Abstract
The peptidylarginine deiminase (PAD) enzymes hydrolyze arginine residues to create the nonnative amino acid citrulline (a process called citrullination or deimination). Five PAD enzymes (1, 2, 3, 4, 6) have been identified, and they exhibit fairly restricted tissue expression patterns. The conversion of arginine to citrulline (referred to as citrullination or deimination) results in only a small change in molecular mass (less than 1 Da) and a loss in a positive charge, which can have dramatic consequences on protein structure and protein–protein interactions. Since citrullination can lead to profound changes in protein function, it is not surprising that citrullination and the activity of the PAD enzymes has been implicated in many diseases, such as rheumatoid arthritis (RA), multiple sclerosis, Alzheimer’s disease, inflammatory bowel disease, psoriasis, and cancer. This chapter provides a general overview of the PAD enzymes and their known functions. Plasma and synovial biopsy specimen from patients with rheumatoid arthritis contain high levels of citrullinated proteins, and, in fact, rheumatoid patients often develop immune reactivity to citrullinated proteins. Here, we also discuss possible physiological pathways that may contribute to the generation of citrullinated self-antigens, which could then prime the development of anti-citrulline peptide autoantibodies in rheumatoid arthritis.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
1.1 The Peptidylarginine Deiminase Family
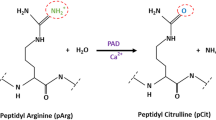
The free amino acid form of citrulline was first isolated from watermelon (Citrullus vulgaris) over 70 years ago (Curis et al. 2005), while the peptidyl form of citrulline was first recognized within the hair follicle (Rogers 1962). Peptidylcitrulline is a noncoding amino acid that is generated through hydrolysis of peptidyl-arginine residues by Ca2+-dependent peptidylarginine deiminase (PAD) enzymes, with ammonia released as a reaction by-product (Fig. 1.1). This process is referred to as deimination or citrullination. The conversion of arginine to citrulline results in only a small increase in molecular mass (less than 1 Da) but also converts the positively charged guanidino group on an arginine residue into the neutrally charged ureido group on the citrulline amino acid. The small mass difference between arginine and citrulline residues has made identifying sites of deimination challenging, especially on proteins isolated from cellular sources (Hao et al. 2009).
Although an approximate 1 Da change in mass may seem like a relatively minor difference, the conversion of charge from an arginine to a citrulline can have dramatic consequences on protein structure, proteolytic susceptibility, and protein–protein interactions (Vossenaar et al. 2003). For example, filaggrin is a highly basic epidermal protein, essential for barrier function, and is first synthesized as a larger pro-filaggrin protein (Smith et al. 2006; Palmer et al. 2006). Deimination of filaggrin by skin-resident PAD proteins facilitates its proteolytic processing into free amino acids and other derivatives that make up “natural moisturizing factor,” a mixture of natural hygroscopic agents that maintain epidermal hydration (Chavanas et al. 2006; Kamata et al. 2009). Additionally, citrullination could alter substrate receptivity to other posttranslational modifications, since arginine residues can be found within many enzyme consensus motifs (Papin et al. 2005). For instance, conventional protein kinase C enzymes can phosphorylate substrates containing serine or threonine with an arginine residue at the −3, −2, and +2 positions (Nishikawa et al. 1997). Thus, one could imagine that conversion of a consensus site arginine into a citrulline could impact a protein’s posttranslational modification landscape. Not surprisingly, conversion of arginine residues to citrulline prevents methylation by members of the protein arginine methyltransferase (PRMT) family (Cuthbert et al. 2004; Wang et al. 2004). Finally, DNA-binding domains are often rich in positively charged arginine residues (Crane-Robinson et al. 2006), and therefore, citrullination could also regulate the association of transcription factors with their DNA response elements.
Since citrullination can lead to profound changes in protein function, it is not surprising that citrullination and the PAD enzymes have been implicated in numerous diseases. PAD enzymes and citrullination have been associated with rheumatoid arthritis (RA), multiple sclerosis, Alzheimer’s disease, inflammatory bowel disease, psoriasis, and cancer (Gyorgy et al. 2006; Chumanevich et al. 2011). This chapter provides a general overview of the PAD enzymes and their known functions. In addition, we discuss possible physiological pathways that may contribute to the generation of citrullinated self-antigens, which could then prime the development of anti-citrulline peptide autoantibodies (ACPA) in RA.
1.1.1 Introduction to the PAD Family
The enzymatic activity capable of generating peptidyl-citrulline was first identified in hair follicle extracts by Rogers and Taylor (Rogers and Taylor 1977). Since this discovery, five PAD enzymes (1,2,3,4/5,6), which are encoded by the PADI loci, have been identified in mammals, and they exhibit fairly high amino acid sequence homology (~41–55 %) (Fig. 1.2) (Chavanas et al. 2004). Mammalian PADI genes are co-localized within a single gene cluster, spanning a region of about 355 kb in length, on chromosome 1 in humans and on chromosome 4 in mice (Zhang et al. 2004). PAD5 was identified in mice and was later revealed to be the orthologue of human PAD4 (Vossenaar et al. 2003). Paralogues of the PAD enzymes appear in birds, amphibians, and bony fish. Phylogenetic analysis suggests that the ancestor of the PADI locus appeared in the last common ancestor shared by teleosteans and mammals (Balandraud et al. 2005). The genome of the bacterium Porphyromonas gingivalis, a pathogen associated with periodontitis, also encodes a PAD enzyme, although the P. gingivalis PAD or PPAD appears to be evolutionarily unrelated to eukaryotic PAD enzymes (Shirai et al. 2001).
Sequence alignment of the human PAD isozymes. Multiple sequence alignment of the human PAD amino acid sequences was created using ClustalW software (Thompson et al. 1994), and Strap (www.bioinformatics.org/strap) was used for the alignment layout. Calcium-binding residues and catalytic residues identified in the PAD4 crystal structure are highlighted in yellow and blue, respectively (Arita et al. 2004). The PAD4 nuclear localization sequence in underlined and in bold type
1.1.2 Regulation of PAD Activity
Members of the PAD family require high Ca2+ concentrations for their activity (Gyorgy et al. 2006). Not surprisingly then, treatment of cells with the Ca2+ ionophores, such as ionomycin, can induce the production of peptidyl-citrulline (Vossenaar et al. 2004; Nakashima et al. 2002). The divalent calcium cation requirement is specific, because other metal ions were unable to substitute for Ca2+ in an in vitro PAD activity assay where deimination of the arginine derivative N-α-benozyl-l-arginine ethyl ester (BAEE) was monitored by colorimetric change (Kearney et al. 2005). However, relatively little is known about physiological stimuli that induce this calcium-dependent PAD activity. Structural analysis of the PAD4 enzyme revealed five Ca2+-binding sites that are fairly conserved amongst all PADs except PAD6 (Fig. 1.2) (Mechin et al. 2007; Arita et al. 2004). Binding of Ca2+ leads to a conformation change, moving the key catalytic C645 residue to the enzyme active site, where it is thought to exert a nucleophilic attack on the guanidium carbon atom of the target arginine (Arita et al. 2004). The thiolate anion C645 is essential for PAD4 activity because mutation of this residue is sufficient to abolish enzymatic activity (Knuckley et al. 2007). Notably, the haloacetamidine-bearing PAD inhibitors F- and Cl-amidine covalently bind to C645 and act as irreversible PAD inhibitors (Knuckley et al. 2007). Using in vitro studies, the optimal Ca2+ concentration for PAD2 and PAD4 is within the high micromolar to millimolar range (Kearney et al. 2005; Nakayama-Hamada et al. 2005). This level is far higher than the micromolar levels achieved following the opening of calcium release-activated channels (CRAC) after T cell receptor ligation, even higher than the sub-micromolar homeostatic intracellular Ca2+ levels (Feske 2007). Since PAD proteins require high micromolar levels of Ca2+, one proposal is that cellular apoptosis exposes PAD proteins to high levels of extracellular Ca2+, which promotes PAD activity. However, apoptosis alone is not sufficient to induce PAD activity, as the apoptosis-inducing agents camptothecin and staurosporine do not induce peptidyl-citrulline generation (Neeli et al. 2009). Perhaps, the PAD enzymes localize to a microenvironment within cells that has access to very high Ca2+ levels. Another possibility is that posttranslational modification of PAD enzymes or the association of the PAD enzymes with accessory proteins could lower the Ca2+ threshold requirement for enzymatic activity.
