Abstract
Carotenoids are an abundant group of isoprenoid pigments present in all photosynthetic organisms and responsible for the typical yellow, orange, or red coloration exhibited by many flowers, fruits, and vegetables. Besides their many functions in plants, these pigments are also essential components in human and animal diet. Within the past three decades, genes encoding all of the enzymes required for the biosynthesis of these indispensable pigments have been identified and characterized in higher plants, primarily as a result of integration of comparative genomics, biochemical genetics, and molecular approaches in the model plant Arabidopsis thaliana and cyanobacterium Synechocystis PCC6803. Mutant analysis and transgenic studies in these and other systems have established a foundation for understanding the function, regulation, and evolution of individual genes and enzymes. The aim of this chapter is to review advances in the structure, function, and evolution of these genes and enzymes, as well as the molecular mechanisms regulating carotenoid biosynthesis in higher plants.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
Introduction
Carotenoids are a family of isoprenoid molecules that are widespread in nature. They are responsible for the typical yellow, orange, and red colors of most fruits, flowers, and vegetables, and for the characteristic colors of many birds, insects, fish, and crustaceous intaking carotenoids through the diet. The basic chemical structure of any carotenoid molecule is the long polyene chain, which may extend from 3 to 15 conjugated double bonds, acting as a chromophore that determines the absorption spectrum of the molecule, and hence its color [1]. This basic structure can be further modified in a number of ways, such as cyclization and oxygenation, to yield a family of more than 600 different carotenoids generally divided into two subgroups, carotenes (hydrocarbon carotenoids) and xanthophylls (oxygenated derivatives) [1]. Their oxidative breakdown products are called apocarotenoids .
Carotenoids perform a broad range of metabolic and ecological functions. Carotenoid pigments are essential components of the photosynthetic membranes in all photosynthetic organisms, including plants, algae, and cyanobacteria, protecting them against photooxidative damage [2, 3]. This protective function is crucial in oxygen-evolving photosynthetic organisms, since an impairment to produce cyclic carotenoids is eventually lethal [1]. In higher plants, carotenoids also act as accessory pigments in the light-harvesting antennae of chloroplasts, transferring energy to chlorophylls, and as precursors for biosynthesis of the phytohormones abscisic acid (ABA), which controls abiotic stress signaling pathways, and strigolactone, which controls lateral shoot growth [3–5]. An additional and important role of carotenoids in higher plants is as coloring agents in flowers and fruits to attract pollinators and agents of seed dispersal [2]. In these tissues, carotenoids accumulate in nonphotosynthetic chromoplasts, where they are found in association with lipid–protein complexes in plastoglobules and/or in carotenoid-accumulating structures of globular, crystalline, membranous, fibrillar, or tubular forms [6].
Since animals are unable to synthesize carotenoids de novo, they rely upon the diet as the source of these compounds. Dietary carotenoids contribute to animal health and behavior, because they stimulate the immune system and aid in the preferential selection by the sexual partner [7, 8]. In mammalians, including humans, carotenoid species containing a β-ring can be converted into retinal (the main visual pigment), retinol (vitamin A) , and retinoic acid (a substance that controls morphogenesis) [9]. Additional beneficial effects of carotenoids in human health are attributed to their antioxidant and anti-inflammatory activities in vivo, which help to prevent certain cancers, cardiovascular diseases, light-induced erythema, and age-related diseases of eye such as cataract and macular degeneration [10, 11].
Here we present the current knowledge about the carotenoid biosynthesis gene families in higher plants from a genomic perspective. This includes information about the basic structure, function, and evolution of the genes and enzymes, as well as the molecular mechanisms regulating carotenoid biosynthesis in different plant tissues.
Carotenoid Biosynthetic Pathway
The biosynthetic pathways involved in carotenoid formation were elucidated during the second half of the twentieth century using both classical biochemical approaches and modern molecular biology techniques [12] . Nevertheless, the major advances in the identification of genes and enzymes of carotenoid biosynthesis occurred in the 1990s. Isolation of carotenoid-defective mutants in plants, and the information resources of the Arabidopsis thaliana EST database and the genome sequence of the cyanobacterium Synechocystis PCC6803 contributed to such advances [9, 12, 13]. Also important was the dissection of carotenoid biosynthesis pathway in bacterial systems, such as Rhodobacter capsulatus, Erwinia uredovora, and Erwinia herbicola. It allowed engineering strains of Escherichia coli accumulating a variety of carotenoid precursors for use as a simple and powerful in vivo system for the assay of enzyme function and substrate specificity [12]. In addition, the different colors exhibited by carotenoid-accumulating E. coli strains were exploited to visually screen complementary DNA (cDNA) and genomic libraries, in a procedure referred to as “color complementation,” enabling the identification of a number of previously unidentified plant, algal, and cyanobacterial carotenogenic genes based on the visualization of color changes in E. coli colonies [14] .
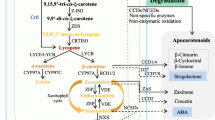
Carotenoids make a part of the plethora of chemical compounds that are produced via the general isoprenoid biosynthetic pathway (Fig. 2.1). As all other isoprenoids, carotenoids are built from the five-carbon (C5) compound isopentenyl diphosphate (IPP) and its allylic isomer, dimethylallyl diphosphate (DMAPP) [10, 13, 15]. Until recently, it was assumed that IPP was synthesized from acetyl-coenzyme A (CoA) via mevalonic acid (MVA) pathway [16]. However, in the early 1990s retro-biosynthetic studies established the presence of an alternative, MVA-independent pathway for the formation of IPP and DMAPP, termed 1-deoxy-D-xylulose-5-phosphate (DXP) or 2-C-methyl-D-erythritol-4-phosphate (MEP) pathway [17]. Several reports have indicated that eubacteria (E. coli and other pathogenic bacteria) and protozoans of apicomplexan phylum (Plasmodium falciparum) synthesize isoprenoids only via the MEP pathway, while archaebacteria, fungi, and animals (including humans) contain only the MVA pathway [18, 19]. The formation of isoprenoids in plants, however, can proceed from both MEP and MVA pathways [15, 18]. IPP is synthesized in plastids through the MEP pathway and in the cytosol through the MVA pathway. Thus, MEP pathway is a potential target for the development of new herbicides and anti-malarian and antimicrobial drugs that, besides the large spectrum of action, are not toxic for humans .
The general isoprenoid carotenoid biosynthetic pathway in plants. a Formation of acyclic carotenoids from the precursor GGPP. b Formation of cyclic carotenoids from the precursor lycopene. Enzyme abbreviations are as follows: IPPI isopentenyl diphosphate isomerase, GGPPS geranylgeranyl diphosphate synthase, PSY phytoene synthase, PDS phytoene desaturase, Z-ISO ζ-carotene isomerase, CrtISO carotenoid isomerase, ZDS ζ-carotene desaturase, LCYE lycopene ε-cyclase, LCYB lycopene β-cyclase, CHYB β-ring hydroxylase, CYP97C ε-ring hydroxylase, CCS capsanthin/capsorubin synthase, ZEP zeaxanthin epoxidase, VDE violaxanthin de-epoxidase, NXS neoxanthin synthase, CCD carotenoid cleavage dioxygenase, ZET β-ring hydroxylase, NCED 9-cis-epoxycarotenoid dioxygenase, IPP isopentenyl diphosphate, DMAPP dimethylallyl diphosphate, GGPP geranylgeranyl diphosphate
The MVA pathway involves a set of six reactions proceeding sequentially from acetyl-CoA to produce IPP and DMAPP [15, 16, 19]. Initially, two acetyl-CoA molecules, obtained through CO2 fixation, are condensed to yield acetoacetyl-CoA, in a reaction catalyzed by acetyl-CoA C-acetyltransferase (AACT, EC 2.3.1.9). Then, a third acetyl-CoA molecule is condensed to acetoacetyl-CoA, forming 3-hydroxy-3-methylglutaryl-CoA (HMG-CoA) by the action of HMG-CoA synthetase (HMGS, EC 4.1.3.5). The nicotinamide adenine dinucleotide phosphate (NADP)-dependent HMG-CoA reductase converts the CoA-derived in (R)-MVA, which is phosphorylated to (R)-MVA 5-diphophate by the sequential action of mevalonate kinase (MK, EC 2.7.1.36) and diphosphomevalonate kinase (PMK, EC 2.7.4.2). (R)-MVA 5-diphophate is further decarboxylated by the mevalonate diphosphate decarboxylase (MDC, EC 4.1.1.33), producing a pool of IPP. Finally, IPP isomerase catalyzes the reversible conversion of IPP into DMAPP, maintaining the equilibrium between these two compounds [16, 20] .
