Abstract
Alzheimer’s disease (AD) is known to be a multifactorial neurodegenerative disorder, and is one of the main causes of dementia in the elderly. Many studies have demonstrated molecules involved in the pathogenesis of AD, however its underlying mechanisms remain obscure. It may be simplistic to try to explain the disease based on the role of a few genes only. Accumulating new, huge amount of information from e.g. genome, proteome and interactome datasets and new knowledge, we are now able to clarify and characterize diseases essentially as a result of dysfunction of molecular networks. Recent studies have indicated that relevant genes affected in human diseases concentrate in a part of the network, often called as “disease module.” In the case of AD, some disease-associated pathways seem different, but some of them are clearly disease-related and coherent. This suggests the existence of a common pathway that negatively drives from healthy state to disease state (i.e., the disease module(s)). Additionally, such disease modules should dynamically change through AD progression. Thus, network-level approaches are indispensable to address unknown mechanisms of AD. In this chapter, we introduce network strategies using gene co-expression and protein interaction networks.
Access provided by CONRICYT – Journals CONACYT. Download protocol PDF
Similar content being viewed by others
Key words
- Alzheimer’s disease
- Systems biology
- Network dysfunction
- Network perturbation
- Disease module
- Gene expression profile
- Gene co-expression network
- Protein interaction network
1 Introduction
Alzheimer’s disease (AD) is neuropathologically characterized by extracellular plaques of amyloid-beta (Aβ) peptide and intra-neuronal accumulation of neurofibrillary tangles (NFTs). However, the molecular mechanisms of AD pathology remain obscure. What molecules accelerate production of Aβ or NFTs? How do those molecules lead to neuronal cell death? It is difficult to take into account pathological mechanisms of AD by only the known molecules. The new approaches need to identify the remaining essential molecules and pathways.
Recently, developments of high-throughput technologies have emerged as a new paradigm for elucidation of biological complexity including complex diseases. Biological molecular interactions (e.g. gene–gene, protein–protein, protein–DNA interactions and others) obtained by comprehensive resources are analyzed using a network representation. In terms of such network biology, there are some attempts to characterize diseases as network perturbations [1–4]. These studies indicate that genes affected in diseases concentrate in a part of the network, often called as “disease module” [5]. AD-associated pathways seem superficially different, but some of them are clearly not incoherent [6, 7]. A common pathway that negatively drives from healthy state to disease state (i.e., the disease module) may exist in AD. Moreover, pursuing the AD-specific modules may help to understand the other neurodegenerative diseases. Actually, a rare mutation in triggering receptor expressed on myeloid cells 2 (TREM2) relates not only to AD [8, 9] but also to Nasu-Hakola disease [10] and frontotemporal dementia [11], which suggests that those diseases share common modules or pathways centered on TREM2. The network-level approaches would shed light on the uncharacterized cellular phenomena within AD brains. In this chapter, we present methods for understanding AD pathology through network-based, but not single molecules analysis. To this end, we introduce available gene expression profiles from AD postmortem brains and the human protein–protein interaction datasets in Subheading 2, and in Subheading 3 we provide the actual approaches from recent studies.
2 Materials
A gene co-expression network is generally reconstructed using available gene expression profiles. On the other hand, the protein interaction network is assembled from protein–protein interaction data in open access repositories. We here provide available gene expression profiles of AD and protein–protein interaction databases.
2.1 Gene Expression Profiles of Alzheimer’s Disease
Many of systems biology studies have yielded important insights into mechanisms underlying AD using gene expression profiles from postmortem brains and autopsied tissues of AD subjects. Public gene expression datasets are basically registered in the Gene Expression Omnibus (GEO) database [12] (see Note 1 ), with GEO accession numbers assigned. Below, we introduce some available gene expression profiles from AD subjects.