The expression pattern of mammalian PAD family members is fairly tissue restricted, suggesting that PAD expression is tightly controlled (see below and Fig. 1.3) (Vossenaar et al. 2003). However, a few studies have addressed the transcriptional regulation of the PADI genes. For example, PAD1 and PAD3 are both expressed in human skin and hair follicles (Mechin et al. 2007). PAD1 expression in normal human epidermal keratinocytes is regulated by Sp1, a transcription factor known to regulate many genes, and by MZF1, a transcription factor with enriched expression in differentiated keratinocytes (Dong et al. 2008; Kamata et al. 2011). The transcription factors Sp1, Sp3, and NY-1 bind to the PADI3 promoter and regulate its expression (Dong et al. 2006). PAD2 is expressed in many tissues and organs and is the most widely expressed mammalian PAD member (Vossenaar et al. 2003). In human keratinocytes, expression of the PADI2 gene is controlled by a minimal GC-rich promoter, which is occupied by the ubiquitous transcription factors Sp1 and Sp3 (Dong et al. 2005). PAD4 expression is largely restricted to immune cells, especially granulocytes, but has also been found in some tumor lines (Jones et al. 2009). Treatment of the MCF-7 breast cancer cell line with estrogen leads to increased levels of the PAD4 protein (Cuthbert et al. 2004). Furthermore, estrogen-induced expression of PAD4 is regulated by cooperative binding of the AP-1, Sp-1, Sp-3, and NF-Y transcription factors to the PAD4 minimal promoter (Dong et al. 2007). The addition of granulocyte-inducing differentiation agents of dimethyl sulfoxide, retinoic acid, and vitamin D3 leads to the appearance of peptidyl-citrulline after several days in culture, but little is known about the factors that induce PAD4 expression in immune cells (Nakashima et al. 1999, 2002). PAD6 transcripts are primarily found in oocytes and embryos (Horibata et al. 2012), and the oocyte-specific, homeobox-containing transcription factor Nobox is critical for expression of PAD6 (Choi et al. 2010). One caveat to studies that focus on the expression of PADI transcripts is that, in some cell types, a disassociation has been occasionally observed between a particular PAD transcript and its protein levels (Vossenaar et al. 2004; Mechin et al. 2010). For example, PAD2 message was found in monocytes and macrophages, but the PAD2 mRNA appeared to be translated only in macrophages (Vossenaar et al. 2004). Conversely, the PAD4 protein was found in macrophages, but PAD4 mRNA was not expressed in detectable levels in these cells (Vossenaar et al. 2004).
Many PAD substrates have been described, including keratins, filaggrin, vimentin, myelin basic protein, fibrinogen, chemokines (CXCL8, CXCL10, CXCL11, CXCL12), p300, and histones (Jones et al. 2009). PAD1, PAD2, PAD3, and PAD4 can also auto-deiminate, which impairs their activity, at least when detected by in vitro assays (Mechin et al. 2010; Andrade et al. 2010). However, only a handful of studies have addressed PAD-mediated substrate recognition. Although PAD family members display a great degree of sequence similarity, direct comparison of PAD2, PAD3, and PAD4 enzymes using HL-60 lysates revealed that each enzyme has distinct substrate preferences (Darrah et al. 2012). Only PAD4 was able to citrullinate histone H3, while PAD2 was able to citrullinate β/γ actin (Darrah et al. 2012). Analysis of sites of PAD2 citrullination on β actin did not reveal a strict consensus sequence for citrullination, but arginines flanked by proline residues were not favored (Darrah et al. 2012). Indeed, the amino acids immediately flanking the arginine appear to be critical in determining whether an arginine can be citrullinated. Sequences containing proline residues adjacent to the target arginine are also unfavored for citrullination by PAD4 (Stensland et al. 2009). Interestingly, systematic examination of fillagrin- and histone H3-derived peptides to reveal favorable and unfavorable amino acid substitutions surrounding the PAD4 target arginine residue yielded a list of some overlapping, but also many non-overlapping, amino acids (Stensland et al. 2009). Co-crystallization of PAD4 with histone H3-1, H3-2, and H4 peptides demonstrated that PAD4 preferred sequences with a highly disordered conformation because the binding of PAD4 to these histone peptides induces a β-turn-like conformation (Arita et al. 2004). Overall, PAD4 has a broad sequence specificity, with a proposed consensus sequence of ϕXRXX, where ϕ denotes amino acids with a small side chain and X denotes any amino acid (Arita et al. 2004). Further studies to define the PAD consensus sites will be useful to identify new PAD substrates and to understand the physiological functions of the PAD enzymes.
1.1.3 Cross Talk Between PADs and Protein Arginine Methyltransferases
Arginine residues can also be subject to posttranslational modification by members of the PRMT family. PRMTs catalyze the addition of a methyl group from S-adenosylmethionine to guanidino nitrogen atoms on arginine residues (Bedford 2007). Three types of PRMTs have been subclassified based on the symmetry of their reaction products. Type I PRMTs (1,3,4—also known as Carm1—6,8) catalyze asymmetric methylation of arginine residues, and type II PRMT5 catalyzes symmetric transfer of methyl groups to arginine residues (Krause et al. 2007). Both type I and type II PRMTs catalyze mono-methylation as a reaction intermediate. The type III PRMT7 catalyzes arginine mono-methylation as its end product (Zurita-Lopez et al. 2012). Modification of arginine residues by both PRMT and PAD family members suggests an intimate regulatory relationship between the two families. Interestingly, PRMT1 has been identified as a possible PAD4 substrate using protein arrays (Guo et al. 2011). Arginine methylation had long been thought to be a permanent posttranslational modification, since no demethylase had been identified. Two groups independently determined that, in addition to arginine, PAD4 could also target mono-methylated arginines on histone proteins from cellular sources. Arginine dimethylation appeared to be a more stable modification, as it is protective against PAD activity (Cuthbert et al. 2004). However, PAD4 acting as a demethylase has been called into question because chemically synthesized mono-methylated histone peptides containing potential target arginines are very poor substrates for PAD4 using in vitro assays (Kearney et al. 2005; Hidaka et al. 2005). Perhaps, the conversion of mono-methyl arginine to citrulline by PAD enzymes is context dependent, requiring associated proteins or additional posttranslational modifications to occur efficiently.
1.2 General Overview of the PAD Family Members
1.2.1 PAD1
PAD1 is cytoplasmic and expressed in all living layers of the epidermis (see Chap. 7), with graded expression increasing in intensity from the basal layer to the granular layer (Nachat et al. 2005a). In hair follicles (see Chap. 8), PAD1 is expressed in the cuticle and in the inner root sheath (Nachat et al. 2005b). Due to its localization, PAD1 is thought to be the primary enzyme responsible for filaggrin and keratin K1 citrullination in the skin epidermis (Senshu et al. 1996). The loss of charge due to citrullination leads to disassembly of the cytokeratin–filaggrin complex and facilitates break down of filaggrin to form “natural moisturizing factor” and maintain epidermal hydration (Chavanas et al. 2006; Kamata et al. 2009). PAD1 is linked to skin disease, because keratin K1 exhibits decreased citrullination levels in the epidermis of psoriasis patients (Ishida-Yamamoto et al. 2000).
1.2.2 PAD2
Of the five PAD enzymes, PAD2 is the most broadly expressed isoform (Vossenaar et al. 2003). PAD2 is also known as “skeletal muscle PAD,” since it was first isolated in large quantities from rabbit skeletal muscle (Takahara et al. 1986). PAD2 mRNA and protein have been detected within the skin epidermis, peripheral nerves, and several hematopoietic cell types (Vossenaar et al. 2004; Ying et al. 2009; Nagata and Senshu 1990; Keilhoff et al. 2008). Genes regulated by the NFκB pathway are involved in a wide range of physiological responses, including cell death, developmental processes, and inflammation (Perkins 2007). PAD2 has also been shown to target IKKγ, a kinase upstream of NFκB activation, and citrullination of IKKγ seems to suppress NFkB activation (Lee et al. 2010). Although PAD2 is expressed across many tissue types and has been shown to regulate NFκB-mediated signal transduction, it is important to note that PAD2 knockout mice are viable and fertile (Raijmakers et al. 2006), as verified by our own unpublished observations.
PAD2 is also expressed in tissues of the reproductive system, including the uterus, pituitary gland, and mammary epithelial cells (Cherrington et al. 2012; Senshu et al. 1989; Takahara et al. 1992). Although PAD2 has been described as a cytoplasmic protein, a fraction of PAD2 was also found to reside in the nucleus of mammary epithelial cells, where it is associated with chromatin and was discovered to citrullinate histone H3 at arginine position 26 (Cherrington et al. 2012; Zhang et al. 2012) (Nakashima et al. 2002). Interference with PAD2 expression using siRNA in the MCF-7 breast cancer cell line impairs estrogen receptor α (ERα)-driven gene expression, supporting the notion that PAD2 facilitates ERα-driven transcription (Cherrington et al. 2012; Zhang et al. 2012). Interestingly, high concentrations (millimolar amounts) of the cancer chemotherapeutic agent paclitaxel, which is used to treat many neoplasms including breast and ovarian cancers, can inhibit the enzymatic activity of PAD2 (Pritzker and Moscarello 1998). In fact, MCF-7 cells transfected with PAD2 siRNA exhibited a reduced proliferation rate in comparison to control cells (Cherrington et al. 2012). These studies suggest that PAD2 may be a target candidate for anticancer therapies. (See Chap. 17 for more information on this topic.)
Protein citrullination in the brain and spinal cord is a hallmark of multiple sclerosis (MS) (Gyorgy et al. 2006) and in the murine model of MS known as experimental autoimmune encephalomyelitis (EAE) (Kidd et al. 2008). In EAE and MS, MBP and glial fibrillary acidic protein (GFAP) are hypercitrullinated (Raijmakers et al. 2006; Nicholas et al. 2004; Moscarello et al. 2002). The current model for the role of citrullination in EAE and MS is that citrullination of the myelin protein components interferes with their association thought to directly contribute to myelin instability and degradation (Gyorgy et al. 2006). PAD activity is up-regulated in DM20 transgenic mice expressing extra copies of the myelin proteolipid protein DM20, and these mice develop spontaneous demyelination (Moscarello et al. 2002). Expression of PAD2 has also been documented in the CNS, including microglia (Asaga et al. 2002), astrocytes (Asaga et al. 2002; Sambandam et al. 2004), and oligodendrocytes (Akiyama et al. 1999). Indeed, PAD2 transgenic mice over-expressing PAD2 under the MBP promoter exhibit increased MBP citrullination and spontaneous demyelinating disease (Musse et al. 2008). While protein citrullination in the CNS of EAE animals is entirely dependent on the presence of PAD2, PAD2-deficient mice remain susceptible to EAE (Raijmakers et al. 2006). These findings indicate that the role of PAD2 in EAE and MS is likely complex, and possibly, in the absence of PAD2, other PAD family members, like PAD4, can participate in disease pathogenesis. A more detailed discussion on the potential role of deimination in MS and EAE is presented in Chaps. 10 and 11.