The MEP pathway consists of seven sequential reactions starting from the condensation of pyruvate and glyceraldehyde 3-phosphate (G3P) to yield 1-deoxy-D-xylulose 5-phosphate (DXP) [10, 17, 18]. This transketolase reaction is catalyzed by DXP synthase (DXS, EC 4.1.3.37), an enzyme that requires thiamine pyrophosphate and a divalent cation (Mg2 + or Mn2 + ) as cofactors [18, 21]. DXP is subsequently rearranged and reduced to MEP in a single step, in a reaction catalyzed by DXP reductoisomerase (DXR, EC 1.1.1.267). DXR requires NADPH and Mn2 + as cofactors [18, 21]. MEP is converted to IPP and DMAPP via 4-(cytidine 5ʹ-diphospho)-2-C-methyl-D-erythritol (CDP-ME), 2-phospho-4-(cytidine 5ʹ-diphospho)-2-C-methyl-D-erythritol (CDP-MEP), 2-C-methyl-D-erythritol-2,4-cyclodiphosphate (MCP), and 1-hydroxy-2-methyl-2-(E)-butenyl 4-phosphate (HMBPP). The enzymes responsible for these reactions are, respectively, MEP cytidyl transferase (MCT, EC 2.7.7.60), CDP-ME kinase (CMK, EC 2.7.1.148), MCP synthase (MCS, EC 4.6.1.12), and HMBPP synthase (HDS) [18, 21]. IPP/DMAPP synthase (IDS) is responsible for the conversion of HMBPP to a 5:1 mixture of IPP and DMAPP [10, 20, 21] .
IPP and DMAPP are subsequently used as blocks in a modular assembly process that produces compounds of 5, 10, 15, 20, or more carbons (in multiples of 5), allowing the biosynthesis of the basic skeletons for the various isoprenoids , including carotenoids, with a relatively small number of basic reaction steps [10, 15, 19]. For instance, the C20 geranylgeranyl diphosphate (GGPP), which serves as the immediate precursor for carotenoids, is formed by the sequential and linear addition of three molecules of IPP to one molecule of DMAPP. The enzyme that catalyzes these reactions, the GGPP synthase (GGPS; EC 2.5.1.29), is encoded by a multigene family of 12 members in the Arabidopsis genome, suggesting the involvement of different isozymes in the production of specific groups of isoprenoids [10, 13].
Basic Structure, Function, and Evolution of Carotenoid Biosynthesis Genes and Enzymes
The C40 skeleton of all plant carotenoids is assembled from the condensation of two molecules of GGPP, which then suffer a series of enzymatic reactions of desaturation, cyclization, and oxidation [10, 19] . Genetic and molecular evidences indicate that all enzymes of carotenoid biosynthetic pathway in plants are encoded by nuclear genes and post-translationally imported to plastids [12, 22]. Here, the genes and enzymes of carotenoid biosynthesis are discussed sequentially in their order within the pathway, giving specific details for one or more examples of each in Arabidopsis and other higher plants whenever possible.
Phytoene Synthase
Phytoene synthase (PSY, EC 2.5.1.32) catalyzes the first committed step in the formation of carotenoids, by the condensation of two GGPP molecules to produce 15-cis phytoene (Fig. 2.1a) [10, 12]. Detailed biochemical characterization of tomato and pepper PSYs has demonstrated that they can be either thylacoid membrane associated, but not integral, or stroma-localized proteins [10, 22]. The catalytic site of PSY, at the carboxy terminus, contains a large central cavity, formed by antiparallel alpha-helices, with two aspartate-rich motifs (DELVD and DVGED) that are positioned on opposite walls of the central cavity [19, 23]. The high degree of sequence conservation of these motifs suggests that they are required for the interaction of enzyme with upstream products [23]. PSY also contains an active site (YAKTF) at the amino terminus and a squalene synthase (SQS) domain type located between the catalytic and active sites (Fig. 2.2) [19, 23]. There is a low sequence similarity at the amino terminus among PSYs of different plant species, partially due to the existence of plastid transit peptide sequences that are known to show a low degree of sequence conservation.
Major functional domains present in some carotenogenic enzymes. 1.I, Active site of PSY; 1.II, squalene synthase domain type; 1.III, catalytic site of PSY. 2.I, Dinucleotide (FAD/NADP)-binding site domain of PDS; 2.II, carotenoid-binding domain of PDS. 3.I, Dinucleotide (FAD/NADP)-binding site domain of ZDS; 3.II, carotenoid-binding domain of ZDS. 4.I, Histidine-binding motif HXXXXXH type of β-CHY; 4.II, histidine-binding motif HXXHH type of β-CHY; 4.III, histidine-binding motif HXXXXXH type of β-CHY; 4.IV, histidine-binding motif HXXHH type of β-CHY. 5.I, Cystein-rich region of VDE and ZEP; 5.II, glutamate-rich region of VDE and ZEP. See text for further details. PSY phytoene synthase, FAD/NADP flavin adenine dinucleotide/nicotinamide adenine dinucleotide phosphate, PDS phytoene desaturase, ZDS ζ-carotene desaturase, VDE violaxanthin de-epoxidase, ZEP zeaxanthin epoxidase
Arabidopsis possesses only one PSY gene, while tomato and tobacco have two PSYs and plants belonging to the Poaceae have three PSYs [24–29]. Scenarios of gene duplication and sub-functionalization can be invoked for the evolution of PSY genes and enzymes from an ancestral PSY gene prototype. The PSY paralogs are involved with the carotenoid synthesis in different plant tissues. For instance, PSY1 encodes a fruit- and flower-specific isoform in tomato, whereas PSY2 encodes an isoform that predominates in photosynthetic tissues [25, 27]. In maize, PSY1 and PSY2 are required for endosperm carotenoid accumulation and photomorphogenesis in photosynthetic tissues, while PSY3 is associated with root carotenogenesis and necessary for drought and salt stress-induced production of ABA [28, 30].
A comparison of the gene structures of PSYs among different plant species shows that the Arabidopsis PSY contains seven exons, as well as Vitis vinifera , while rice and maize PSYs show a loss of exon 1 (Fig. 2.3a). The gene structures at the 5ʹ-untranslated region (UTR) of all PSY genes analyzed show differences in length and sequence, even among the different genes within a species, such as OsPSY1 (227 bp), OsPSY2 (147 bp), and OsPSY3 (193 bp; Fig. 2.3a). In contrast, the lengths of exons 3, 4, 5, and 6 are comparable among the different plant species, with 45–51, 173, 236, and 193–211 bp, respectively. Such conservation may be associated to the presence of nucleotide sequences encoding conserved domains of enzyme, such as the catalytic site located at the exon 5 of Arabidopsis. The dendrogram of similarity reveals that PSYs of rice and maize clustered together, in a separated clade from PSYs of dicots, which includes the tomato PSY1 and PSY2. It suggests that the duplication event of PSY occurred separately in Poaceae and Solanaceae (Fig. 2.3b). Furthermore, the proximity between AtPSY and OsPSY1 represents the existence of an ancient PSY constituting a common ancestor of monocots and dicots [29].