The Braak stage is used as the neuropathological staging in AD, which is diagnosed based on expansion of neurofibrillary tangles (NFTs) across brain regions. NFTs deposit in the following order; the transentorhinal region (Braak stage I–II), the limbic system (Braak stage III–IV) and the isocortical region (Braak stage V–VI) [13]. Liang et al. provided gene expression profiles from postmortem brains of 14 healthy subjects (Braak stage 0–II) and 34AD-affected subjects (Braak stage III–VI) (GEO accession number: GSE5281) [14, 15]. Postmortem brains were laser-captured in six brain regions (entorhinal cortex, hippocampus, medial temporal gyrus, posterior cingulate, superior frontal gyrus and primary visual cortex). The gene expression profiles were obtained with Affymetrix Human Genome U133 Plus 2.0 microarrays (Affymetrix Inc., Santa Clara, CA, USA).
The MiniMental State Examination (MMSE) test is a clinical assessment for cognitive function [16]. Blalock et al. stratified 35 subjects by MMSE score into four groups, “Control” (MMSE score > 25), “Incipient AD” (MMSE score 20–25), “Moderate AD” (MMSE score 14–19), and “Severe AD” (MMSE score < 14) (GSE1297) [17]. The CA1 and CA3 regions were dissected from the frozen hippocampal tissues and they were profiled on Affymetrix Human Genome U133A Array.
In order to construct gene regulatory network in late-onset alzheimer’s disease (LOAD) and non-demented healthy controls, Zhang et al. collected 690 autopsied tissues from dorsolateral prefrontal cortex BA9, visual cortex BA17 and cerebellum in brains of LOAD patients, and utilized custom microarrays manufactured by Agilent Technologies (GSE44772) [18].
2.2 The Human Protein–Protein Interaction Data
Each interaction between proteins has been identified by established methods (i.e. two-hybrid system, immunoprecipitation method, and others). Over the past decade, high-throughput technologies including large-scale yeast two-hybrid screenings and mass spectrometry have enabled to obtain comprehensive protein–protein interaction (PPI) datasets in human [19–21]. At present, PPI datasets curated from published studies regardless of small- or large-scale experiments are integrated in databases as those in Table 1.
3 Methods
3.1 Construction of the Gene Co-expression Network
In order to build the gene co-expression network, associations between genes are determined by Pearson correlation coefficient (PCC) using gene expressions across samples. PCC ranges from −1 (negative correlation) to 1 (positive correlation). When PCC between gene i and gene j (PCC i,j ) exceeds a threshold, two genes are linked (co-expression). The PCC value (e.g., |PCC| > 0.5) and p-value can be used directly as test for no correlation. However, these thresholds depend on sample size and are often arbitrary. To overcome these difficulties, the weighted gene co-expression network analysis (WGCNA) (see Note 2 ) [22, 23], which is widely applied in some studies including AD [18, 24, 25], determines a threshold based on the fact that biological networks are essentially scale-free (see Note 3 ). First, PCC i,j is transformed into “similarity,” s i,j , taking from 0 to 1 (see Note 4 ):
If you preserved the sign of PCC i,j ,
is used (see Note 5 ). Next, s i,j is assigned into the power function:
where β is the parameter. The parameter β should be set to be higher than the scale-free topology model fit (R 2) that is the slope between log10(p(k)) and log10(k) (see Note 3 about p(k)). A stringent parameter brings the higher R 2, but it may lead to networks with very few interactions because of trade-off relationships between R 2 and the number of interactions.
3.2 Construction of the Protein Interaction Network
Some studies analyze PPI datasets that combine data from several databases and repositories, however the curation policies of each database are different. In addition, registered proteins are often maintained with different identifiers (e.g. Entrez gene ID and UniProt ID). The International Molecular Exchange (IMEx) consortium recently developed common strategies and attempts to provide a nonredundant dataset through the participating databases [26]. To avoid problems in some efforts at unifying IDs for example, iRefIndex provides an index across 13 primary databases [27].
3.3 Module Detection
The enormous amount of information in biological networks makes it difficult to be analyzed. Therefore, networks are usually divided into modules, which are defined as subsets of nodes (genes or proteins) that densely interact with each other (represented as links or edges).
There are mainly two methods for module detection. Basically, either a node belongs to only one module or to multiple modules. We here present the Infomap algorithm and the topological overlap as the first method, and the link clustering algorithm as the second method.