1.2.3 PAD3
PAD3 expression is principally limited to the medullary and inner root sheath of the hair follicle with a localization overlapping that of its substrate trichohyalin (THH) (Nachat et al. 2005b). THH is a high-molecular-weight, α-helix-rich, structural protein of the hair follicle (see Chap. 8). After it is first synthesized, THH resides within soluble vacuoles, where it is stabilized by interactions between its α-helices (Gyorgy et al. 2006). Citrullination of THH is thought to promote its solubility and facilitate its cross-linking with cytokeratins and other THH molecules by transglutaminase, leading to directional hair growth (Gyorgy et al. 2006). PAD3 is also found in the granular and lower stratum corneum of the skin epidermis (see Chap. 7), where it may serve to collaborate with PAD1 to citrullinate filaggrin (Nachat et al. 2005a). Finally, the PAD3 transcript and protein are found in cells of the peripheral nervous system (Keilhoff et al. 2008) (see Chap. 9).
1.2.4 PAD4
PAD4 has been implicated in regulating inflammation (Nakayama-Hamada et al. 2005; Foulquier et al. 2007) and exhibits an expression pattern largely restricted to immune cell types, particularly macrophages and granulocytes (Foulquier et al. 2007; Asaga et al. 2001). Indeed, the addition of the granulocyte-inducing differentiation agents such as dimethylsulfoxide (DMSO) to the human promyelocytic HL-60 cell line leads to the acquisition of mature neutrophil properties and to the expression of PAD4 (Foulquier et al. 2007). Importantly, animals homozygous for the PAD deletion are viable, are fertile, and have no gross anatomical abnormalities (Hemmers et al. 2011). In addition, we have observed no perturbations in the development of any of the analyzed immune subsets in the absence of PAD4 (our unpublished observations).
Neutrophils are a critical component of the innate antimicrobial immune response (Nathan 2006; Borregaard 2010). The primary mission of the neutrophils is to seek and destroy pathogens. Upon recruitment to the site of infection, neutrophils can kill invading pathogens by phagocytosis, by release of preformed microbicidal granules, and by generation of reactive oxygen species (Flannagan et al. 2009; Nauseef 2007). Alternatively, neutrophils can kill extracellular pathogens by weaponizing their nuclear contents and releasing neutrophil extracellular traps (NETs) (Brinkmann et al. 2004).
NET structures are composed of decondensed chromatin decorated with antimicrobial mediators such as defensins, histones, neutrophil elastase, and myeloperoxidase (Urban et al. 2009; Wartha et al. 2007). In response to inflammatory stimuli, neutrophils can decondense their chromatin and actively expel their DNA, producing NETs that are decorated with granular and nuclear proteins, including citrullinated histones (Brinkmann and Zychlinsky 2007; Wang et al. 2009). Work from our lab and others have shown that PAD4 is essential for the production of NETs and NET-associated histone citrullination (Hemmers et al. 2011; Wang et al. 2009; Buono et al. 2005). In contrast, neutrophil phagocytic and chemotactic responses are unimpaired in PAD4-deficient mice (Hemmers et al. 2011; Li et al. 2010). PAD4-mediated histone citrullination is a hallmark of NET formation and is thought to play a mechanical role in NET configuration, where the conversion of positively charged arginine residues into the neutral citrulline amino acid by PAD4 promotes chromatin decondensation (Wang et al. 2009). PAD4-mediated NET formation is critical for controlling at least a subset of bacterial infections, because PAD4-deficient mice are more susceptible to infectious disease in a necrotizing fasciitis model (Buono et al. 2005). Through an unknown mechanism, the formation of NETs is coupled with the phagocytosis pathway because both myeloperoxidase and neutrophil elastase are essential for NET formation, though their exact function in NET formation is unclear. NET-mediated killing has been described for gram-positive and gram-negative bacteria as well as fungi. Targets include Staphylococcus aureus, Group A streptococci, Salmonella enterica, and Candida albicans (Brinkmann et al. 2004; Urban et al. 2009; Ermert et al. 2009). Proteomic studies have identified that PAD4 can be found in neutrophil-specific granules (Lominadze et al. 2005); however, it is still not known if PAD4 also contributes to neutrophil function and NET generation through other substrates.
Using in vitro studies, incubation of neutrophils with PMA, ionomycin, H2O2, lipopolysaccharide (LPS), and bacteria induces PAD4-mediated histone citrullination and NET formation (Hemmers et al. 2011; Wang et al. 2009; Buono et al. 2005; Neeli et al. 2008). Additionally, histone citrullination is sensitive to NADPH inhibitors, suggesting that PAD4 activation is downstream of ROS generation, but the precise signaling pathways by which ROS potentially activates PAD4 are unknown (Neeli et al. 2009; Denis et al. 2009). In contrast to published reports, we have been unable to demonstrate consistently that LPS stimulation induces PAD4 activity in murine bone marrow-derived neutrophils (BMDN) (Fig. 1.4), which is in line with several other studies reporting that LPS is incapable of inducing NET formation (Clark et al. 2007; Remijsen et al. 2011). We also detected citrullinated histone H4 in BMDN that were purified by gradient separation, but LPS stimulation did not increase histone H4 citrullination over the unstimulated, Ca2+-incubated control sample (Fig. 1.4a). We speculated that the histone H4 citrullination we detected in BMDN, isolated using density gradient separation, was linked to the purification process. Thus, we purified BMDN using a gentler, magnetic bead separation method, where we could isolate largely untouched neutrophils via negative selection. Using these cells, PMA/ionomycin stimulation induced histone H4 citrullination, but LPS did not (Fig. 1.4b). To insure that our LPS preparation was properly stimulating our neutrophils, we examined the activation of p38 by immunoblotting for phospho-p38 (Fig. 1.4c). These results do differ from previous studies, in terms of culture conditions and neutrophil populations used. In an earlier study with murine BMDN, the untreated negative control was prepared just after cell separation rather than incubating the untreated control samples in the presence of CaCl2 for the entire culture period, as we have done (Li et al. 2010). Radic and colleagues performed their studies on LPS-induced histone citrullination using human neutrophils isolated from peripheral blood (Neeli et al. 2008, 2009). It is important to note that there are differences in the efficiency and kinetics of NET formation in human and murine neutrophils (Ermert et al. 2009). Thus, it is important to regard the sample preparation and stimulation conditions when comparing the results from different studies on LPS-stimulated neutrophils and histone citrullination. Perhaps, LPS binding to TLR4 provides a second signal that, by itself, is not sufficient to activate PAD4 in neutrophils. As a result, our findings suggest that another innate immunity sensor may be more relevant for activation of PAD4 in neutrophils.
LPS stimulation alone is not sufficient to induce PAD4-mediated histone H4 citrulline 3 generation in murine neutrophils. Murine bone marrow-derived neutrophils (BMDN) were isolated by density gradient sedimentation (a) or by negative selection using magnetic bead separation (b) and (c). BMDN were stimulated with LPS (1 μg/mL) in the presence of 2 mM of CaCl2 in Lock’s buffer (10 mM Hepes pH 7.4, 150 mM NaCl, 5 mM KCl, 0.1 % glucose) for the indicated times. As a control, cells were incubated in Lock’s buffer in the absence (−Ca2+) or in the presence (+Ca2+) for 3 h. Cells were stimulated with PMA/ionomycin (P/I) as a control. (c) The activation of the TLR4 signaling pathway by LPS was determined by examining the lysates from (b) for phospho-p38
PAD4 is unique in that it contains a classical nuclear localization sequence (NLS) and therefore can be found primarily within the nucleus, where it is a well-documented transcriptional regulator (Nakashima et al. 2002; Jones et al. 2009). In fact, PAD4 has been shown to associate with the transcriptional regulators HDAC1 and p300/CBP proteins (Denis et al. 2009; Lee et al. 2005). In general, PAD4 and PAD4-mediated histone deimination have been linked with transcriptional repression (Cuthbert et al. 2004; Wang et al. 2004). However, the interaction between PAD4 and p300 may also promote p300 activity and presumably enhance transcription (Jones et al. 2009). PAD4 is also recruited by p53 to repress the expression of select p53 target genes, and inhibition of PAD4 by chemical inhibitors or depletion via siRNA leads to cell cycle arrest and apoptosis (Yao et al. 2008; Li et al. 2008). These results and the expression of PAD4 in many tumor cells have led some to speculate that PAD4 may be a target for cancer therapeutics (Chang and Han 2006; Slack et al. 2011). The potential regulation of cellular transformation is likely complex. Indeed, PAD4 also targets the tumor suppressor and p53-binding partner inhibitor of growth 4 (ING4). Citrullination of ING4 increases its susceptibility, thereby inhibiting p53-driven gene expression. Thus, more investigation will be necessary to understand the function of PAD4 in cellular growth and survival pathways.
1.2.5 PAD6
PAD6 was first identified as a highly abundant protein found in murine oocytes and embryos (Fig. 1.3). Hence, its original moniker was ePAD for “egg or embryonic PAD” (Wright et al. 2003). However, transcripts for the human PAD6 orthologue were also detected in ovary, testis, small intestine, spleen, lung, liver, skeletal muscle, fetal tissue, and peripheral blood leukocytes (Chavanas et al. 2004; Zhang et al. 2004). On the amino acid level, PAD6 shares ~42 % homology with the rest of the PAD family (Chavanas et al. 2004) and is missing several Ca2+-binding residues that are conserved in the other PAD family members (Fig. 1.2) (Arita et al. 2004). In fact, evidence of PAD6 enzymatic activity in vitro has yet to be demonstrated (Snow et al. 2008).
Expression of PAD6 within the ovary is regulated by the oocyte-specific transcription factor, Nobox, which binds to regulatory elements within the PAD6 promoter and drives PAD6 expression during oogenesis (Choi et al. 2010). Although male PAD6−/− mice are fertile, PAD6 is a maternal effect gene and essential for female fertility. Zygotes derived from fertilized PAD6−/− oocytes arrest at the two-cell stage of embryonic development, prior to implantation (Esposito et al. 2007). In the oocyte and zygote, PAD6 localizes to cytoplasmic lattice structures and is essential for lattice formation (Esposito et al. 2007; Yurttas et al. 2008). PAD6-containing cytoplasmic lattices seem to be important for de novo protein synthesis, embryonic gene activation, and microtubule-mediated organelle reorganization within the zygote (Yurttas et al. 2008; Kan et al. 2011). An antibody against histone H4, with a citrullinated moiety at position 3, recognized a nonnuclear and unidentified deiminated protein within oocytes, and this reactivity was not present in PAD6−/− oocytes (Esposito et al. 2007). Since the protein expression profile between wild-type and PAD6−/− oocytes is similar (Yurttas et al. 2008), these findings suggest that PAD6 is active in vivo and either directly deiminates this substrate or PAD6 is required to activate another PAD present in oocytes. Interestingly, PAD6 is phosphorylated in the mature egg, and PAD6 phosphorylation is required for its interaction with tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein (YWHA), a molecule known to aid oocyte maturation in amphibians (Snow et al. 2008). Perhaps, the association with phosphorylation-dependent binding partners or a phosphorylation-induced conformational change facilitates enzymatic activity of PAD6 in vivo. In fact, the recombinant form of the arginine methyltransferase PRMT5 is several hundredfold times less active than PRMT5 isolated from mammalian cells, likely due to the association of cellular factors that can promote PRMT5 enzymatic activity (Rho et al. 2001). Further enzymology and crystallography studies of PAD6 will be useful in determining whether PAD6 is an active deiminase and, if so, whether Ca2+ is required for its activity. Although it is unclear whether PAD6 is an enzymatically active PAD family member, collectively these findings demonstrate that PAD6 is essential for mammalian development.
1.3 PADs and RA
1.3.1 Anti-citrullinated Autoantibodies and RA
RA is a chronic, systemic inflammatory disease affecting approximately 2 % of the world population and is discussed in great detail in Chaps. 2–6. Briefly, this disorder is characterized by leukocyte invasion of the normally acellular synovial fluid and membrane, which thereby activates resident macrophages, mast cells, and synoviocytes, resulting in cell division and thickening of the joint lining (Firestein 2003). Plasma and synovial biopsy specimen from patients with RA contain high levels of citrullinated proteins (Chang et al. 2005; Makrygiannakis et al. 2006). In fact, anti-citrullinated peptide antibodies (ACPA) exhibit high specificity and sensitivity as diagnostic markers of the disease, suggesting that RA patients have defects in tolerance generated to citrullinated epitopes (Suzuki et al. 2007). Furthermore, the presence of intracellular and extracellular synovial citrullinated proteins correlates with inflammatory arthritis, as citrullinated proteins are found in RA patients but not in osteoarthritis patients (Foulquier et al. 2007; Kinloch et al. 2008; Lundberg et al. 2005). ACPA also develop in the murine CIA model of RA, and immunization with citrullinated collagen or administration of ACPA contributes to disease pathogenesis in mouse models (Lundberg et al. 2005; Kuhn et al. 2006).
It is now well known that PAD enzymes are activated during the inflammation process (Klareskog et al. 2008). Indeed, the presence of citrullinated proteins in the affected tissues of patients with RA, inflammatory bowel disease, and polymyositis, but not in healthy controls, supports the notion that deimination is linked with inflammation (Makrygiannakis et al. 2006). So far, PAD2 and PAD4 have been the only PADs detected in hematopoietic cells and in RA synovium (Nakayama-Hamada et al. 2005; Foulquier et al. 2007). Most PAD2- and PAD4-expressing cells within the RA synovium are positive for CD68, a marker of macrophages, neutrophils, and mast cells (Foulquier et al. 2007; Chang et al. 2005). Though variants of PAD4 are linked to RA in several Japanese and Korean cohorts, this association has not held true in most North American and European study groups (van der Helm-van Mil and Huizinga 2008). Despite the conflicting data regarding disease-contributing PAD4 haplotypes amongst different ethnic groups, the prevalence of ACPA in all ethnic groups supports the notion that aberrant PAD activity may contribute to RA pathogenesis (Klareskog et al. 2008).
It is possible not only that PAD enzymes could contribute to RA through self-antigen generation but also that the action of PAD enzymes could also contribute to the effector mechanisms of disease, such as immune cell recruitment and joint destruction. Recently, a subclass of ACPA that was especially reactive to citrullinated vimentin was also shown to stimulate osteoclastogenesis and bone resorptive activity, suggesting that ACPA might directly contribute to RA disease pathogenesis (Harre et al. 2012). The F1 generation between the KRN TCR transgenic mouse specific for bovine RNase (Stensland et al. 2009; Bedford 2007; Krause et al. 2007; Zurita-Lopez et al. 2012; Guo et al. 2011; Hidaka et al. 2005; Nachat et al. 2005a, b; Senshu et al. 1996; Ishida-Yamamoto et al. 2000; Takahara et al. 1986; Ying et al. 2009; Nagata and Senshu 1990; Keilhoff et al. 2008; Perkins 2007) I-Ak and the nonobese diabetic (NOD) background spontaneously develop a progressive, inflammatory joint disease that is very similar to human RA (Kouskoff et al. 1996). The autoantigen in this model is glucose-6-phosphate isomerase (GPI), a ubiquitous cytoplasmic enzyme. Serum or purified anti-GPI autoantibodies are sufficient to transfer disease to healthy, wild-type mice, providing a model for the RA effector phase (Maccioni et al. 2002; Matsumoto et al. 1999). The region of murine chromosome 4 containing all PAD genes is linked to arthritis severity in the K/B×N model, with the highest associated SNPs being within the PAD2 gene (Johnsen et al. 2011). Indeed, increased splenic expression of both PAD2 and PAD4 correlated with disease severity in the K/B×N model (Johnsen et al. 2011). Using PAD4-deficient mice generated in our laboratory, we found that K/B×N serum transfer arthritis is independent of PAD4 (Rohrbach et al. 2012). Perhaps the loss of PAD2 and PAD4 together may produce a more apparent phenotype in the K/B×N model. However, Willis et al. showed that the PAD inhibitor Cl-amidine provided therapeutic benefit in the collagen-induced arthritis model but had no benefit when arthritic disease was induced by the administration of anti-collagen antibodies (another model of the arthritis effector phase). Since the Padi locus is linked to disease severity in the K/B×N serum transfer model, it may be necessary to eliminate several PAD family members, either by targeting multiple locations within the PAD locus or by combining treatment with specific PAD inhibitors with targeted PAD alleles. Further studies will be necessary to fully dissect the role of the PAD enzymes in the effector phase of arthritis.
1.3.2 Citrullinated Epitope Generation
ACPAs can appear before the onset of disease and correlate with the most erosive form of RA (Raptopoulou et al. 2007). Several candidate anti-citrulline autoantigens have been identified, including citrullinated fillagrin, fibrin, vimentin, and α-enolase (Klareskog et al. 2008). There is a strong association between ACPA and the RA susceptibility major histocompatibility complex (MHC) II HLA-DRB1 alleles (Klareskog et al. 2008). Indeed, conversion of arginine to citrulline increased peptide–MHC binding affinity in one of the HLA-DRB1 genes (Fig. 1.5) (Klareskog et al. 2008). Though autoreactivity to protein citrullination is strongly associated with RA, the mechanism by which a healthy immune system tolerates citrullinated epitopes is unknown.