PSY gene structure and phylogeny. a Exon/intron structure of PSY in different plant species. Red arrow denotes the catalytic site of PSY. White, gray, and black thin bars indicate UTR, intron and exon regions, respectively. b Similarity dendrogram of plant PSYs and squalene synthases (SQSs). Amino acid sequences were aligned using ClustalW and a neighbor-joining tree was constructed with a 1000-bootstrap replication support. Abbreviations for the name of plant species are as follows: At Arabidopsis thaliana, Os Oryza sativa, Zm Zea mays, Nt Nicotiana tabacum, Cs Citrus sinensis, Po Populus trichocarpa, Sl Solanum lycopersicum, Ca Capsicum annum, Vv Vitis vinifera, PSY phytoene synthase, UTR untranslated region
Desaturases
Colorless phytoene undergoes a series of four desaturation reactions in plants that results in the formation of the red-colored carotenoid lycopene (Fig. 2.1a). These reactions are catalyzed by two related enzymes in plants: phytoene desaturase (PDS, EC 1.3.5.5) and ζ-carotene desaturase (ZDS, EC 1.3.5.6). PDS converts phytoene in phytofluene and then in ζ-carotene, while ZDS converts ζ-carotene in neurosporene and then in lycopene [10, 19]. In contrast with plants, bacteria and fungi contain only a single desaturase, CrtI, which catalyzes the four desaturation steps [19].
PDS and ZDS are found to be associated with other enzymes of carotenoid biosynthetic pathway, forming multimeric complexes of about 350 kDa [12, 31]. It has been proposed that two molecules of each PDS and ZDS, associated with one molecule of lycopene β-cyclase (β-LCY) and another of lycopene ε-cyclase (ε-LCY), form a multienzymatic complex responsible for the synthesis of α-carotene [12, 32]. A similar association of PDS and ZDS with two molecules of β-LCY would be responsible for the synthesis of β-carotene [12, 32]. PDS may be also associated with chloroplastic chaperonins (Cpn60) or heat-shock proteins (Hsp70) when located in stroma [31, 33, 34].
The active form of PDS is tightly bound to thylacoid membranes, whereas the stroma free form is inactive [31, 33]. Besides the association with membranes, the desaturases require cofactors for their complete activity. The removal of two hydrogen atoms during each desaturation step suggests the involvement of an electron transport chain for the regeneration of reductants [10]. A plastid terminal oxidase (PTOX) was identified in Arabidopsis mutants as one component of this electron transport chain required for the desaturation of phytoene [35]. PTOX is a plastoquinone oxidoreductase that regenerates the reduced plastoquinone formed during the desaturation of phytoene and ζ-carotene, using oxygen (O2) as a terminal acceptor. Thus, PTOX and O2 are considered as the main cofactors involved in the desaturation of phytoene, in both photosynthetic and nonphotosynthetic tissues [35].
The genomes of higher plants apparently contain only one copy of desaturase genes, PDS and ZDS [12, 26, 33, 36, 37]. A unique exception has been recently discovered in sweet orange (Citrus sinensis), in which 2 PDS and 12 ZDS members were found to be clustered, respectively, at one and three loci [38]. A high degree of similarity is observed in the deduced amino acid sequences of all plant desaturases [10]. All contain a conserved dinucleotide (FAD/NADP)-binding site domain at the amino terminus (Fig. 2.2). The carboxy terminus contains another conserved region, the carotenoid-binding domain.
Isomerases
In higher plants, phytoene occurs predominantly as the 15-cis isomer, while the predominant isomer of lycopene is all-trans, suggesting the existence of an enzymatic step mediating the cis–trans isomerization of lycopene precursors (Fig. 2.1a). Map-based cloning of the gene responsible for the tangerine tomato fruit phenotype, which accumulates 7,9,7ʹ,9ʹ-tetra-cis lycopene (prolycopene) and traces of its poly-cis precursors, resulted in the isolation of carotenoid isomerase (CrtISO). CrtISO encodes a carotenoid isomerase (CrtISO, EC 5.2.1.13) catalyzing the isomerization of prolycopene to all-trans lycopene [39]. A CrtISO homolog termed Ccr-2 was also isolated in Arabidopsis [40].
Both tomato and Arabidopsis isomerases contain a dinucleotide (FAD/NADP)-binding domain, like PDS and ZDS. However, the isomerases show more identities to bacterial phytoene desaturases (CrtI) than the plant desaturases [39, 40]. In fact, the bacterial desaturase CrtI also possesses the function of isomerization in combination with that of desaturation, converting 15-cis phytoene to all-trans lycopene [10].
Cyclases
Cyclization of the linear carotenoid lycopene marks an important branching point in the carotenoid pathway: one branch leads to β-carotene and its derivative xanthophylls , whereas the other leads to α-carotene and lutein (Fig. 2.1b). These carotenoids differ in the type of cyclic end group that is added. It can be a ε- or β-ionone ring, depending on the position of a double bond within the cyclohexane ring. Carotenoids with two β-rings, such as β-carotene and zeaxanthin , are primarily involved in protection against photooxidative damage and dissipation of the excess of light energy in the photosynthetic membranes [12]. Carotenoids with one β-ring and one ε-ring, such as lutein, act as accessory pigments in light-harvesting antennae of the chloroplasts [12]. Carotenoids with two ε-rings, such as lactucaxanthin in lettuce, are rare [41].
The type of end group produced depends on the nature of cyclase enzyme. In higher plants, there are two major cyclases: β-LCY (EC 5.5.1.19), which introduces β-rings, and ε-LCY (EC 5.5.1.18) that introduces ε-rings. The formation of β-carotene requires the introduction of two β-rings by β-LCY, whereas α-carotene requires the interaction of both β-LCY and ε-LCY (Fig. 2.1b) [32]. In contrast with β-LCY, ε-LCY is able to incorporate only one ε-ring to the symmetrical lycopene , forming δ-carotene [32]. Lettuce ε-LCY is the only example of cyclase that can introduce two ε-rings to the lycopene molecule [41]. All lycopene cyclases, irrespective of class, proceed via a carbocationic mechanism [19].
A membrane-associated multienzymatic complex involving the association of β-LCY and ε-LCY with PDS and ZDS is postulated to act in the synthesis of α- and β-carotene (see the desaturases section). These carotenogenic complexes possibly are associated to other enzymes and cofactors that regulate their catalytic activity [12]. Flux directing towards the β,β- or β,ε-branch of the pathway seems to be determined by the relative amounts of enzymatic activity and/or substrate specificity of β-LCY and ε-LCY [32, 42].
The lycopene cyclases contain a dinucleotide (FAD/NADP)-binding site domain, apparently involved in allosteric activation, and two characteristic conserved motifs: cyclase I and cyclase II (Fig. 2.4) [43, 44]. The FAD/NADP-binding site domain is composed by a typical secondary structure (β-sheet/α-helice/β-sheet) present in all plant enzymes with lycopene cyclase activity [44]. The cyclization reaction seems to be a simple rearrangement that does not involve any change in the oxidation level of lycopene molecule [19]. Thus, the involvement NAD(P)H in the reaction is not expected. NAD(P)H seems to have an indirect action, participating in the enzymatic reaction as an allosteric activator [44].
