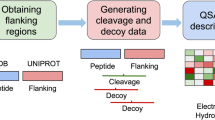
Abstract
T cell epitopes presented on the surface of mammalian cells are subjected to a complex network of antigen processing and presentation. Among them, C-terminal antigen processing constitutes one of the main bottlenecks for the generation of epitopes, as it defines the C-terminal end of the final epitope and delimits the peptidome that will be presented downstream. Previously (Amengual-Rigo and Guallar, Sci Rep 111(11):1–8, 2021), we demonstrated that NetCleave stands out as one of the best algorithms for the prediction of C-terminal processing, which in its turn can be crucial to design peptide-based vaccination strategies. In this chapter, we provide a pipeline to exploit the full capabilities of NetCleave, an open-source and retrainable algorithm for predicting the C-terminal antigen processing for the MHC-I and MHC-II pathways.
Roc Farriol-Duran and Marina Vallejo-Vallés are co-first authors.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Komanduri KV (2018) Divining T-cell targets for cancer immunotherapy. Blood 132:1861–1863. https://doi.org/10.1182/blood-2018-09-873588
Ali Awadelkareem E, Osman Mohammed N, Bakor Mohammed Gaafar B, AwadElkariem Ali S (2020) Epitope-based peptide vaccine design against spike protein (S) of novel coronavirus (2019-nCoV): an immunoinformatics approach. https://doi.org/10.21203/rs.3.rs-30076/v1
Bhattacharya M, Sharma AR, Patra P, Ghosh P, Sharma G, Patra BC, Lee S-S, Chakraborty C (2020) Development of epitope-based peptide vaccine against novel coronavirus 2019 (SARS-COV-2): immunoinformatics approach. J Med Virol 92:618. https://doi.org/10.1002/jmv.25736
Moise L, Buller RM, Schriewer J, Lee J, Frey SE, Weiner DB, Martin W, De Groot AS (2011) VennVax, a DNA-prime, peptide-boost multi-T-cell epitope poxvirus vaccine, induces protective immunity against vaccinia infection by T cell response alone. Vaccine 29:501–511. https://doi.org/10.1016/j.vaccine.2010.10.064
Walter S (2012) Multipeptide immune response to cancer vaccine IMA901 after single-dose cyclophosphamide associates with longer patient survival. Nat Med 18:1254. https://doi.org/10.1038/nm.2883
Lázaro S, Gamarra D, Del Val M (2015) Proteolytic enzymes involved in MHC class I antigen processing: a guerrilla army that partners with the proteasome. Mol Immunol 68:72–76. https://doi.org/10.1016/j.molimm.2015.04.014
Brutkiewicz RR (2016) Cell signaling pathways that regulate antigen presentation. J Immunol 197:2971–2979. https://doi.org/10.4049/jimmunol.1600460
Gfeller D, Bassani-Sternberg M (2018) Predicting antigen presentation—what could we learn from a million peptides? Front Immunol 9:1716. https://doi.org/10.3389/fimmu.2018.01716
Mei S, Li F, Leier A, Marquez-Lago TT, Giam K, Croft NP, Akutsu T, Smith AI, Li J, Rossjohn J, Purcell AW, Song J (2020) A comprehensive review and performance evaluation of bioinformatics tools for HLA class I peptide-binding prediction. Brief Bioinform 21:1119–1135. https://doi.org/10.1093/bib/bbz051
Reynisson B, Alvarez B, Paul S, Peters B, Nielsen M (2020) NetMHCpan-4.1 and NetMHCIIpan-4.0: improved predictions of MHC antigen presentation by concurrent motif deconvolution and integration of MS MHC eluted ligand data. Nucleic Acids Res 48:W449–W454. https://doi.org/10.1093/nar/gkaa379