Potential pathways that may contribute to the generation of citrullinated self-antigens. Citrullinated (cit) peptides could be shuttled to the MHC I or the MHC II antigen presentation compartments. (a) PAD-mediated protein citrullination could change the susceptibility to, or pattern of, cleavage by proteases. Cleaved peptides could then be shuttled into the ER for loading onto MHC I. (b) Extracellular proteins that are taken up by the lysosome could be citrullinated following fusion with the autophagosome, which is possible due to PAD residence in the autophagosome. Citrullinated proteins could then be cleaved and presented via the MHC II pathway. (c) NET formation, which can be triggered by immune complex recognition via Fcγ receptors, could allow for the release of active PAD enzymes and citrullinated self-proteins
On average, each protein encoded by the human genome bears approximately 2.5 posttranslational modifications (Papin et al. 2005). To maintain tolerance, the immune system must consider the added diversity of potential self-antigens by posttranslational modifications (Doyle and Mamula 2012). Since citrullination leads to a loss of a positive charge and potentially changes protease cleavage sites, inflammation-induced protein citrullination could result in the presentation of new self-epitopes (Fig. 1.5) (Vossenaar et al. 2003). In fact, deimination of filaggrin increases its susceptibility by the protease bleomycin hydrolase, an enzyme that also contributes to peptide generation for antigen presentation by MHC I (Kamata et al. 2009; Stoltze et al. 2000). Unanue and colleagues have elegantly shown that a CD4+ T cell response to citrullinated epitopes naturally develops following immunization with the exogenous non-citrullinated antigen hen egg lysozyme (HEL) (Ireland et al. 2006). Autophagy is a process by which cytoplasmic proteins are engulfed by the membrane phagophore to be shuttled to the lysosome for degradation. One outcome of autophagy is the generation of peptides for MHC presentation (Kuballa et al. 2012). The presentation of citrullinated epitopes, at least from exogenous sources, is blocked by chemical inhibition of autophagy (Ireland and Unanue 2011). Investigation into intracellular and extracellular pathways that lead to the generation of citrullinated self-antigens and their presentation by MHC I and MHC II will be important for understanding the role of ACPA in RA.
Chronic joint inflammation in RA involves the influx of large number of inflammatory cells, including macrophages, mast cells, T cells, B cells, and neutrophils (Firestein 2003). NET formation by neutrophils, although critical for the full activation of the innate immune response (Buono et al. 2005), has also been implicated in inflammatory disease pathogenesis, including the autoimmune disorder lupus (Garcia-Romo et al. 2011), cystic fibrosis (Manzenreiter et al. 2012; Papayannopoulos et al. 2011; Marcos et al. 2010), sepsis (Clark et al. 2007), and thrombosis (Fuchs et al. 2007). Since PAD2, PAD4, and citrullinated proteins are found in the synovial fluid of RA patients (Kinloch et al. 2008), these enzymes can presumably act on extracellular proteins. For example, collagen and fibrinogen are both PAD substrates (Zhao et al. 2008; Yoshida et al. 2006), but the mechanism by which PAD molecules gain access to the extracellular space is unknown. Interestingly, it has been suggested that NETs offer a possible mechanism by which PAD4 may be liberated from the cell to generate citrullinated antigens and exacerbate inflammation (Jones et al. 2009; Dwivedi et al. 2012). Recently, Dwievedi et al. (2012) described hypercitrullination in neutrophils from arthritic patients as well as the specific reactivity of arthritic serum to activated neutrophils and citrullinated histones. The stimulus that induces PAD activity during autoimmune-mediated inflammation is currently unknown.
Because PAD4 activation requires Ca2+ and ROS generation, signaling through Fc receptors, which induce phagosome/granule fusion as well as NADPH oxidase assembly, may provide the necessary signals to induce PAD4 activity (Nimmerjahn and Ravetch 2008). This possibility would suggest that the relevant stimulus for PAD4 activation during an autoimmune response could be immune complex driven. Indeed, we have recently found that PAD4 activity and NETs are readily detected within the affected arthritic joint in a murine model of arthritis, which is induced by immune complexes (Rohrbach et al. 2012). In fact, the presence of deiminated histones in this model corresponded primarily to the infiltrating cells of the joint sublining, which is consistent with the expression pattern of PAD4 found in RA patients (Nakayama-Hamada et al. 2005; Rohrbach et al. 2012; Stahl et al. 2010). In a recent study, the interaction of neutrophil FcγRIIA with immune complexes induces NET formation (Chen et al. 2012). Furthermore, in autoimmune small-vessel vasculitis, anti-neutrophil cytoplasm antibodies (ANCA) triggered the formation of NETs, which promoted necrotic inflammation of blood vessels in this condition (Kessenbrock et al. 2009). Systemic lupus erythematous (SLE) is a systemic autoimmune disease characterized by the formation of pathogenic immune complexes. When activated by autoantibodies, neutrophils isolated from SLE patients also produce NETs, exposing immune-stimulatory proteins and potential autoantigens, leading to the induction of type I interferons by plasmacytoid dendritic cells (Garcia-Romo et al. 2011; Hakkim et al. 2010; Villanueva et al. 2011). Collectively, these results support the notion that NET production can contribute to disease pathogenesis in inflammatory conditions and that immune complexes may drive NET formation during inflammation (Fig. 1.5). In a chronic setting, NETs might play a role in propagating an inflamed state, similar to what has been reported for NETs in chronic lung inflammation (Marcos et al. 2010). This is of special interest, since PAD4 dysregulation has been implicated in diseases like rheumatoid arthritis (Anzilotti et al. 2010; Suzuki et al. 2003), multiple sclerosis (Wood et al. 2008), and malignant tumors (Chang and Han 2006).
1.4 Conclusion
PAD enzymes contribute to RA through self-antigen generation, but PAD enzymes could also contribute to the effector mechanisms of this disease. In addition to RA, PAD enzymes and citrullination are associated with multiple sclerosis, Alzheimer’s disease, psoriasis, and cancer (Gyorgy et al. 2006). Thus, the generation of isozyme-specific inhibitors and additional mouse models will be important to fully appreciate the physiological functions of PAD enzymes. Understanding the regulation of PAD activity may have broader implications in understanding the inflammation-linked disease states and may also shed light on the etiology of anti-citrulline autoantibodies.
References
Akiyama K, Sakurai Y, Asou H, Senshu T (1999) Localization of peptidylarginine deiminase type II in a stage-specific immature oligodendrocyte from rat cerebral hemisphere. Neurosci Lett 274(1):53–55
Andrade F, Darrah E, Gucek M, Cole RN, Rosen A, Zhu X (2010) Autocitrullination of human peptidylarginine deiminase 4 regulates protein citrullination during cell activation. Arthritis Rheum 62(6):1630–1640
Anzilotti C, Pratesi F, Tommasi C, Migliorini P (2010) Peptidylarginine deiminase 4 and citrullination in health and disease. Autoimmun Rev 9(3):158–160
Arita K, Hashimoto H, Shimizu T, Nakashima K, Yamada M, Sato M (2004) Structural basis for Ca(2+)-induced activation of human PAD4. Nat Struct Mol Biol 11(8):777–783
Asaga H, Nakashima K, Senshu T, Ishigami A, Yamada M (2001) Immunocytochemical localization of peptidylarginine deiminase in human eosinophils and neutrophils. J Leukoc Biol 70(1):46–51
Asaga H, Akiyama K, Ohsawa T, Ishigami A (2002) Increased and type II-specific expression of peptidylarginine deiminase in activated microglia but not hyperplastic astrocytes following kainic acid-evoked neurodegeneration in the rat brain. Neurosci Lett 326(2):129–132
Balandraud N, Gouret P, Danchin EG, Blanc M, Zinn D, Roudier J et al (2005) A rigorous method for multigenic families’ functional annotation: the peptidyl arginine deiminase (PADs) proteins family example. BMC Genomics 6:153
Bedford MT (2007) Arginine methylation at a glance. J Cell Sci 120(Pt 24):4243–4246
Borregaard N (2010) Neutrophils, from marrow to microbes. Immunity 33(5):657–670
Brinkmann V, Zychlinsky A (2007) Beneficial suicide: why neutrophils die to make NETs. Nat Rev Microbiol 5(8):577–582