The Infomap algorithm that proposed by Rosvall and Bergstrom, which detects modules based on the random walk [28]. The algorithm divides a network into m modules with an optimal number of modules, M. Here, the module is defined as the region in which the random walker tends to stay for a long time. The efficiency on M is assessed by the map equation [29]:
where q ↷ and \( H(Q) \) are the probability and the entropy of the movement of the random walker between modules, p i↻ and \( H\left({P}^i\right) \) are the fraction and the entropy of the movement within module i. This equation takes/results in a low value when a random walker has less module transitions and less within-module movements. It seeks the best number of modules to minimize the map equation over all possible partitions. The Infomap algorithm is reported to have the best-performance compared to several algorithms [30].
Next, the topological overlap is the method focused on a link similarity between node i and j (ω i,j ), which is given by the formula below:
where l i,j is the number of common nodes connected between node i and j, a i,j is adjacency function (a i,j = 1, if i and j are linked. and a i,j = 0, otherwise), and k i is the connection degree (the number of interacting partners) of i [31]. The topological overlap calculated across all nodes is displayed as a matrix. Hierarchical clustering is implemented to its matrix, and the classified clusters are regarded as modules. This method can be applied to unweighted and weighted networks. In practice, the weighted gene co-expression network analysis (WGCNA) detects modules by the topological overlap matrix (TOM) from a constructed gene co-expression network.
The two methods above essentially assign a node to a module, whereas in real networks a node could participate/belong to multiple modules. For instance, proteins that have a lot of functions may associate with several protein complexes in the biological network. In particular, such proteins are called as “date hubs” in the context of systems biology [32]. The link clustering is a method to classify links into distinct modules [33]. The originality of this method is to calculate similarity between links e ik and e jk that share a node k as:
where \( {n}_{+}(i) \) is the node set of node i and the neighbors. Calculated similarities are reordered by the application of hierarchical clustering and the results are represented as a dendrogram. In order to determine the best threshold to cut branches in a dendrogram, the partition density, D, is used:
where M is the number of links in the network, c is the number of the modules, m c is the number of links in a module and n c is the number of the nodes in a module. The partition density, D, indicates the average density across each module and takes the value from 0 (sparse) to 1 (dense). D is computed at each height of the dendrogram. The height at which D takes the maximum value is adopted as the cutting threshold.
3.4 Application Using a Genetic Interaction Network
Zhang and coworkers analyzed the gene expression profiles of three brain regions (dorsolateral prefrontal cortex (PFC), visual cortex (VC) and cerebellum (CB)) from LOAD and non-demented individuals described in Subheading 2.1 (GSE44772) [22]. They first obtained 13,193 (one-third) of the most variable probesets in each brain. The probesets were assigned a unique identifier, combined probeset ID and brain region name, and those expression data were merged. Based on these multi-tissue expression data sets containing each 39,579 probesets in LOAD and non-demented brains, multi-tissue co-expression networks were constructed by WGCNA. From the topological overlap matrices (see Subheading 3.3), 111 and 89 modules were identified in LOAD and non-demented brains, respectively. Next, they measured the modular differential connectivity (MDC) to compare the connectivity among modules in LOAD and normal healthy brains. MDC is defined by the following:
where N is the number of genes in a module, k ij is the connectivity between genes i and j. Here, k ij equals to a i,j in Subheading 3.1. The modules with MDC > 1 indicate gain of connectivity (GOC), in contrast, those with MDC <1 indicate loss of connectivity (LOC). The GOC modules were found more than ten times greater than the LOC modules. In GOC modules with at least 100 genes, the immune/microglia module was identified, and 99.5 % of genes in this module were differentially expressed in PFC, which is commonly affected in AD. Interestingly, expressions of genes in the PFC immune/microglia module correlated with atrophy levels in several brain regions. Furthermore, expression quantitative trait loci (eQTL) analyses were performed to identify SNPs associated with gene expressions (eSNPs). Many genes in the PFC immune/microglia module were significantly enriched cis-eSNPs located within around 1 Mb of the gene body. Finally, the directed Bayesian networks for the immune/microglia module were constructed. As a result of calculation of the combined score, based on the number of downstream genes and differential expression, TYRO protein tyrosine kinase-binding protein (TYROBP) was ranked the highest score, indicating TYROBP is a key causal regulator. TYROBP is also known as DNAX-activating protein of 12 kD (DAP12), and works as a signaling adaptor protein of TREM2. A rare variant of TREM2 was recently reported increases the risk to develop LOAD in cohorts from North America and Europe [6, 7].