O’Donnell TJ, Rubinsteyn A, Laserson U (2020) MHCflurry 2.0: improved pan-allele prediction of MHC class I-presented peptides by incorporating antigen processing. Cell Syst 11:42–48.e7. https://doi.org/10.1016/j.cels.2020.06.010
Bassani-Sternberg M, Chong C, Guillaume P, Solleder M, Pak HS, Gannon PO, Kandalaft LE, Coukos G, Gfeller D (2017) Deciphering HLA-I motifs across HLA peptidomes improves neo-antigen predictions and identifies allostery regulating HLA specificity. PLoS Comput Biol 13:e1005725. https://doi.org/10.1371/journal.pcbi.1005725
Harndahl M, Rasmussen M, Roder G, Dalgaard Pedersen I, Sørensen M, Nielsen M, Buus S (2012) Peptide-MHC class I stability is a better predictor than peptide affinity of CTL immunogenicity. Eur J Immunol 42:1405–1416. https://doi.org/10.1002/eji.201141774
Gomez-Perosanz M, Ras-Carmona A, Reche PA (2020) PCPS: a web server to predict proteasomal cleavage sites. Methods Mol Biol 2131:399–406. https://doi.org/10.1007/978-1-0716-0389-5_23
Jørgensen KW, Rasmussen M, Buus S, Nielsen M (2014) Net MHC stab – predicting stability of peptide-MHC-I complexes; impacts for cytotoxic T lymphocyte epitope discovery. Immunology 141:18–26. https://doi.org/10.1111/imm.12160
Rasmussen M, Fenoy E, Harndahl M, Kristensen AB, Nielsen IK, Nielsen M, Buus S (2016) Pan-specific prediction of peptide–MHC class I complex stability, a correlate of T cell immunogenicity. J Immunol 197:1517–1524. https://doi.org/10.4049/jimmunol.1600582
Besser H, Louzoun Y (2018) Cross-modality deep learning-based prediction of TAP binding and naturally processed peptide. Immunogenetics 70:419–428. https://doi.org/10.1007/s00251-018-1054-6
Murata S, Takahama Y, Kasahara M, Tanaka K (2018) The immunoproteasome and thymoproteasome: functions, evolution and human disease. Nat Immunol 19:923–931. https://doi.org/10.1038/s41590-018-0186-z
Neefjes J, Jongsma MLM, Paul P, Bakke O (2011) Towards a systems understanding of MHC class I and MHC class II antigen presentation. Nat Rev Immunol 11:823–836. https://doi.org/10.1038/nri3084
Nielsen M, Lundegaard C, Lund O, Keşmir C (2005) The role of the proteasome in generating cytotoxic T-cell epitopes: insights obtained from improved predictions of proteasomal cleavage. Immunogenetics 57:33–41. https://doi.org/10.1007/s00251-005-0781-7
Hakenberg J, Nussbaum AK, Schild H, Rammensee H-G, Kuttler C, Holzhütter H-G, Kloetzel P-M, Kaufmann SHE, Mollenkopf H-J (2003) MAPPP: MHC class I antigenic peptide processing prediction. Appl Bioinformatics 2:155–158
Nussbaum AK, Kuttler C, Hadeler KP, Rammensee HG, Schild H (2001) PAProC: a prediction algorithm for proteasomal cleavages available on the WWW. Immunogenetics 53:87–94. https://doi.org/10.1007/s002510100300
Amengual-Rigo P, Guallar V (2021) NetCleave: an open-source algorithm for predicting C-terminal antigen processing for MHC-I and MHC-II. Sci Rep 111(11):1–8. https://doi.org/10.1038/s41598-021-92632-y
Vita R, Mahajan S, Overton JA, Dhanda SK, Martini S, Cantrell JR, Wheeler DK, Sette A, Peters B (2019) The immune epitope database (IEDB): 2018 update. Nucleic Acids Res 47:D339–D343. https://doi.org/10.1093/nar/gky1006
Rizk JG, Lippi G, Henry BM, Forthal DN, Rizk Y (2022) Prevention and treatment of monkeypox. Drugs 82:957–963. https://doi.org/10.1007/s40265-022-01742-y
Marcu A, Bichmann L, Kuchenbecker L, Kowalewski DJ, Freudenmann LK, Backert L, Mühlenbruch L, Szolek A, Lübke M, Wagner P, Engler T, