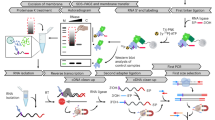
Abstract
Hundreds of RNA binding proteins posttranscriptionally regulate gene expression, but relatively few have been characterized in plants. One successful approach to determine protein function has been to identify interacting molecules and the conditions of their association. The ribonucleoprotein immunopurification (RIP) assay facilitates the identification and quantitative comparison of RNA association to specific proteins under different experimental conditions. A variety of molecular techniques can be used to analyze the enriched RNAs, whether few as in the case of highly specific interactions, or many. Identification of associated RNAs can inform hypothesis generation about the processes or pathways regulated by the target protein. Downstream analysis of associated RNA sequences can lead to the identification of candidate motifs or features that mediate the protein–RNA interaction. We present a rapid method for RIP from tissues of plants that is suitable for experiments that require immediate tissue cryopreservation, such as monitoring a rapid response to an environmental stimulus.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281–297
Dinger ME, Pang KC, Mercer TR et al (2008) Differentiating protein-coding and noncoding RNA: challenges and ambiguities. PLoS Comput Biol 4:e1000176
Jen C-H, Michalopoulos I, Westhead DR et al (2005) Natural antisense transcripts with coding capacity in Arabidopsis may have a regulatory role that is not linked to double-stranded RNA degradation. Genome Biol 6:R51
Zhu Q-H, Wang M-B (2012) Molecular functions of long non-coding RNAs in plants. Genes (Basel) 3:176–190
Bailey-Serres J, Sorenson R, Juntawong P (2009) Getting the message across: cytoplasmic ribonucleoprotein complexes. Trends Plant Sci 14:443–453
Sorenson R, Bailey-Serres J (2014) Selective mRNA sequestration by OLIGOURIDYLATE-BINDING PROTEIN 1 contributes to translational control during hypoxia in Arabidopsis. Proc Natl Acad Sci 111:2373–2378
Juntawong P, Sorenson R, Bailey-Serres J (2013) Cold shock protein 1 chaperones mRNAs during translation in Arabidopsis thaliana. Plant J Cell Mol Biol 74:1016–1028
Ji L, Liu X, Yan J et al (2011) ARGONAUTE10 and ARGONAUTE1 regulate the termination of floral stem cells through two microRNAs in Arabidopsis. PLoS Genet 7:e1001358
Schmitz-Linneweber C, Williams-Carrier R, Barkan A (2005) RNA immunoprecipitation and microarray analysis show a chloroplast Pentatricopeptide repeat protein to be associated with the 5′ region of mRNAs whose translation it activates. Plant Cell 17:2791–2804
Zanetti ME, Chang I-F, Gong F et al (2005) Immunopurification of polyribosomal complexes of Arabidopsis for global analysis of gene expression. Plant Physiol 138:624–635
Mustroph A, Zanetti ME, Jang CJH et al (2009) Profiling translatomes of discrete cell populations resolves altered cellular priorities during hypoxia in Arabidopsis. Proc Natl Acad Sci U S A 106:18843–18848
Bailey T, Elkan C (1994) Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Sec Int Conf on Intell Syst Mol Biol 2:28–36, Menlo Park, CA: AAAI Press
Ho JJD, Marsden PA (2014) Competition and collaboration between RNA-binding proteins and microRNAs. Wiley Interdiscip Rev RNA 5:69–86
Terzi LC, Simpson GG (2009) Arabidopsis RNA immunoprecipitation. Plant J Cell Mol Biol 59:163–168
Barkan A (2009) Genome-wide analysis of RNA-protein interactions in plants. In: Belostotsky DA (ed) Plant systems biology. Humana Press, Totowa, NJ, pp 13–37
Köster T, Staiger D (2014) RNA-binding protein immunoprecipitation from whole-cell extracts. In: Sanchez-Serrano JJ, Salinas J (eds) Arabidopsis protocols. Humana Press, Totowa, NJ, pp 679–695
Mustroph A, Juntawong P, Bailey-Serres J (2009) Isolation of plant polysomal mRNA by differential centrifugation and ribosome immunopurification methods. Methods Mol Biol 553:109–126
Acknowledgments
We thank Piyada Juntawong for many helpful discussions. This work was supported by the US National Science Foundation grants IOS-0750811 and MCB-1021969 (to J.B.-S.) and an Integrative Graduate Education and Research Traineeship DGE-0504249 award (to R.S.).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2015 Springer Science+Business Media New York
About this protocol
Cite this protocol
Sorenson, R., Bailey-Serres, J. (2015). Rapid Immunopurification of Ribonucleoprotein Complexes of Plants. In: Alonso, J., Stepanova, A. (eds) Plant Functional Genomics. Methods in Molecular Biology, vol 1284. Humana Press, New York, NY. https://doi.org/10.1007/978-1-4939-2444-8_10
Download citation
DOI: https://doi.org/10.1007/978-1-4939-2444-8_10
Published:
Publisher Name: Humana Press, New York, NY
Print ISBN: 978-1-4939-2443-1
Online ISBN: 978-1-4939-2444-8
eBook Packages: Springer Protocols