3.5 Application Using a Protein Interaction Network
The biggest risk for AD is aging. AD progresses slowly over years or decades, rather than a rapid transition from healthy to disease state. We therefore have to consider dynamic, temporal changes of the AD-associated networks and modules.
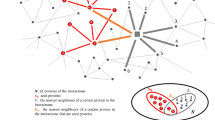
We recently identified modules disrupted with the progression of AD by combining a protein interaction network with gene expression profiles of brains from AD and normal aging individuals [34]. The AD gene expression profiles used were from postmortem brains of AD subjects (GSE5281), introduced in Subheading 2.1. We also used the gene expression profiles from postmortem brains (entorhinal cortex, hippocampus, superior frontal gyrus and postcentral gyrus) of cognitively intact subjects aged 60–99 years as normal aging [35]. Normal aging subjects were classified into the following four age groups: 60–69, 70–79, 80–89, and 90–99 years old. We analysed gene expression profiles from common three brain regions (entorhinal cortex (EC), hippocampus (HIP) and superior frontal gyrus (SFG)) between two datasets. First, gene expression datasets were normalized using the MAS 5.0 algorithm (Affymetrix, Santa Clara, CA). Then, we used probe sets marked as “present” by the detection call algorithm (Affymetrix) and averaged their expression values through samples in same brain region in same stage (Braak stage or age group). Here, we considered that a gene is expressed if the average expression values exceeded 200, and assumed direct protein expression from gene expression (RNA expression) datasets (see Note 6 ). We next retrieved the human interaction dataset from BioGRID [36]. Adding physical interactions between expressed proteins, the expressed protein interaction networks (PINs) were constructed in each stage, and they were divided into modules using the Infomap algorithm (see Subheading 3.3). To observe trajectories of modules through AD progression (Braak stages), we performed the brute-force approach to compute similarities of interactions (C L) and cellular functions of proteins (C GO) between modules in a stage and the next stage. The similarity was defined as follows:
where A(t) is a set of interactions to obtain similarities of interactions (C L), or cellular functions, to obtain similarities of cellular functions (C GO), in a module at time t (i.e., Braak stage or age group) (Fig. 1). A similarity takes 1 when the modules in time t and t + 1 have same interactions or same cellular functions (see Note 7 ). To estimate whether two modules in time t and t + 1 are conserved, we considered that the both modules were conserved if a module pair has the highest C L and their C L and C GO exceed 0.5 (see Note 8 ). Otherwise, they are not conserved. Repeating this procedure, the conserved relationships between modules in consecutive stages were linked as a module lineage. Next, we sought AD-specific disrupted module lineages, which are defined as module lineages that are fully conserved across all age groups in normal aging but are not conserved across Braak stages in AD. AD-specific, disrupted module lineages are classified into the early-disrupted type and the late-disrupted type. In entorthinal cortex (EC), affected in the incipient Braak stage, 4.0 % of all module lineages indicated early-disrupted type, and 40.0 % of all module lineages indicated late-disrupted type (see Note 9 ). Of the late-disrupted type in EC, we found a module that lost the most interactions across Braak stages. The members in the module are significantly associated with the histone acetyltransferase (HAT) complex. We also found that the HAT module tightly interacted with the proteasome module via the deubiquitinating enzyme UCHL5 in Braak stage I (Fig. 2). However, interactions between UCHL5 and some members in the HAT module (INO80B/C, NFRKB and others) were beginning to disappear in Braak stage II, and fully collapsed in Braak stage IV. UCHL5 has been reported to interact with the INO80 complex via NFRKB [37]. This complex could alter chromatin conformation and regulate gene transcription or DNA repair [38]. Furthermore, the deubiquitinating enzyme UCHL5 is also associated with the 26S proteasome. In healthy cells, abnormal toxic proteins (e.g., Aβ in AD) are decomposed by protein quality control systems such as the ubiquitin-proteasome system (UPS). However, the degradation of toxic proteins does not seem to work efficiently in AD compared to healthy subject. Indeed, an impairment in ubiquitin-proteasome system function has recently been observed in AD [39, 40]. Our findings suggest that down-regulated UCHL5 and affected network interactions may disturb proteolysis, with also presence of aberrant gene expression in AD.