Brinkmann V, Reichard U, Goosmann C, Fauler B, Uhlemann Y, Weiss DS et al (2004) Neutrophil extracellular traps kill bacteria. Science 303(5663):1532–1535
Buono C, Binder CJ, Stavrakis G, Witztum JL, Lichtman AH (2005) T-bet deficiency reduces atherosclerosis and alters plaque antigen-specific immune responses. Proc Natl Acad Sci U S A 102:1596–1601
Chang X, Han J (2006) Expression of peptidylarginine deiminase type 4 (PAD4) in various tumors. Mol Carcinog 45(3):183–196
Chang X, Yamada R, Suzuki A, Sawada T, Yoshino S, Tokuhiro S et al (2005) Localization of peptidylarginine deiminase 4 (PADI4) and citrullinated protein in synovial tissue of rheumatoid arthritis. Rheumatology (Oxford) 44(1):40–50
Chavanas S, Mechin MC, Takahara H, Kawada A, Nachat R, Serre G et al (2004) Comparative analysis of the mouse and human peptidylarginine deiminase gene clusters reveals highly conserved non-coding segments and a new human gene, PADI6. Gene 330:19–27
Chavanas S, Mechin MC, Nachat R, Adoue V, Coudane F, Serre G et al (2006) Peptidylarginine deiminases and deimination in biology and pathology: relevance to skin homeostasis. J Dermatol Sci 44(2):63–72
Chen K, Nishi H, Travers R, Tsuboi N, Martinod K, Wagner DD et al (2012) Endocytosis of soluble immune complexes leads to their clearance by FcgammaRIIIB but induces neutrophil extracellular traps via FcgammaRIIA in vivo. Blood 120:4421–4431
Cherrington BD, Zhang X, McElwee JL, Morency E, Anguish LJ, Coonrod SA (2012) Potential role for PAD2 in gene regulation in breast cancer cells. PLoS One 7(7):e41242
Choi M, Lee OH, Jeon S, Park M, Lee DR, Ko JJ et al (2010) The oocyte-specific transcription factor, Nobox, regulates the expression of Pad6, a peptidylarginine deiminase in the oocyte. FEBS Lett 584(16):3629–3634
Chumanevich AA, Causey CP, Knuckley BA, Jones JE, Poudyal D, Chumanevich AP et al (2011) Suppression of colitis in mice by Cl-amidine: a novel peptidylarginine deiminase inhibitor. Am J Physiol Gastrointest Liver Physiol 300(6):G929–G938
Clark SR, Ma AC, Tavener SA, McDonald B, Goodarzi Z, Kelly MM et al (2007) Platelet TLR4 activates neutrophil extracellular traps to ensnare bacteria in septic blood. Nat Med 13(4):463–469
Crane-Robinson C, Dragan AI, Privalov PL (2006) The extended arms of DNA-binding domains: a tale of tails. Trends Biochem Sci 31(10):547–552
Curis E, Nicolis I, Moinard C, Osowska S, Zerrouk N, Benazeth S et al (2005) Almost all about citrulline in mammals. Amino Acids 29(3):177–205
Cuthbert GL, Daujat S, Snowden AW, Erdjument-Bromage H, Hagiwara T, Yamada M et al (2004) Histone deimination antagonizes arginine methylation. Cell 118(5):545–553
Darrah E, Rosen A, Giles JT, Andrade F (2012) Peptidylarginine deiminase 2, 3 and 4 have distinct specificities against cellular substrates: novel insights into autoantigen selection in rheumatoid arthritis. Ann Rheum Dis 71(1):92–98
Denis H, Deplus R, Putmans P, Yamada M, Metivier R, Fuks F (2009) Functional connection between deimination and deacetylation of histones. Mol Cell Biol 29(18):4982–4993
Dong S, Kojima T, Shiraiwa M, Mechin MC, Chavanas S, Serre G et al (2005) Regulation of the expression of peptidylarginine deiminase type II gene (PADI2) in human keratinocytes involves Sp1 and Sp3 transcription factors. J Invest Dermatol 124(5):1026–1033
Dong S, Kanno T, Yamaki A, Kojima T, Shiraiwa M, Kawada A et al (2006) NF-Y and Sp1/Sp3 are involved in the transcriptional regulation of the peptidylarginine deiminase type III gene (PADI3) in human keratinocytes. Biochem J 397(3):449–459
Dong S, Zhang Z, Takahara H (2007) Estrogen-enhanced peptidylarginine deiminase type IV gene (PADI4) expression in MCF-7 cells is mediated by estrogen receptor-alpha-promoted transfactors activator protein-1, nuclear factor-Y, and Sp1. Mol Endocrinol 21(7):1617–1629
Dong S, Ying S, Kojima T, Shiraiwa M, Kawada A, Mechin MC et al (2008) Crucial roles of MZF1 and Sp1 in the transcriptional regulation of the peptidylarginine deiminase type I gene (PADI1) in human keratinocytes. J Invest Dermatol 128(3):549–557
Doyle HA, Mamula MJ (2012) Autoantigenesis: the evolution of protein modifications in autoimmune disease. Curr Opin Immunol 24(1):112–118
Dwivedi N, Upadhyay J, Neeli I, Khan S, Pattanaik D, Myers L et al (2012) Felty’s syndrome autoantibodies bind to deiminated histones and neutrophil extracellular traps. Arthritis Rheum 64(4):982–992
Ermert D, Urban CF, Laube B, Goosmann C, Zychlinsky A, Brinkmann V (2009) Mouse neutrophil extracellular traps in microbial infections. J Innate Immun 1(3):181–193
Esposito G, Vitale AM, Leijten FP, Strik AM, Koonen-Reemst AM, Yurttas P et al (2007) Peptidylarginine deiminase (PAD) 6 is essential for oocyte cytoskeletal sheet formation and female fertility. Mol Cell Endocrinol 273(1–2):25–31
Feske S (2007) Calcium signalling in lymphocyte activation and disease. Nat Rev Immunol 7(9):690–702
Firestein GS (2003) Evolving concepts of rheumatoid arthritis. Nature 423(6937):356–361
Flannagan RS, Cosio G, Grinstein S (2009) Antimicrobial mechanisms of phagocytes and bacterial evasion strategies. Nat Rev Microbiol 7(5):355–366
Foulquier C, Sebbag M, Clavel C, Chapuy-Regaud S, Al Badine R, Mechin MC et al (2007) Peptidyl arginine deiminase type 2 (PAD-2) and PAD-4 but not PAD-1, PAD-3, and PAD-6 are expressed in rheumatoid arthritis synovium in close association with tissue inflammation. Arthritis Rheum 56(11):3541–3553
Fuchs TA, Abed U, Goosmann C, Hurwitz R, Schulze I, Wahn V et al (2007) Novel cell death program leads to neutrophil extracellular traps. J Cell Biol 176(2):231–241
Garcia-Romo GS, Caielli S, Vega B, Connolly J, Allantaz F, Xu Z et al (2011) Netting neutrophils are major inducers of type I IFN production in pediatric systemic lupus erythematosus. Sci Transl Med 3(73):73ra20
Guo Q, Bedford MT, Fast W (2011) Discovery of peptidylarginine deiminase-4 substrates by protein array: antagonistic citrullination and methylation of human ribosomal protein S2. Mol Biosyst 7(7):2286–2295
Gyorgy B, Toth E, Tarcsa E, Falus A, Buzas EI (2006) Citrullination: a posttranslational modification in health and disease. Int J Biochem Cell Biol 38(10):1662–1677
Hakkim A, Furnrohr BG, Amann K, Laube B, Abed UA, Brinkmann V et al (2010) Impairment of neutrophil extracellular trap degradation is associated with lupus nephritis. Proc Natl Acad Sci U S A 107(21):9813–9818
Hao G, Wang D, Gu J, Shen Q, Gross SS, Wang Y (2009) Neutral loss of isocyanic acid in peptide CID spectra: a novel diagnostic marker for mass spectrometric identification of protein citrullination. J Am Soc Mass Spectrom 20(4):723–727
Harre U, Georgess D, Bang H, Bozec A, Axmann R, Ossipova E et al (2012) Induction of osteoclastogenesis and bone loss by human autoantibodies against citrullinated vimentin. J Clin Invest 122(5):1791–1802
Hemmers S, Teijaro JR, Arandjelovic S, Mowen KA (2011) PAD4-mediated neutrophil extracellular trap formation is not required for immunity against influenza infection. PLoS One 6(7):e22043
Hidaka Y, Hagiwara T, Yamada M (2005) Methylation of the guanidino group of arginine residues prevents citrullination by peptidylarginine deiminase IV. FEBS Lett 579(19):4088–4092
Horibata S, Coonrod SA, Cherrington BD (2012) Role for peptidylarginine deiminase enzymes in disease and female reproduction. J Reprod Dev 58(3):274–282
Ireland JM, Unanue ER (2011) Autophagy in antigen-presenting cells results in presentation of citrullinated peptides to CD4 T cells. J Exp Med 208(13):2625–2632
Ireland J, Herzog J, Unanue ER (2006) Cutting edge: unique T cells that recognize citrullinated peptides are a feature of protein immunization. J Immunol 177(3):1421–1425
Ishida-Yamamoto A, Senshu T, Takahashi H, Akiyama K, Nomura K, Iizuka H (2000) Decreased deiminated keratin K1 in psoriatic hyperproliferative epidermis. J Invest Dermatol 114(4):701–705
Johnsen AK, Valdar W, Golden L, Ortiz-Lopez A, Hitzemann R, Flint J et al (2011) Genome-wide and species-wide dissection of the genetics of arthritis severity in heterogeneous stock mice. Arthritis Rheum 63(9):2630–2640
Jones JE, Causey CP, Knuckley B, Slack-Noyes JL, Thompson PR (2009) Protein arginine deiminase 4 (PAD4): current understanding and future therapeutic potential. Curr Opin Drug Discov Devel 12(5):616–627
Kamata Y, Taniguchi A, Yamamoto M, Nomura J, Ishihara K, Takahara H et al (2009) Neutral cysteine protease bleomycin hydrolase is essential for the breakdown of deiminated filaggrin into amino acids. J Biol Chem 284(19):12829–12836