Alignment of the partial amino acid sequences of β-LCY and ε-LCY from different plant species. Conserved amino acid sequences of β-LCY and ε-LCY in a given position are in white text on a blue and black background, respectively. Dashes denote a gap in the amino acid sequence. The conserved regions are plant ε-cyclase conserved region (I), plant β-cyclase conserved region (II), dinucleotide (FAD/NADP)-binding site domain (III), cyclase I motif (IV and V), cyclase II motif (VI) and cyclase activity region (VII). Asterisk (*), colon (:) and dot (.) symbols denote identical, conserved and similar amino acids, respectively. Abbreviations for the species names are as follows: At Arabidopsis thaliana, Ca Capsicum annum, Cs Citrus sinensis, Ls Lactuca sativa, Os Oryza sativa, Pt Populus trichocarpa, Sl Solanum lycopersicum, Vv Vitis vinifera, Zm Zea mays, FAD/NADP flavin adenine dinucleotide/nicotinamide adenine dinucleotide phosphate, LCY lycopene cyclase
The first searches for homology carried out with plant ε-LCYs revealed the existence of a conserved region (VQMQQ), which was termed “ε-cyclase conserved region” (Fig. 2.4) [32]. Similarly, β-LCYs contain a conserved region (PLYD) that has been identified as “β-cyclase conserved region.” All lycopene cyclases also contain a conserved region that shows similarity to motifs of β-cyclase [32]. Since this region is present in cyclases that introduce β-, ε- or κ-ring, it has been termed “cyclase activity region” (Fig. 2.4).
Only a copy of ε-LCY gene has been identified in the genome of Arabidopsis and tomato [32, 45]. Arabidopsis also contains a copy of β-LCY [32], but two β-LCY copies, Crtl-B and Cyc-B, were identified in tomato [46, 47]. Crtl-B is active in photosynthetic tissues, whereas Cyc-B functions only in chromoplast-containing tissues [22]. The presence of two β-LCY genes, one with a chromoplast-specific expression, has been also reported in carotenogenic fruits other than tomato, including watermelon [48], orange, and grapefruit [49, 50].
Plant lycopene cyclases are also related to two other carotenoid cyclase enzymes: the capsanthin–capsorubin synthase (CCS, EC 5.3.99.8) of pepper [32] and the neoxanthin synthase (NSY, EC 5.3.99.9) of tomato [51] and potato [52]. CCS catalyzes the formation of the unusual five-carbon κ-ring [53], converting antheraxanthin or violaxanthin to capsanthin or capsorubin, respectively. In addition, CCS exhibits a β-LCY activity when lycopene is provided as a substrate [43]. NSY also modifies violaxanthin to the allenic product via a carbocation with a structure similar to the intermediate in the CCS-catalyzed reaction [54]. Although NSY operates mechanistically like CCS, its cryptic LCY activity has not been demonstrated [19].
Conservation of amino-acid sequences and their similar mechanisms of catalysis suggest that all plant cyclases, including CCS and NSY, have evolved from a common ancestor, most probably the cyanobacterial CrtL [55]. Since cyanobacteria do not contain any ε-LCY activity, it is presumed that this enzyme evolved by gene duplication of a β-LCY in prochlorophytes, where carotenoids with an ε-end group appear. In contrast with higher plants, Prochlorococcus marinus can synthesize α-carotene from lycopene by a single enzyme [55]. Thus, the P. marinus ε-LCY can be regarded as a premature form of the plant ε-LCY, which has not yet lost its β-cyclase activity [55].
Hydroxylases
Xanthophylls are oxidation products of α- and β-carotene in higher plants [10]. Hydroxylation of the number three carbon of each ring (C-3 and C-3ʹ) of α-carotene and β-carotene results in the formation of lutein and zeaxanthin via α-cryptoxanthin and β-cryptoxanthin, respectively (Fig. 2.1b). These reactions are carried out by two types of enzymes, one specific for ε-rings (ε-hydroxylase, ε-CHY, EC 1.14.99.45) and the other for β-rings (β-CHY, EC 1.14.13.129) [22].
Two genes encoding β-CHY (CHB1, CHB2) were first cloned from Arabidopsis [56] and pepper [57]. The deduced amino acid sequences of β-CHY indicate the presence of transmembrane helices, suggesting that they are integral membrane enzymes. Pepper β-CHY displays histidine motifs, HXXXXH and HXXHH (Fig. 2.2) that play a vital role in coordinating the Fe ions in the active site [58]. Thus, β-CHY is a nonheme-di-iron monooxygenase that uses activated molecular oxygen to break the C–H bond, with the concomitant formation of unsaturated bonds and the retention of hydroxyl groups [57].
Although the β-ring of α-carotene is hydroxylated by a nonheme-di-iron monooxygenase, β-CHY, the ε-ring is not. Molecular analysis of the Arabidopsis mutant lut1, which shows up to a 95 % decrease in lutein content, led to the identification of a cytochrome P450-type oxygenase (CYP), CYP97C1, involved in the hydroxylation of the ε-ring of lutein [59]. Thus, the introduction of hydroxy group into the β- and ε-rings of α-carotene, to form lutein, is catalyzed by structurally unrelated enzymes such as nonheme-di-iron monooxygenases and cytochrome P450-type oxygenases.
Epoxidases and De-epoxidases
Zeaxanthin is converted into antheraxanthin and violaxanthin by the introduction of, respectively, one or two 5,6-epoxygroups into the 3-hydroxy-β-rings (Fig. 2.1b). This reaction is catalyzed by zeaxanthin epoxidase (ZEP, EC 1.14.13.90) [10]. The resulting antheraxanthin and violaxanthin are subjected to de-epoxidation by violaxanthin de-epoxidase (VDE, EC 1.10.99.3), which converts violaxanthin and antheraxanthin back to zeaxanthin [10]. These reactions of interconversion are known as the xanthophyll cycle, which has a key role in protecting the photosynthetic membranes against excess light by dissipating light energy as heat [19]. Zeaxanthin is converted into antheraxanthin and violaxanthin under light conditions that do not saturate photosynthesis or in the darkness [60]. Under light conditions that exceed the leaf photosynthetic capacity, violaxanthin is converted back to zeaxanthin [60].
ZEP and VDE are members of a group of proteins known as lipocalins, which bind and transport small hydrophobic molecules [61]. Lipocalins show a conserved secondary structure containing eight antiparallel β-sheets, which encompass three major motifs: motif I, first of the eight β-sheets, preceded by a stretch of α-helice; motif II, segments of sixth and seventh β-sheets, along with a loop between the sheets; and motif III, part of eighth β-sheet along with a carboxy terminal α-helice fragment and a loop between these two structures [61]. ZEP differs from VDE on the number of amino acid between the motifs I and II [62].
In addition, ZEP and VDE also contain a cystein-rich amino terminal region and a glutamate-rich carboxy terminal region (Fig. 2.2), probably involved in the formation of α-helices [62]. ZEP also contains a dinucleotide (FAD/NADP)-binding motif, like those in the carotenoid desaturases and cyclases. ZEP activity is controlled by the availability of β,β-xanthophyll precursors and requires the presence of FAD and ferredoxin [19]. VDE activity depends on the availability of violaxanthin, acidic pH of the lumen and protonated ascorbate [19]. During the light period, VDE binds to thylakoid membranes due to the acidification of lumen. In the dark, pH is neutral and VDE is released from thylakoid membranes.
VDE seems to be encoded by single copy genes in plants, while small gene families encode ZEP in plants such as maize, rice, and Vitis [26, 63, 64].