Calculation for similarities between modules stage t and t + 1. Similarities of interactions (C L) and cellular functions (C GO) are calculated over all possible module pairs between stage t and t + 1. We considered that the both modules were conserved if a module pair has the highest C L and their C L and C GO exceed 0.5
Dynamics of module interactions in the entorhinal cortex during AD progression. The upper yellow and lower green nodes are components of the histone acetyltransferase and proteasome modules respectively. Hub proteins disappearing with Braak stage are depicted as large nodes. Figure obtained, adapted from studies/data in [34]
4 Notes
-
1.
The Gene Expression Omnibus (GEO) is provided at the National Center for Biotechnology Information (NCBI), and is freely accessible at (http://www.ncbi.nlm.nih.gov/geo/).
-
2.
WGCNA is implemented based on the R project for statistical computing package (http://www.r-project.org).
-
3.
A network is composed of nodes (e.g., genes or proteins) and edges/links (e.g., co-expression relationships or physical interactions). In a scale-free network, the frequency of connection degree (number of partners a node interacts with) is p(k) ~ k −γ, where k is the connection degree and γ is the degree exponent. This indicates the presence of many nodes with a few interactions and a few nodes with many interactions. Many biological networks are scale-free [41]. In WGCNA, the users can determine the parameter β to conserve scale-free topology.
-
4.
Besides Pearson correlation coefficient, the other measurements (e.g. biweight midcorrelation, mutual information) are calculable.
-
5.
The users can select “unsigned” or “signed” from the variables in corresponding functions (“type” and “networkType”).
-
6.
To determine whether the gene is expressed or not, we adopted a 200 threshold based on the method proposed by Bossi et al. [42]. An expression value of 200 represents approximately 3–5 copies per cell [43].
-
7.
Based on the “biological process” functions of the Gene Ontology Annotation (GOA), we assigned proteins with cellular functions. Note that one protein can have several functions. Next, we assigned an interaction with the GOA common to both proteins constituting the interaction. Using interaction sets with the GOA functions, we next sought significantly enriched functions within each module by hypergeometric test. If the probability by hypergeometric distribution was less than 0.05 and the ratio to expected value was greater than 2, we assigned the GOA function to the module. As an example of calculation of C GO, we consider a module with functions A, B and C at time t (M 1 t ), and a module with functions B and D at time t + 1 \( \left({M}_{t+1}^1\right) \). The common function is B, and the union of functions is A, B, C and D, therefore the C GO is 1/4.
-
8.
This criterion has two steps: (1) filtering module pairs with the highest C L and, (2) extracting module pairs with C L and C GO > 0.5 from module pairs filtered in step (1). In the first step, if the modules at time t and t + 1 are conserved, each interaction that constitute the two modules has to be highly shared. For instance, when a module at time t M 1 t shows the highest C L with a module at time t + 1 \( {M}_{t+1}^1 \) and \( {M}_{t+1}^1 \) also shows the highest C L with M 1 t , \( {M}_t^1-{M}_{t+1}^1 \) pair moves to the next step. On the other hand, if \( {M}_{t+1}^1 \) shows the highest C L with a different module at time t M 2 t , \( {M}_t^1-{M}_{t+1}^1 \) pair is omitted from this criterion. Note that the highest C L can be same value (e.g., when M 1 t equally splits into \( {M}_{t+1}^1 \) and \( {M}_{t+1}^2 \) at time t + 1). The second step is a process to filter out pairs with same highest C L and lowest conserved pairs. Summation of C L of a module is ≤1. From this, it follows that with a threshold >0.5, the pair satisfying this threshold is determined uniquely. Conversely, summation of C GO of a module can be >1 because cellular functions can be redundant. The threshold of C GO is therefore arbitrary.
