifetime risk for the trait. The genealogy of the internal isolate is well documented and the individuals form a ‘megapedigree’ reaching back to the seventeenth century.
Samples were also supplied from families outside of Kuusamo, all of which had at least two affected siblings. All diagnoses are based on DSM-IV and for a large fraction of cases there is cognitive data.
UK10K_NEURO_ASD_FI
(EGA study ID: EGAS00001000110)
These samples are a subset of a nationwide collection of Finnish autism spectrum disorder (ASD) samples. The samples were collected from Central Hospitals across Finland in collaboration with the University of Helsinki and consisted of individuals with a diagnosis of autistic disorder or Asperger syndrome from families with at least two affected individuals. All diagnoses were based on ICD-10 and DSM-IV diagnostic criteria for ASDs.
UK10K_NEURO_IOP_COLLIER
(EGA study ID: EGAS00001000121)
This set was made up of samples taken from three different studies (all of UK origin).
The Genetics and Psychosis (GAP) samples, taken from subjects with schizophrenia ascertained as a new-onset case.
The Maudsley twin series consisting of probands ascertained from the Maudsley Twin Register, and defined as patients of multiple births who had suffered psychotic symptoms.
The Maudsley family study (MFS) consisting of over 250 families with a history of schizophrenia or bipolar disorder.
UK10K_NEURO_UKSCZ
(EGA study ID: EGAS00001000123)
These samples were collected from throughout the UK and Ireland, and fell into two main categories: cases with a positive family history of schizophrenia, either collected as sib-pairs or from multiplex kindred's—and samples that were systematically collected within South Wales, and in addition to a full diagnostic work up also underwent detailed cognitive testing. All samples obtained a DSM IV diagnosis of schizophrenia or schizoaffective disorder.
UK10K_NEURO_IMGSAC
(EGA study ID: EGAS00001000120)
Samples of UK origin were supplied from the IMGSAC cohort; an international collection of families containing children ascertained for autism spectrum disorders. Affected individuals were phenotyped using ADI-R and ADOS. Individuals with a past or current medical disorder of probable etiological significance or TSC were excluded. Where possible, the IMGSAC study performed karyotyping on one affected individual per family to exclude Fragile X syndrome.
UK10K_NEURO_ASD_GALLAGHER
(EGA study ID: EGAS00001000112)
Individuals in this Irish sample set were diagnosed with ADI/ADOS, measures of cognition/adaptive function, and approximately 50 % also presented with comorbid intellectual disability. This group represented a more severe, narrowly defined cohort of ASD subjects for the UK10K project.
UK10K_NEURO_GURLING
(EGA study ID: EGAS00001000225)
This sample set consisted of DNA from multiply affected schizophrenia families, diagnosed using the SADS-L and DSMIIIR criteria. All families were collected to ensure uni-lineal transmission of schizophrenia (i.e. families only had one affected parent with schizophrenia, or a relative of only one transmitting/obligate carrier parent with schizophrenia). Families with bi-lineal transmission of schizophrenia (i.e. with both parents being affected) were not sampled for this study. All families had multiple cases of schizophrenia and related disorders, and were selected to ensure an absence of cases of bipolar disorder both within the family and in any relatives on either side of the family.
UK10K_NEURO_ABERDEEN
(EGA study ID: EGAS00001000109)
This sample set comprises cases of schizophrenia with additional cognitive measurements, collected in Aberdeen, Scotland.
The Rare Diseases Group
There are no constraints attached to the use of the SIR, Neuromuscular, Thyroid, and Familial Hypercholesterolemia datasets, which may be used for control purposes in analyses.
UK10K_RARE_SIR
(EGA study ID: EGAS00001000130)
The Severe Insulin Resistance (SIR) sample set was supplied by the Cambridge Severe Insulin Resistance Study Cohort.
UK10K_RARE_NEUROMUSCULAR
(EGA study ID: EGAS00001000101)
These samples were taken from the Molecular Genetics of Neuromuscular Disorders Study, and fell into the following groups:
-
1.
Congenital muscular dystrophies and congenital myopathies.
-
2.
Neurogenic conditions.
-
3.
Mitochondrial disorders.
-
4.
Periodic paralysis.
UK10K_RARE_COLOBOMA
(EGA study ID: EGAS00001000127)
Ocular coloboma is the most common significant developmental eye defect with an incidence of approximately 1 in every 5,000 live births, resulting from the failure of optic fissure closure during embryogenesis. The samples used in UK10K mostly comprised isolated coloboma cases without systemic involvement (aka ‘non-syndromal coloboma’). There is strong evidence from family studies that coloboma has a major genetic component with autosomal dominance being the most common pattern of inheritance. However, many cases are isolated or show complex patterns of familial clustering. The genes responsible for isolated coloboma are largely unknown, but in a small number of families mutations in SHH, CHX10, and PAX6 have been identified indicating marked genetic heterogeneity. Thus, in addition to the clinical benefits of achieving a molecular diagnosis there are also major scientific advantages to identifying coloboma genes, as these are likely to provide insights into the complex process of optic fissure closure, that is critical to normal eye development. In the longer term, understanding the molecular basis of the disease may provide clues to therapeutic strategies.