Neoxanthin Synthase
The conversion of violaxanthin into neoxanthin, by the rearrangement of a single epoxy group to 5-hydroxy, is the final step in the β,β-branch of carotenoid biosynthetic pathway (Fig. 2.1b). This reaction is catalyzed by NSY [10]. Neoxanthin can be cleaved to form a C15 precursor of ABA [4].
NSY has been characterized in potato and tomato [52, 54]. Interestingly, the amino acid sequence of NSY is homolog to CYC-B, the chromoplast-targeted isoform of β-LCY in tomato [47], and to CCS [43], which converts antheraxanthin and violaxanthin to capsanthin and capsorubin, respectively, in pepper. However, in contrast to β-LCY, which carries out the cyclization of end groups in the linear carotenoid lycopene, NSY acts by means of epoxidation, carbocation neutralization, and intramolecular rearrangement of the violaxanthin precursor [19]. NSY activity is similar to that of CCS, which also catalyzes an epoxidation reaction in the 5,6-epoxy cyclohexenyl end groups [19].
NSY is encoded by a single-copy gene in potato and tomato [52, 54]. Surprisingly, no NSY homologous gene has been found in the Arabidopsis genome yet. Its extensive homology with CYC-B has led to speculation that the NSY activity may be played by bifunctional β-LCYs, capable of converting both lycopene to β-carotene and violaxanthin to neoxanthin [65].
Regulation of Carotenoid Biosynthesis in Higher Plants
Since carotenoids play a central role in plant development and adaptation, their synthesis must be coordinated with the developmental processes, such as plastid differentiation, flowering and fruit development, and related metabolic pathways . Several mechanisms are used to regulate the carotenoid biosynthesis in higher plants, defining the types and amounts of the diverse carotenoids that will be synthesized in the different tissues [66]. Transcriptional regulation of carotenoid biosynthesis genes appears to be the major regulatory mechanism controlling carotenoid accumulation in the different plant tissues. However, other regulatory mechanisms have been also observed to control carotenoid biosynthesis, including posttranscriptional regulation, natural genetic variation and carotenoid degradation, turnover, and sequestration.
A key regulatory mechanism during the formation of isoprenoids is the control of precursor partitioning into the branches of the pathway. This has led to the concept of metabolic channeling among each branch of the pathway [10]. Considering the precursor supply as a limiting factor for carotenoid biosynthesis in plants, the enzymatic steps of MEP pathway involved in the synthesis of IPP and DMAPP would be the most restrictive ones to the flux into the pathway. Consistent with the idea of rate-limiting steps in the MEP pathway, changes in levels of DXS messenger RNA (mRNA) expression result in alterations in the contents of several isoprenoids, such as chlorophylls, tocopherols, carotenoids, and ABA [66]. For instance, expression analysis of DXS showed a developmental regulation during tomato fruit ripening, which correlated with an increase in PSY mRNA transcripts and carotenoid accumulation [67]. Furthermore, transgenic Arabidopsis seedlings overexpressing DXS and DXR showed increased levels of several isoprenoids , while antisense silencing of DXS significantly reduced isoprenoid formation [68, 69] .
PSY catalyzes the most important rate-limiting step in the carotenoid biosynthetic pathway. PSY genes respond transcriptionally to ABA, high light, photoperiod, development cues, abiotic stresses, and posttranscriptional feedback regulation [66]. Some plant species possess more than one PSY, which show tissue-specific expression and different responses to environmental stimuli. For example, the mRNA expression of a flower- and fruit-specific PSY (PSY-1) significantly increases during tomato fruit ripening [45], contributing to the accumulation of lycopene. The induction of PSY3 transcription in maize and rice under salt and drought stresses correlates with increased carotenoid flux and ABA synthesis in their roots [28, 29]. Allelic variation in PSY has been also described as a mechanism that may change PSY enzymatic activity. Alternative splicing of PSY-A1 allele was considered to be a major quantitative trait locus (QTL) determinant of flour color in bread wheat [70]. The accumulation of phytoene has been demonstrated to involve negative feedback regulation of upstream genes, including PSY, as well as downstream genes such as ZDS and β-LCY, which were all downregulated in the pds3 mutant [71] .
CrtISO has recently emerged as a regulatory node in the carotenoid biosynthetic pathway. CRTISO mutants, such as ccr2 and tangerine, accumulates cis-carotenes like 7,7ʹ9,9ʹ-tetra-cis-lycopene in etioplasts (dark-grown plastids) of seedlings and chromoplasts of fruit [39, 40]. In chloroplasts of the mutants, the biosynthetic pathway proceeds via photoisomerization, but there is delayed greening and substantial reduction in lutein in Arabidopsis and varying degrees of chlorosis in tomato and rice [39, 40, 72] .
The branching point in the carotenoid pathway proceeding after lycopene has a major regulatory role in modulating the partitioning of β,β- and β,ε-ring carotenoids. Flux through the two branches can be controlled by ε-LCY and β-LCY [66]. The massive accumulation of lycopene during tomato fruit development is a result of developmentally controlled downregulation of mRNA expression of β-LCY and ε-LCY, besides the upregulation of PSY [45]. A similar mechanism of carotenoid gene expression regulation during fruit development has been found in other plant species. In citrus, such as oranges, lemons, mandarins, and grapefruit, there is a decrease in the lutein content after the green stage of fruit development and color appears in various tones, from light yellow to deep orange, due to the massive accumulation of β,β-xanthophylls in the flavedo and juice sacs [73, 74]. During this process, the mRNA levels of PSY, PDS, ZDS, β-LCY2, and β-CHY increase while that of ε-LCY disappears [49, 50, 73–75]. Natural genetic variation at the ε-LCY locus in maize was shown to be another mechanism regulating the flux down β,ε- or β,β-branch of the carotenoid pathway [76] . Four natural ε-LCY polymorphisms explained 58 % of the variation in lutein and β,β-carotenoids [76].
Several studies have demonstrated that a pool of carotenoids is also regulated, in part, by the rate of degradation by carotenoid cleavage dioxygenases (CCDs) [66]. Members of this family not only synthesize important apocarotenoid molecules, such as the phytohormones ABA and strigolactone, but also aid in the maintenance of suitable carotenoid levels in different plant tissues. In Arabidopsis, the high carotenoid levels observed in maturing seeds have been attributed to a loss of CCD activity [77]. In strawberry , there was a correlation between the increase in FaCCD1 expression levels during ripening and a decrease in lutein content [78]. Petal color in chrysanthemum can also be regulated by CCD activity. Elevated transcript levels of CmCCD4a were observed in white petals, which correlated with a breakdown of the yellow carotenoid pigments [79] .
Plastid biogenesis is another important regulatory mechanism providing a sink for carotenoid accumulation. The high-pigment-2 (hp-2) and hp-3 tomato mutants contain higher fruit pigmentation caused by an enlargement of the plastid compartment size, enabling greater pigment biosynthesis and storage capacity for carotenoids in mature fruits [80, 81]. The cauliflower Orange (Or) mutant accumulates a high level of β-carotene in various normally white tissues of the plant, turning them orange, since the Or gene triggers the differentiation of proplastids and other colorless plastids into chromoplasts, creating a metabolic sink for carotenoid accumulation [82] .
The sequestration of carotenoids within the various plastid types is another important form of regulation. Chloroplasts and chromoplasts appear to differ considerably in the way they sequestrate end-product carotenoids. In chloroplasts, end-product carotenoids are associated with light-harvesting complexes [10]. The unbound carotenoids are likely to be associated with specific proteins as small plastoglobuli. For instance, end-product carotenoids are typically esterified and associated with the fibrillin protein as fibrils in chromoplasts of pepper [83]. Tomato chromoplasts appear to sequestrate lycopene as crystals [84]. The plastid ultrastructure may be altered in response to changes in carotenoid content [84, 85]. Esterification of carotenoids appears to be an effective mechanism used by flowers of sunflower, daffodil, and marigold for a high-level carotenoid accumulation [10] .