-
9.
We did not verify the statistical significance of the disrupted modules in [34]. To do this here, we propose bootstrap analysis as a useful approach. More specifically, we randomly resample protein sets (e.g., 1,000) with the same number of proteins as the observed module from expressed proteins (i.e. “resampling set” and “observed set”). We compare statistics (e.g., number of interactions lost across Braak stages) between the observed set and the resampling sets. If the statistics of the observed set are significantly different with those of the resampling sets, we evaluate that the observed module is a disrupted module.
References
Lewis NE, Schramm G, Bordbar A et al (2010) Large-scale in silico modeling of metabolic interactions between cell types in the human brain. Nat Biotechnol 28:1279–1285
Mine KL, Shulzhenko N, Yambartsev A et al (2013) Gene network reconstruction reveals cell cycle and antiviral genes as major drivers of cervical cancer. Nat Commun 4:1806
Pichlmair A, Kandasamy K, Alvisi G et al (2012) Viral immune modulators perturb the human molecular network by common and unique strategies. Nature 487:486–490
Rozenblatt-Rosen O, Deo RC, Padi M et al (2012) Interpreting cancer genomes using systematic host network perturbations by tumour virus proteins. Nature 487:491–495
Barabasi AL, Gulbahce N, Loscalzo J (2011) Network medicine: a network-based approach to human disease. Nat Rev Genet 12:56–68
Mizuno S, Iijima R, Ogishima S et al (2012) AlzPathway: a comprehensive map of signaling pathways of Alzheimer’s disease. BMC Syst Biol 6:52
Huang Y, Mucke L (2012) Alzheimer mechanisms and therapeutic strategies. Cell 148:1204–1222
Jonsson T, Stefansson H, Steinberg S et al (2013) Variant of TREM2 associated with the risk of Alzheimer’s disease. N Engl J Med 368:107–116
Guerreiro R, Wojtas A, Bras J et al (2013) TREM2 variants in Alzheimer’s disease. N Engl J Med 368:117–127
Paloneva J, Manninen T, Christman G et al (2002) Mutations in two genes encoding different subunits of a receptor signaling complex result in an identical disease phenotype. Am J Hum Genet 71:656–662
Guerreiro RJ, Lohmann E, Bras JM et al (2013) Using exome sequencing to reveal mutations in TREM2 presenting as a frontotemporal dementia-like syndrome without bone involvement. JAMA Neurol 70:78–84
Edgar R, Domrachev M, Lash AE (2002) Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30:207–210
Braak H, Braak E (1991) Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol 82:239–259
Liang WS, Dunckley T, Beach TG et al (2007) Gene expression profiles in anatomically and functionally distinct regions of the normal aged human brain. Physiol Genomics 28:311–322
Liang WS, Dunckley T, Beach TG et al (2008) Altered neuronal gene expression in brain regions differentially affected by Alzheimer’s disease: a reference data set. Physiol Genomics 33:240–256
Folstein MF, Folstein SE, McHugh PR (1975) “Mini-mental state.” A practical method for grading the cognitive state of patients for the clinician. J Psychiatr Res 12:189–198
Blalock EM, Geddes JW, Chen KC et al (2004) Incipient Alzheimer’s disease: microarray correlation analyses reveal major transcriptional and tumor suppressor responses. Proc Natl Acad Sci U S A 101:2173–2178
Zhang B, Gaiteri C, Bodea LG et al (2013) Integrated systems approach identifies genetic nodes and networks in late-onset alzheimer’s disease. Cell 153:707–720
Rual JF, Venkatesan K, Hao T et al (2005) Towards a proteome-scale map of the human protein-protein interaction network. Nature 437:1173–1178
Stelzl U, Worm U, Lalowski M et al (2005) A human protein-protein interaction network: a resource for annotating the proteome. Cell 122:957–968
Ewing RM, Chu P, Elisma F et al (2007) Large-scale mapping of human protein-protein interactions by mass spectrometry. Mol Syst Biol 3:89