UK10K_RARE_CHD
(EGA study ID: EGAS00001000125)
The Congenital Heart Disease (CHD) samples used for UK10K were supplied from the Genetic Origins of Congenital Heart Disease Study (GOCHD Study).
UK10K_RARE_CILIOPATHIES
(EGA study ID: EGAS00001000126)
The ciliopathies are an emerging group of disorders that arise from dysfunction of cilia (both motile or immotile forms). It is predicted that over 100 known conditions are likely to fall under this category, but only a handful have thus far been studied in any depth. Most individual ciliopathies are rare with just a small number of cases having been reported, thereby presenting researchers with often insurmountable difficulties for causative gene identification. Samples were supplied from the Cilia in Disease and Development study (CINDAD).
UK10K_RARE_FIND
(EGA study ID: EGAS00001000128)
Familial INtellectual Disability (FIND) is a cohort of families with intellectual impairment. Affected family members are at the extreme end of the spectrum with the majority having moderate to severe mental retardation where the recurrence risks suggests most are likely to have monogenic causes. A subset of the cohort underwent detailed analysis of the X chromosome by Sanger sequence analysis of exomes and more recently by detailed high resolution aCGH of the X chromosome. Samples from the first study where no causal variant could be identified were selected for inclusion in UK10K. The sample set comprised mostly non-syndromic cases, selected for bias towards families with male sib-pairs to enrich for non-X linked disease genes.
UK10K_RARE_THYROID
(EGA study ID: EGAS00001000131)
Samples were supplied from two different cohorts of subjects: ‘Individuals with Congenital Hypothyroidism (CH)’ due either to dysgenesis or dyshormonogenesis; and patients with ‘Resistance to Thyroid hormone (RTH)’, a disorder characterized by elevated thyroid hormones and variable tissue refractoriness to hormone action. The CH group was enriched for genetic aetiologies by recruiting cases that were familial, on a consanguineous background or syndromic. The RTH cohort consisted of cases in which candidate gene analyses were negative.
UK10K_RARE_HYPERCHOL
(EGA study ID: EGAS00001000129)
Familial Hypercholesterolemia is a condition where the affected person has a consistently high level of LDL, which can lead to early clogging of the coronary arteries. All patients selected for this study did not to carry the common APOB and PCSK9 mutations, and had no detectable LDLR mutations (tested for screening for 18 common mutations and SSCP, HRM, and MLPA screening for gross deletions/insertions.
The Obesity Group
There are no constraints attached to the use of the Generation Scotland and obese TwinsUK datasets, which may be used for control purposes in analyses.
UK10K_OBESITY_SCOOP
(EGA study ID: EGAS00001000124)
The Severe Childhood Onset Obesity Project (SCOOP) cohort is composed of Caucasian patients of UK origin with severe early onset obesity (all patients have a BMI Standard Deviation Score > 3 and obesity onset before the age of 10 years), in whom all known monogenic causes of obesity have been excluded.
UK10K_OBESITY_GS
(EGA study ID: EGAS00001000242)
The Generation Scotland: Scottish Family Health Study (GS:SFHS) is a family-based genetic study with more than 24,000 volunteers across Scotland, consisting of DNA, clinical, and socio-demographic data. This sample set consists of individuals from families with extreme obese subjects, including trios of extreme obese subjects with non-obese patients and multiple obese subjects within the same family.
UK10K_OBESITY_TWINSUK
(EGA study ID: EGAS00001000306)
This sample set consisted of extremely obese individuals from the TwinsUK study, with a BMI > 40.
References
DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C et al (2011) A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 43(5):491–498
Genomes Project Consortium (2010) A map of human genome variation from population-scale. Nature 467(7319):1061–1073
Lee S, Teslovich TM, Boehnke M, Lin X (2013) General framework for meta-analysis of rare variants in sequencing association studies. Am J Hum Genet 93:42–53
Li H (2011) A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27(21):2987–2993
Muddyman D et al (2013) Implementing a successful data-management framework: the UK10K managed access model. Genome Med 5:100
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2015 Springer Science+Business Media New York
About this chapter
Cite this chapter
Muddyman, D. (2015). The UK10K Project: 10,000 UK Genome Sequences—Accessing the Role of Rare Genetic Variants in Health and Disease. In: Zeggini, E., Morris, A. (eds) Assessing Rare Variation in Complex Traits. Springer, New York, NY. https://doi.org/10.1007/978-1-4939-2824-8_7
Download citation
DOI: https://doi.org/10.1007/978-1-4939-2824-8_7
Publisher Name: Springer, New York, NY
Print ISBN: 978-1-4939-2823-1
Online ISBN: 978-1-4939-2824-8
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)