Transcriptional Co-regulation of Carotenoid Biosynthesis Genes
Transcriptional co-regulation has shown to play a central role in coordinating the cellular responses, which involve multiple genes and their products. Some studies have shown that many genes encoding metabolic enzymes that function within the same or functionally related pathways form co-expression modules [86, 87] . Thus, it is believed that the synthesis of functionally related isoprenoid molecules may be mediated by their transcriptional co-regulation. However, it is important to emphasize that changes in gene transcription do not necessarily reflect in altered protein abundance and functional activity due to the existence of posttranscriptional regulatory mechanisms of gene expression . Nevertheless, as the transcriptional regulation is the first level of protein synthesis regulation, changes in gene transcription in response to specific stimuli can be considered a primary regulatory response that reflects a change in requirement for specific proteins at a specific time period.
An expression correlation analysis using PSY and β-LCY as driver genes was carried out in order to determine the level of co-expression that they share with all of the other genes represented on the Arabidopsis co-expression tool (ACT) (http://web.archive.org/20051222113921/www.arabidopsis.leeds.ac.uk/act/coexpanalyser.php). PSY and β-LCY were chosen as driver genes for this analysis as their transcriptions are known to be positively correlated with carotenoid accumulation, as discussed above. It is believed that genes that are highly co-expressed with PSY and β-LCY will have closely associated functional roles. A co-correlation scatterplot between PSY and β-LCY reveals that both genes have a high level of co-expression with other genes related to photosynthesis , including those encoding the photosystem II protein PsbP and chlorophyll a/b-binding proteins. The scatterplot also shows that expression of PSY and β-LCY is highly correlated with the expression of many genes involved in MEP and carotenoid biosynthetic pathways, with Pearson correlation coefficient (r-value) ranging from 0.52 to 0.87 (Fig. 2.5). These findings provide additional evidence that the coordinated transcriptional regulation of these biosynthesis genes is critical in regulating and coordinating the synthesis of functionally related carotenoids .
Co-correlation scatter plot representing the level of co-expression of all Arabidopsis genes (blue squares) relative to PSY (x-axis) and β-LCY (LCY-B) (y-axis). Carotenogenesis-related genes are highlighted in red. Data were obtained using the Arabidopsis coexpression tool (ACT) (http://web.archive.org/20051222113921/www.arabidopsis.leeds.ac.uk/act/coexpanalyser.php). See text and figure legends for abbreviations. PSY phytoene synthase, LCY lycopene cyclase
Concluding Comments
The carotenoid biosynthetic pathway is a wonderful example of successful interdisciplinary approach applied to clarify the fundamental reaction steps of the pathway and their associated genes and enzymes. The biochemical reaction sequences involved in the biosynthesis of carotenoids are now known, as are all the encoding genes. A significant progress has also been made in understanding the regulation of carotenoid biosynthesis accumulation in higher plants. Although most of the carotenoid-related research has focused on model species such as Arabidopsis, tomato, maize and rice, other exciting reports have emerged from vegetables and fruits such as orange. These studies along with more in-depth research on the field of genomics will strengthen knowledge about the carotenoid pathway and its regulation in the future. The recent progress in the technological capacities in genomics, including the emergence of next-generation sequencing technologies, can lead to more fascinating discoveries.
References
Britton G (1995) Structure and properties of carotenoids in relation to function. Faseb J 9(15):1551–1558
Goodwin TW (1986) Metabolism, nutrition, and function of carotenoids. Annu Rev Nutr 6:273–297
Li Z, Wakao S, Fischer BB, Niyogi KK (2009) Sensing and responding to excess light. Annu Rev Plant Biol 60:239–260
Nambara E, Marion-Poll A (2005) Abscisic acid biosynthesis and catabolism. Ann Rev of Plant Biol 56:165–185
Umehara M, Hanada A, Yoshida S, Akiyama K, Arite T, Takeda-Kamiya N et al (2008) Inhibition of shoot branching by new terpenoid plant hormones. Nature 455(7210):195–200
Vishnevetsky M, Ovadis M, Vainstein A (1999) Carotenoid sequestration in plants: the role of carotenoid-associated proteins. Trends Plant Sci 4(6):232–235
Toomey MB, McGraw KJ (2012) Mate choice for a male carotenoid-based ornament is linked to female dietary carotenoid intake and accumulation. BMC Evol Biol 12:3
Butler MW, McGraw KJ (2012) Differential effects of early- and late-life access to carotenoids on adult immune function and ornamentation in mallard ducks (Anas platyrhynchos). PLoS One 7(5):e38043
DellaPenna D, Pogson BJ (2006) Vitamin synthesis in plants: tocopherols and carotenoids. Annu Rev Plant Biol 57:711–738
Fraser PD, Bramley PM (2004) The biosynthesis and nutritional uses of carotenoids. Prog Lipid Res 43(3):228–265
Krinsky NI, Johnson EJ (2005) Carotenoid actions and their relation to health and disease. Mol Aspects Med 26(6):459–516
Cunningham FX, Gantt E (1998) Genes and enzymes of carotenoid biosynthesis in plants. Annu Rev Plant Phys 49:557–583
Ruiz-Sola MA, Rodriguez-Concepcion M (2012) Carotenoid biosynthesis in Arabidopsis: a colorful pathway. Arabidopsis Book 10:e0158
Cunningham FX Jr, Gantt E (2007) A portfolio of plasmids for identification and analysis of carotenoid pathway enzymes: Adonis aestivalis as a case study. Photosynth Res 92(2):245–259
Rodriguez-Concepcion M (2010) Supply of precursors for carotenoid biosynthesis in plants. Arch Biochem Biophys 504(1):118–122
McGarvey DJ, Croteau R (1995) Terpenoid metabolism. Plant Cell 7(7):1015–1026
Eisenreich W, Rohdich F, Bacher A (2001) Deoxyxylulose phosphate pathway to terpenoids. Trends Plant Sci 6(2):78–84
Kuzuyama T, Seto H (2012) Two distinct pathways for essential metabolic precursors for isoprenoid biosynthesis. Proc Jpn Acad Ser B Phys Biol Sci 88(3):41–52
Bouvier F, Rahier A, Camara B (2005) Biogenesis, molecular regulation and function of plant isoprenoids. Prog Lipid Res 44(6):357–429
Kirby J, Keasling JD (2009) Biosynthesis of plant isoprenoids: perspectives for microbial engineering. Annu Rev Plant Biol 60:335–355
Hunter WN (2007) The non-mevalonate pathway of isoprenoid precursor biosynthesis. J Biol Chem 282(30):21573–21577
Hirschberg J (2001) Carotenoid biosynthesis in flowering plants. Curr Opin Plant Biol 4(3):210–218