Kamata Y, Yamamoto M, Kawakami F, Tsuboi R, Takeda A, Ishihara K et al (2011) Bleomycin hydrolase is regulated biphasically in a differentiation- and cytokine-dependent manner: relevance to atopic dermatitis. J Biol Chem 286(10):8204–8212
Kan R, Yurttas P, Kim B, Jin M, Wo L, Lee B et al (2011) Regulation of mouse oocyte microtubule and organelle dynamics by PADI6 and the cytoplasmic lattices. Dev Biol 350(2):311–322
Kearney PL, Bhatia M, Jones NG, Yuan L, Glascock MC, Catchings KL et al (2005) Kinetic characterization of protein arginine deiminase 4: a transcriptional corepressor implicated in the onset and progression of rheumatoid arthritis. Biochemistry 44(31):10570–10582
Keilhoff G, Prell T, Langnaese K, Mawrin C, Simon M, Fansa H et al (2008) Expression pattern of peptidylarginine deiminase in rat and human Schwann cells. Dev Neurobiol 68(1):101–114
Kessenbrock K, Krumbholz M, Schonermarck U, Back W, Gross WL, Werb Z et al (2009) Netting neutrophils in autoimmune small-vessel vasculitis. Nat Med 15(6):623–625
Kidd BA, Ho PP, Sharpe O, Zhao X, Tomooka BH, Kanter JL et al (2008) Epitope spreading to citrullinated antigens in mouse models of autoimmune arthritis and demyelination. Arthritis Res Ther 10(5):R119
Kinloch A, Lundberg K, Wait R, Wegner N, Lim NH, Zendman AJ et al (2008) Synovial fluid is a site of citrullination of autoantigens in inflammatory arthritis. Arthritis Rheum 58(8):2287–2295
Klareskog L, Ronnelid J, Lundberg K, Padyukov L, Alfredsson L (2008) Immunity to citrullinated proteins in rheumatoid arthritis. Annu Rev Immunol 26:651–675
Knuckley B, Bhatia M, Thompson PR (2007) Protein arginine deiminase 4: evidence for a reverse protonation mechanism. Biochemistry 46(22):6578–6587
Kouskoff V, Korganow AS, Duchatelle V, Degott C, Benoist C, Mathis D (1996) Organ-specific disease provoked by systemic autoimmunity. Cell 87(5):811–822
Krause CD, Yang ZH, Kim YS, Lee JH, Cook JR, Pestka S (2007) Protein arginine methyltransferases: evolution and assessment of their pharmacological and therapeutic potential. Pharmacol Ther 113(1):50–87
Kuballa P, Nolte WM, Castoreno AB, Xavier RJ (2012) Autophagy and the immune system. Annu Rev Immunol 30:611–646
Kuhn KA, Kulik L, Tomooka B, Braschler KJ, Arend WP, Robinson WH et al (2006) Antibodies against citrullinated proteins enhance tissue injury in experimental autoimmune arthritis. J Clin Invest 116(4):961–973
Lee YH, Coonrod SA, Kraus WL, Jelinek MA, Stallcup MR (2005) Regulation of coactivator complex assembly and function by protein arginine methylation and demethylimination. Proc Natl Acad Sci U S A 102(10):3611–3616
Lee HJ, Joo M, Abdolrasulnia R, Young DG, Choi I, Ware LB et al (2010) Peptidylarginine deiminase 2 suppresses inhibitory {kappa}B kinase activity in lipopolysaccharide-stimulated RAW 264.7 macrophages. J Biol Chem 285(51):39655–39662
Li P, Yao H, Zhang Z, Li M, Luo Y, Thompson PR et al (2008) Regulation of p53 target gene expression by peptidylarginine deiminase 4. Mol Cell Biol 28(15):4745–4758
Li P, Li M, Lindberg MR, Kennett MJ, Xiong N, Wang Y (2010) PAD4 is essential for antibacterial innate immunity mediated by neutrophil extracellular traps. J Exp Med 207(9):1853–1862
Lominadze G, Powell DW, Luerman GC, Link AJ, Ward RA, McLeish KR (2005) Proteomic analysis of human neutrophil granules. Mol Cell Proteomics 4(10):1503–1521
Lundberg K, Nijenhuis S, Vossenaar ER, Palmblad K, van Venrooij WJ, Klareskog L et al (2005) Citrullinated proteins have increased immunogenicity and arthritogenicity and their presence in arthritic joints correlates with disease severity. Arthritis Res Ther 7(3):R458–R467
Maccioni M, Zeder-Lutz G, Huang H, Ebel C, Gerber P, Hergueux J et al (2002) Arthritogenic monoclonal antibodies from K/B×N mice. J Exp Med 195(8):1071–1077
Makrygiannakis D, af Klint E, Lundberg IE, Lofberg R, Ulfgren AK, Klareskog L et al (2006) Citrullination is an inflammation-dependent process. Ann Rheum Dis 65(9):1219–1222
Manzenreiter R, Kienberger F, Marcos V, Schilcher K, Krautgartner WD, Obermayer A et al (2012) Ultrastructural characterization of cystic fibrosis sputum using atomic force and scanning electron microscopy. J Cyst Fibros 11(2):84–92
Marcos V, Zhou Z, Yildirim AO, Bohla A, Hector A, Vitkov L et al (2010) CXCR2 mediates NADPH oxidase-independent neutrophil extracellular trap formation in cystic fibrosis airway inflammation. Nat Med 16(9):1018–1023
Matsumoto I, Staub A, Benoist C, Mathis D (1999) Arthritis provoked by linked T and B cell recognition of a glycolytic enzyme. Science 286(5445):1732–1735
Mechin MC, Sebbag M, Arnaud J, Nachat R, Foulquier C, Adoue V et al (2007) Update on peptidylarginine deiminases and deimination in skin physiology and severe human diseases. Int J Cosmet Sci 29(3):147–168
Mechin MC, Coudane F, Adoue V, Arnaud J, Duplan H, Charveron M et al (2010) Deimination is regulated at multiple levels including auto-deimination of peptidylarginine deiminases. Cell Mol Life Sci 67(9):1491–1503
Moscarello MA, Pritzker L, Mastronardi FG, Wood DD (2002) Peptidylarginine deiminase: a candidate factor in demyelinating disease. J Neurochem 81(2):335–343
Musse AA, Li Z, Ackerley CA, Bienzle D, Lei H, Poma R et al (2008) Peptidylarginine deiminase 2 (PAD2) overexpression in transgenic mice leads to myelin loss in the central nervous system. Dis Model Mech 1(4–5):229–240
Nachat R, Mechin MC, Takahara H, Chavanas S, Charveron M, Serre G et al (2005a) Peptidylarginine deiminase isoforms 1-3 are expressed in the epidermis and involved in the deimination of K1 and filaggrin. J Invest Dermatol 124(2):384–393
Nachat R, Mechin MC, Charveron M, Serre G, Constans J, Simon M (2005b) Peptidylarginine deiminase isoforms are differentially expressed in the anagen hair follicles and other human skin appendages. J Invest Dermatol 125(1):34–41
Nagata S, Senshu T (1990) Peptidylarginine deiminase in rat and mouse hemopoietic cells. Experientia 46(1):72–74
Nakashima K, Hagiwara T, Ishigami A, Nagata S, Asaga H, Kuramoto M et al (1999) Molecular characterization of peptidylarginine deiminase in HL-60 cells induced by retinoic acid and 1alpha,25-dihydroxyvitamin D(3). J Biol Chem 274(39):27786–27792
Nakashima K, Hagiwara T, Yamada M (2002) Nuclear localization of peptidylarginine deiminase V and histone deimination in granulocytes. J Biol Chem 277(51):49562–49568
Nakayama-Hamada M, Suzuki A, Kubota K, Takazawa T, Ohsaka M, Kawaida R et al (2005) Comparison of enzymatic properties between hPADI2 and hPADI4. Biochem Biophys Res Commun 327(1):192–200
Nathan C (2006) Neutrophils and immunity: challenges and opportunities. Nat Rev Immunol 6(3):173–182
Nauseef WM (2007) How human neutrophils kill and degrade microbes: an integrated view. Immunol Rev 219:88–102
Neeli I, Khan SN, Radic M (2008) Histone deimination as a response to inflammatory stimuli in neutrophils. J Immunol 180(3):1895–1902
Neeli I, Dwivedi N, Khan S, Radic M (2009) Regulation of extracellular chromatin release from neutrophils. J Innate Immun 1(3):194–201
Nicholas AP, Sambandam T, Echols JD, Tourtellotte WW (2004) Increased citrullinated glial fibrillary acidic protein in secondary progressive multiple sclerosis. J Comp Neurol 473(1):128–136
Nimmerjahn F, Ravetch JV (2008) Fcgamma receptors as regulators of immune responses. Nat Rev Immunol 8(1):34–47
Nishikawa K, Toker A, Johannes FJ, Songyang Z, Cantley LC (1997) Determination of the specific substrate sequence motifs of protein kinase C isozymes. J Biol Chem 272(2):952–960
Palmer CN, Irvine AD, Terron-Kwiatkowski A, Zhao Y, Liao H, Lee SP et al (2006) Common loss-of-function variants of the epidermal barrier protein filaggrin are a major predisposing factor for atopic dermatitis. Nat Genet 38(4):441–446
Papayannopoulos V, Staab D, Zychlinsky A (2011) Neutrophil elastase enhances sputum solubilization in cystic fibrosis patients receiving DNase therapy. PLoS One 6(12):e28526
Papin JA, Hunter T, Palsson BO, Subramaniam S (2005) Reconstruction of cellular signalling networks and analysis of their properties. Nat Rev Mol Cell Biol 6(2):99–111
Perkins ND (2007) Integrating cell-signalling pathways with NF-kappaB and IKK function. Nat Rev Mol Cell Biol 8(1):49–62
Pritzker LB, Moscarello MA (1998) A novel microtubule independent effect of paclitaxel: the inhibition of peptidylarginine deiminase from bovine brain. Biochim Biophys Acta 1388(1):154–160
Raijmakers R, Vogelzangs J, Raats J, Panzenbeck M, Corby M, Jiang H et al (2006) Experimental autoimmune encephalomyelitis induction in peptidylarginine deiminase 2 knockout mice. J Comp Neurol 498(2):217–226