Zhang B, Horvath S (2005) A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol 4:17
Langfelder P, Horvath S (2008) WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9:559
Miller JA, Oldham MC, Geschwind DH (2008) A systems level analysis of transcriptional changes in Alzheimer’s disease and normal aging. J Neurosci 28:1410–1420
Miller JA, Horvath S, Geschwind DH (2010) Divergence of human and mouse brain transcriptome highlights Alzheimer disease pathways. Proc Natl Acad Sci U S A 107:12698–12703
Orchard S, Kerrien S, Abbani S et al (2012) Protein interaction data curation: the International Molecular Exchange (IMEx) consortium. Nat Methods 9:345–350
Razick S, Magklaras G, Donaldson IM (2008) iRefIndex: a consolidated protein interaction database with provenance. BMC Bioinformatics 9:405
Rosvall M, Bergstrom CT (2008) Maps of random walks on complex networks reveal community structure. Proc Natl Acad Sci U S A 105:1118–1123
Rosvall M, Axelsson D, Bergstrom CT (2008) The map equation. Eur Phys J Spec Top 178:13–23
Lancichinetti A, Fortunato S (2009) Community detection algorithms: a comparative analysis. Phys Rev E 80:056117
Ravasz E, Somera AL, Mongru DA et al (2002) Hierarchical organization of modularity in metabolic networks. Science 297:1551–1555
Han JD, Bertin N, Hao T et al (2004) Evidence for dynamically organized modularity in the yeast protein-protein interaction network. Nature 430:88–93
Ahn YY, Bagrow JP, Lehmann S (2010) Link communities reveal multiscale complexity in networks. Nature 466:761–764
Kikuchi M, Ogishima S, Miyamoto T et al (2013) Identification of unstable network modules reveals disease modules associated with the progression of Alzheimer’s disease. PLoS One 8:e76162
Berchtold NC, Cribbs DH, Coleman PD et al (2008) Gene expression changes in the course of normal brain aging are sexually dimorphic. Proc Natl Acad Sci U S A 105:15605–15610
Stark C, Breitkreutz BJ, Chatr-Aryamontri A et al (2011) The BioGRID interaction database: 2011 update. Nucleic Acids Res 39:D698–D704
Yao T, Song L, Jin J et al (2008) Distinct modes of regulation of the Uch37 deubiquitinating enzyme in the proteasome and in the Ino80 chromatin-remodeling complex. Mol Cell 31:909–917
Zediak VP, Berger SL (2008) Hit and run: transient deubiquitylase activity in a chromatin-remodeling complex. Mol Cell 31:773–774
Keller JN, Hanni KB, Markesbery WR (2000) Impaired proteasome function in Alzheimer’s disease. J Neurochem 75:436–439
Lam YA, Pickart CM, Alban A et al (2000) Inhibition of the ubiquitin-proteasome system in Alzheimer’s disease. Proc Natl Acad Sci U S A 97:9902–9906
Barabasi AL, Oltvai ZN (2004) Network biology: understanding the cell’s functional organization. Nat Rev Genet 5:101–113
Bossi A, Lehner B (2009) Tissue specificity and the human protein interaction network. Mol Syst Biol 5:260
Su AI, Cooke MP, Ching KA et al (2002) Large-scale analysis of the human and mouse transcriptomes. Proc Natl Acad Sci U S A 99:4465–4470
Acknowledgement
We thank Drs. Takeshi Ikeuchi and Kensaku Kasuga of Niigata University for helpful discussions.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2016 Springer Science+Business Media New York
About this protocol
Cite this protocol
Kikuchi, M. et al. (2016). Network-Based Analysis for Uncovering Mechanisms Underlying Alzheimer’s Disease. In: Castrillo, J., Oliver, S. (eds) Systems Biology of Alzheimer's Disease. Methods in Molecular Biology, vol 1303. Humana Press, New York, NY. https://doi.org/10.1007/978-1-4939-2627-5_29
Download citation
DOI: https://doi.org/10.1007/978-1-4939-2627-5_29
Publisher Name: Humana Press, New York, NY
Print ISBN: 978-1-4939-2626-8
Online ISBN: 978-1-4939-2627-5
eBook Packages: Springer Protocols