Fu Z, Yan J, Zheng Y, Warburton ML, Crouch JH, Li JS (2010) Nucleotide diversity and molecular evolution of the PSY1 gene in Zea mays compared to some other grass species. Theor Appl Genet 120(4):709–720
Bartley GE, Scolnik PA (1993) cDNA cloning, expression during development, and genome mapping of PSY2, a second tomato gene encoding phytoene synthase. J Biol Chem 268(34):25718–25721
Fraser PD, Kiano JW, Truesdale MR, Schuch W, Bramley PM (1999) Phytoene synthase-2 enzyme activity in tomato does not contribute to carotenoid synthesis in ripening fruit. Plant Mol Biol 40(4):687–698
Lange BM, Ghassemian M (2003) Genome organization in Arabidopsis thaliana: a survey for genes involved in isoprenoid and chlorophyll metabolism. Plant Mol Biol 51(6):925–948
Giorio G, Stigliani AL, D’Ambrosio C (2008) Phytoene synthase genes in tomato (Solanum lycopersicum L.)—new data on the structures, the deduced amino acid sequences and the expression patterns. FEBS J 275(3):527–535
Li FQ, Vallabhaneni R, Wurtzel ET (2008) PSY3, a new member of the phytoene synthase gene family conserved in the Poaceae and regulator of abiotic stress-induced root carotenogenesis. Plant Physiol 146(3):1333–1345
Welsch R, Wust F, Bar C, Al-Babili S, Beyer P (2008) A third phytoene synthase is devoted to abiotic stress-induced abscisic acid formation in rice and defines functional diversification of phytoene synthase genes. Plant Physiol 147(1):367–380
Li FQ, Vallabhaneni R, Yu J, Rocheford T, Wurtzel ET (2008) The maize phytoene synthase gene family: overlapping roles for carotenogenesis in endosperm, photomorphogenesis, and thermal stress tolerance. Plant Physiol 147(3):1334–1346
Lopez AB, Yang Y, Thannhauser TW, Li L (2008) Phytoene desaturase is present in a large protein complex in the plastid membrane. Physiol Plant 133(2):190–198
Cunningham FX Jr, Pogson B, Sun Z, McDonald KA, DellaPenna D, Gantt E (1996) Functional analysis of the beta and epsilon lycopene cyclase enzymes of Arabidopsis reveals a mechanism for control of cyclic carotenoid formation. Plant Cell 8(9):1613–1626
Al-Babili S, von Lintig J, Haubruck H, Beyer P (1996) A novel, soluble form of phytoene desaturase from Narcissus pseudonarcissus chromoplasts is Hsp70-complexed and competent for flavinylation, membrane association and enzymatic activation. Plant J 9(5):601–612
Bonk M, Hoffmann B, Von Lintig J, Schledz M, Al-Babili S, Hobeika E et al (1997) Chloroplast import of four carotenoid biosynthetic enzymes in vitro reveals differential fates prior to membrane binding and oligomeric assembly. Eur J Biochem 247(3):942–950
Carol P, Kuntz M (2001) A plastid terminal oxidase comes to light: implications for carotenoid biosynthesis and chlororespiration. Trends Plant Sci 6(1):31–36
Romer S, Hugueney P, Bouvier F, Camara B, Kuntz M (1993) Expression of the genes encoding the early carotenoid biosynthetic enzymes in capsicum annuum. Biochem Biophys Res Commun 196(3):1414–1421
Matthews PD, Luo R, Wurtzel ET (2003) Maize phytoene desaturase and zeta-carotene desaturase catalyse a poly-Z desaturation pathway: implications for genetic engineering of carotenoid content among cereal crops. J Exp Bot 54(391):2215–2230
Chen CX, Costa MGC, Yu QB, Moore GA, Gmitter FG (2010) Identification of novel members in sweet orange carotenoid biosynthesis gene families. Tree Genet Genomes 6(6):905–914
Isaacson T, Ronen G, Zamir D, Hirschberg J (2002) Cloning of tangerine from tomato reveals a carotenoid isomerase essential for the production of beta-carotene and xanthophylls in plants. Plant Cell 14(2):333–342
Park H, Kreunen SS, Cuttriss AJ, DellaPenna D, Pogson BJ (2002) Identification of the carotenoid isomerase provides insight into carotenoid biosynthesis, prolamellar body formation, and photomorphogenesis. Plant Cell 14(2):321–332
Cunningham FX Jr, Gantt E (2001) One ring or two? Determination of ring number in carotenoids by lycopene epsilon-cyclases. Proc Natl Acad Sci U S A 98(5):2905–2910
Bramley PM (2002) Regulation of carotenoid formation during tomato fruit ripening and development. J Exp Bot 53(377):2107–2113
Hugueney P, Badillo A, Chen HC, Klein A, Hirschberg J, Camara B et al (1995) Metabolism of cyclic carotenoids: a model for the alteration of this biosynthetic pathway in Capsicum annuum chromoplasts. Plant J 8(3):417–424
Hornero-Mendez D, Britton G (2002) Involvement of NADPH in the cyclisation reaction of carotenoid biosynthesis. FEBS Lett 515(1–3):133–136
Ronen G, Cohen M, Zamir D, Hirschberg J (1999) Regulation of carotenoid biosynthesis during tomato fruit development: expression of the gene for lycopene epsilon-cyclase is down-regulated during ripening and is elevated in the mutant delta. Plant J 17(4):341–351
Pecker I, Gabbay R, Cunningham FX Jr, Hirschberg J (1996) Cloning and characterization of the cDNA for lycopene beta-cyclase from tomato reveals decrease in its expression during fruit ripening. Plant Mol Biol 30(4):807–819
Ronen G, Carmel-Goren L, Zamir D, Hirschberg J (2000) An alternative pathway to beta-carotene formation in plant chromoplasts discovered by map-based cloning of beta and old-gold color mutations in tomato. Proc Natl Acad Sci U S A 97(20):11102–11107
Lewinsohn E, Sitrit Y, Bar E, Azulay Y, Meir A, Zamir D et al (2005) Carotenoid pigmentation affects the volatile composition of tomato and watermelon fruits, as revealed by comparative genetic analyses. J Agric Food Chem 53(8):3142–3148
Alquezar B, Zacarias L, Rodrigo MJ (2009) Molecular and functional characterization of a novel chromoplast-specific lycopene beta-cyclase from Citrus and its relation to lycopene accumulation. J Exp Bot 60(6):1783–1797
Mendes AF, Chen C, Gmitter FG Jr, Moore GA, Costa MG (2011) Expression and phylogenetic analysis of two new lycopene beta-cyclases from Citrus paradisi. Physiol Plant 141(1):1–10
Bouvier F, D’Harlingue A, Backhaus RA, Kumagai MH, Camara B (2000) Identification of neoxanthin synthase as a carotenoid cyclase paralog. Eur J Biochem 267(21):6346–6352
Al-Babili S, Hugueney P, Schledz M, Welsch R, Frohnmeyer H, Laule O et al (2000) Identification of a novel gene coding for neoxanthin synthase from Solanum tuberosum. FEBS Lett 485(2/3):168–172
Bouvier F, Hugueney P, d’Harlingue A, Kuntz M, Camara B (1994) Xanthophyll biosynthesis in chromoplasts: isolation and molecular cloning of an enzyme catalyzing the conversion of 5,6-epoxycarotenoid into ketocarotenoid. Plant J 6(1):45–54
Bouvier F, D’Harlingue A, Backhaus RA, Kumagai MH, Camara B (2000) Identification of neoxanthin synthase as a carotenoid cyclase paralog. Eur J Biochem 267(21):6346–6352
Sandmann G (2002) Molecular evolution of carotenoid biosynthesis from bacteria to plants. Physiol Plant 116(4):431–440
Sun Z, Gantt E, Cunningham FX Jr (1996) Cloning and functional analysis of the beta-carotene hydroxylase of Arabidopsis thaliana. J Biol Chem 271(40):24349–24352