Raptopoulou A, Sidiropoulos P, Katsouraki M, Boumpas DT (2007) Anti-citrulline antibodies in the diagnosis and prognosis of rheumatoid arthritis: evolving concepts. Crit Rev Clin Lab Sci 44(4):339–363
Remijsen Q, Vanden Berghe T, Wirawan E, Asselbergh B, Parthoens E, De Rycke R et al (2011) Neutrophil extracellular trap cell death requires both autophagy and superoxide generation. Cell Res 21(2):290–304
Rho J, Choi S, Seong YR, Cho WK, Kim SH, Im DS (2001) Prmt5, which forms distinct homo-oligomers, is a member of the protein-arginine methyltransferase family. J Biol Chem 276(14):11393–11401
Rogers GE (1962) Occurrence of citrulline in proteins. Nature 194:1149–1151
Rogers GE, Taylor LD (1977) The enzymic derivation of citrulline residues from arginine residues in situ during the biosynthesis of hair proteins that are cross-linked by isopeptide bonds. Adv Exp Med Biol 86A:283–294
Rohrbach AS, Hemmers S, Arandjelovic S, Corr M, Mowen KA (2012) PAD4 is not essential for disease in the K/B×N murine autoantibody-mediated model of arthritis. Arthritis Res Ther 14(3):R104
Sambandam T, Belousova M, Accaviti-Loper MA, Blanquicett C, Guercello V, Raijmakers R et al (2004) Increased peptidylarginine deiminase type II in hypoxic astrocytes. Biochem Biophys Res Commun 325(4):1324–1329
Senshu T, Akiyama K, Nagata S, Watanabe K, Hikichi K (1989) Peptidylarginine deiminase in rat pituitary: sex difference, estrous cycle-related changes, and estrogen dependence. Endocrinology 124(6):2666–2670
Senshu T, Kan S, Ogawa H, Manabe M, Asaga H (1996) Preferential deimination of keratin K1 and filaggrin during the terminal differentiation of human epidermis. Biochem Biophys Res Commun 225(3):712–719
Shirai H, Blundell TL, Mizuguchi K (2001) A novel superfamily of enzymes that catalyze the modification of guanidino groups. Trends Biochem Sci 26(8):465–468
Slack JL, Causey CP, Thompson PR (2011) Protein arginine deiminase 4: a target for an epigenetic cancer therapy. Cell Mol Life Sci 68(4):709–720
Smith FJ, Irvine AD, Terron-Kwiatkowski A, Sandilands A, Campbell LE, Zhao Y et al (2006) Loss-of-function mutations in the gene encoding filaggrin cause ichthyosis vulgaris. Nat Genet 38(3):337–342
Snow AJ, Puri P, Acker-Palmer A, Bouwmeester T, Vijayaraghavan S, Kline D (2008) Phosphorylation-dependent interaction of tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein (YWHA) with PADI6 following oocyte maturation in mice. Biol Reprod 79(2):337–347
Stahl EA, Raychaudhuri S, Remmers EF, Xie G, Eyre S, Thomson BP et al (2010) Genome-wide association study meta-analysis identifies seven new rheumatoid arthritis risk loci. Nat Genet 42(6):508–514
Stensland ME, Pollmann S, Molberg O, Sollid LM, Fleckenstein B (2009) Primary sequence, together with other factors, influence peptide deimination by peptidylarginine deiminase-4. Biol Chem 390(2):99–107
Stoltze L, Schirle M, Schwarz G, Schroter C, Thompson MW, Hersh LB et al (2000) Two new proteases in the MHC class I processing pathway. Nat Immunol 1(5):413–418
Suzuki A, Yamada R, Chang X, Tokuhiro S, Sawada T, Suzuki M et al (2003) Functional haplotypes of PADI4, encoding citrullinating enzyme peptidylarginine deiminase 4, are associated with rheumatoid arthritis. Nat Genet 34(4):395–402
Suzuki A, Yamada R, Yamamoto K (2007) Citrullination by peptidylarginine deiminase in rheumatoid arthritis. Ann N Y Acad Sci 1108:323–339
Takahara H, Okamoto H, Sugawara K (1986) Affinity chromatography of peptidylarginine deiminase from rabbit skeletal muscle on a column of soybean trypsin inhibitor (Kunitz)-Sepharose. J Biochem (Tokyo) 99(5):1417–1424
Takahara H, Kusubata M, Tsuchida M, Kohsaka T, Tagami S, Sugawara K (1992) Expression of peptidylarginine deiminase in the uterine epithelial cells of mouse is dependent on estrogen. J Biol Chem 267(1):520–525
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22(22):4673–4680
Urban CF, Ermert D, Schmid M, Abu-Abed U, Goosmann C, Nacken W et al (2009) Neutrophil extracellular traps contain calprotectin, a cytosolic protein complex involved in host defense against Candida albicans. PLoS Pathog 5(10):e1000639
van der Helm-van Mil AH, Huizinga TW (2008) Advances in the genetics of rheumatoid arthritis point to subclassification into distinct disease subsets. Arthritis Res Ther 10(2):205
Villanueva E, Yalavarthi S, Berthier CC, Hodgin JB, Khandpur R, Lin AM et al (2011) Netting neutrophils induce endothelial damage, infiltrate tissues, and expose immunostimulatory molecules in systemic lupus erythematosus. J Immunol 187(1):538–552
Vossenaar ER, Zendman AJ, van Venrooij WJ, Pruijn GJ (2003) PAD, a growing family of citrullinating enzymes: genes, features and involvement in disease. Bioessays 25(11):1106–1118
Vossenaar ER, Radstake TR, van der Heijden A, van Mansum MA, Dieteren C, de Rooij DJ et al (2004) Expression and activity of citrullinating peptidylarginine deiminase enzymes in monocytes and macrophages. Ann Rheum Dis 63(4):373–381
Wang Y, Wysocka J, Sayegh J, Lee YH, Perlin JR, Leonelli L et al (2004) Human PAD4 regulates histone arginine methylation levels via demethylimination. Science 306(5694):279–283
Wang Y, Li M, Stadler S, Correll S, Li P, Wang D et al (2009) Histone hypercitrullination mediates chromatin decondensation and neutrophil extracellular trap formation. J Cell Biol 184(2):205–213
Wartha F, Beiter K, Normark S, Henriques-Normark B (2007) Neutrophil extracellular traps: casting the NET over pathogenesis. Curr Opin Microbiol 10(1):52–56
Wood DD, Ackerley CA, Brand B, Zhang L, Raijmakers R, Mastronardi FG et al (2008) Myelin localization of peptidylarginine deiminases 2 and 4: comparison of PAD2 and PAD4 activities. Lab Invest 88(4):354–364
Wright PW, Bolling LC, Calvert ME, Sarmento OF, Berkeley EV, Shea MC et al (2003) ePAD, an oocyte and early embryo-abundant peptidylarginine deiminase-like protein that localizes to egg cytoplasmic sheets. Dev Biol 256(1):73–88
Yao H, Li P, Venters BJ, Zheng S, Thompson PR, Pugh BF et al (2008) Histone Arg modifications and p53 regulate the expression of OKL38, a mediator of apoptosis. J Biol Chem 283(29):20060–20068
Ying S, Dong S, Kawada A, Kojima T, Chavanas S, Mechin MC et al (2009) Transcriptional regulation of peptidylarginine deiminase expression in human keratinocytes. J Dermatol Sci 53(1):2–9
Yoshida M, Tsuji M, Kurosaka D, Yasuda J, Ito Y, Nishizawa T et al (2006) Autoimmunity to citrullinated type II collagen in rheumatoid arthritis. Mod Rheumatol 16(5):276–281
Yurttas P, Vitale AM, Fitzhenry RJ, Cohen-Gould L, Wu W, Gossen JA et al (2008) Role for PADI6 and the cytoplasmic lattices in ribosomal storage in oocytes and translational control in the early mouse embryo. Development 135(15):2627–2636
Zhang J, Dai J, Zhao E, Lin Y, Zeng L, Chen J et al (2004) cDNA cloning, gene organization and expression analysis of human peptidylarginine deiminase type VI. Acta Biochim Pol 51(4):1051–1058
Zhang X, Bolt M, Guertin MJ, Chen W, Zhang S, Cherrington BD et al (2012) Peptidylarginine deiminase 2-catalyzed histone H3 arginine 26 citrullination facilitates estrogen receptor alpha target gene activation. Proc Natl Acad Sci U S A 109(33):13331–13336
Zhao X, Okeke NL, Sharpe O, Batliwalla FM, Lee AT, Ho PP et al (2008) Circulating immune complexes contain citrullinated fibrinogen in rheumatoid arthritis. Arthritis Res Ther 10(4):R94
Zurita-Lopez CI, Sandberg T, Kelly R, Clarke SG (2012) Human protein arginine methyltransferase 7 (PRMT7) is a type III enzyme forming omega-NG-monomethylated arginine residues. J Biol Chem 287(11):7859–7870
Acknowledgements
We are grateful to Katherine McKenney and Myles Dillon for useful conversations and the National Institutes of Health for support (AI099728-01A1 and AI067460). Dr. Arandjelovic was supported by the Philip S. Magaram, Esq. Research Award from the Arthritis Foundation. We apologize to investigators whose important contributions were not included due to space limitations.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2014 Springer Science+Business Media New York
About this chapter
Cite this chapter
Rohrbach, A.S., Arandjelovic, S., Mowen, K.A. (2014). Physiological Pathways of PAD Activation and Citrullinated Epitope Generation. In: Nicholas, A., Bhattacharya, S. (eds) Protein Deimination in Human Health and Disease. Springer, New York, NY. https://doi.org/10.1007/978-1-4614-8317-5_1
Download citation
DOI: https://doi.org/10.1007/978-1-4614-8317-5_1
Published:
Publisher Name: Springer, New York, NY
Print ISBN: 978-1-4614-8316-8
Online ISBN: 978-1-4614-8317-5
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)