Bouvier F, Keller Y, d’Harlingue A, Camara B (1998) Xanthophyll biosynthesis: molecular and functional characterization of carotenoid hydroxylases from pepper fruits (Capsicum annuum L.). Biochim Biophys Acta 1391(3):320–328
Shanklin J, Cahoon EB (1998) Desaturation and related modifications of fatty acids1. Annu Rev Plant Physiol Plant Mol Biol 49:611–641
Tian L, Musetti V, Kim J, Magallanes-Lundback M, DellaPenna D (2004) The Arabidopsis LUT1 locus encodes a member of the cytochrome p450 family that is required for carotenoid epsilon-ring hydroxylation activity. Proc Natl Acad Sci U S A 101(1):402–407
Muller P, Li XP, Niyogi KK (2001) Non-photochemical quenching. A response to excess light energy. Plant Physiol 125(4):1558–1566
Flower DR (1996) The lipocalin protein family: structure and function. Biochem J 318(Pt 1):1–14
Hieber AD, Bugos RC, Yamamoto HY (2000) Plant lipocalins: violaxanthin de-epoxidase and zeaxanthin epoxidase. Biochim Biophys Acta 1482(1/2):84–91
Vallabhaneni R, Wurtzel ET (2009) Timing and biosynthetic potential for carotenoid accumulation in genetically diverse germplasm of maize. Plant Physiol 150(2):562–572
Young PR, Lashbrooke JG, Alexandersson E, Jacobson D, Moser C, Velasco R et al (2012) The genes and enzymes of the carotenoid metabolic pathway in Vitis vinifera L. BMC Genomics 13(1):243
North HM, De Almeida A, Boutin JP, Frey A, To A, Botran L et al (2007) The Arabidopsis ABA-deficient mutant aba4 demonstrates that the major route for stress-induced ABA accumulation is via neoxanthin isomers. Plant J 50(5):810–824
Cazzonelli CI, Pogson BJ (2010) Source to sink: regulation of carotenoid biosynthesis in plants. Trends Plant Sci 15(5):266–274
Lois LM, Rodriguez-Concepcion M, Gallego F, Campos N, Boronat A (2000) Carotenoid biosynthesis during tomato fruit development: regulatory role of 1-deoxy-D-xylulose 5-phosphate synthase. Plant J 22(6):503–513
Estevez JM, Cantero A, Reindl A, Reichler S, Leon P (2001) 1-Deoxy-D-xylulose-5-phosphate synthase, a limiting enzyme for plastidic isoprenoid biosynthesis in plants. J Biol Chem 276(25):22901–22909
Carretero-Paulet L, Cairo A, Botella-Pavia P, Besumbes O, Campos N, Boronat A et al (2006) Enhanced flux through the methylerythritol 4-phosphate pathway in Arabidopsis plants overexpressing deoxyxylulose 5-phosphate reductoisomerase. Plant Mol Biol 62(4/5):683–695
Howitt CA, Cavanagh CR, Bowerman AF, Cazzonelli C, Rampling L, Mimica JL et al (2009) Alternative splicing, activation of cryptic exons and amino acid substitutions in carotenoid biosynthetic genes are associated with lutein accumulation in wheat endosperm. Funct Integr Genomics 9(3):363–376
Qin G, Gu H, Ma L, Peng Y, Deng XW, Chen Z et al (2007) Disruption of phytoene desaturase gene results in albino and dwarf phenotypes in Arabidopsis by impairing chlorophyll, carotenoid, and gibberellin biosynthesis. Cell Res 17(5):471–482
Fang J, Chai C, Qian Q, Li C, Tang J, Sun L et al (2008) Mutations of genes in synthesis of the carotenoid precursors of ABA lead to pre-harvest sprouting and photo-oxidation in rice. Plant J 54(2):177–189
Kato M (2004) Accumulation of carotenoids and expression of carotenoid biosynthetic genes during maturation in citrus fruit. Plant Physiol 134(2):824–837
Rodrigo MJ, Marcos JF, Zacarias L (2004) Biochemical and molecular analysis of carotenoid biosynthesis in flavedo of orange (Citrus sinensis L.) during fruit development and maturation. J Agric Food Chem 52(22):6724–6731
Costa MG, Moreira CD, Melton JR, Otoni WC, Moore GA (2012) Characterization and developmental expression of genes encoding the early carotenoid biosynthetic enzymes in Citrus paradisi macf. Mol Biol Rep 39(2):895–902
Harjes CE, Rocheford TR, Bai L, Brutnell TP, Kandianis CB, Sowinski SG et al (2008) Natural genetic variation in lycopene epsilon cyclase tapped for maize biofortification. Science 319(5861):330–333
Auldridge ME, Block A, Vogel JT, Dabney-Smith C, Mila I, Bouzayen M et al (2006) Characterization of three members of the Arabidopsis carotenoid cleavage dioxygenase family demonstrates the divergent roles of this multifunctional enzyme family. Plant J 45(6):982–993
Garcia-Limones C, Schnabele K, Blanco-Portales R, Luz Bellido M, Caballero JL, Schwab W et al (2008) Functional characterization of FaCCD1: a carotenoid cleavage dioxygenase from strawberry involved in lutein degradation during fruit ripening. J Agric Food Chem 56(19):9277–9285
Ohmiya A, Kishimoto S, Aida R, Yoshioka S, Sumitomo K (2006) Carotenoid cleavage dioxygenase (CmCCD4a) contributes to white color formation in chrysanthemum petals. Plant Physiol 142(3):1193–1201
Kolotilin I, Koltai H, Tadmor Y, Bar-Or C, Reuveni M, Meir A et al (2007) Transcriptional profiling of high pigment-2dg tomato mutant links early fruit plastid biogenesis with its overproduction of phytonutrients. Plant Physiol 145(2):389–401
Galpaz N, Wang Q, Menda N, Zamir D, Hirschberg J (2008) Abscisic acid deficiency in the tomato mutant high-pigment 3 leading to increased plastid number and higher fruit lycopene content. Plant J 53(5):717–730
Lu S, Van Eck J, Zhou X, Lopez AB, O’Halloran DM, Cosman KM et al (2006) The cauliflower or gene encodes a DnaJ cysteine-rich domain-containing protein that mediates high levels of beta-carotene accumulation. Plant Cell 18(12):3594–3605
Deruere J, Romer S, d’Harlingue A, Backhaus RA, Kuntz M, Camara B (1994) Fibril assembly and carotenoid overaccumulation in chromoplasts: a model for supramolecular lipoprotein structures. Plant Cell 6(1):119–133
Romer S, Fraser PD, Kiano JW, Shipton CA, Misawa N, Schuch W et al (2000) Elevation of the provitamin a content of transgenic tomato plants. Nat Biotechnol 18(6):666–669
Shewmaker CK, Sheehy JA, Daley M, Colburn S, Ke DY (1999) Seed-specific overexpression of phytoene synthase: increase in carotenoids and other metabolic effects. Plant J 20(4):401–412X
Ihmels J, Bergmann S, Barkai N (2004) Defining transcription modules using large-scale gene expression data. Bioinformatics 20(13):1993–2003
Wei H, Persson S, Mehta T, Srinivasasainagendra V, Chen L, Page GP et al (2006) Transcriptional coordination of the metabolic network in Arabidopsis. Plant Physiol 142(2):762–774
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2015 Springer Science+Business Media New York
About this chapter
Cite this chapter
Mendes, A., Soares, V., Costa, M. (2015). Carotenoid Biosynthesis Genomics. In: Chen, C. (eds) Pigments in Fruits and Vegetables. Springer, New York, NY. https://doi.org/10.1007/978-1-4939-2356-4_2
Download citation
DOI: https://doi.org/10.1007/978-1-4939-2356-4_2
Published:
Publisher Name: Springer, New York, NY
Print ISBN: 978-1-4939-2355-7
Online ISBN: 978-1-4939-2356-4
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)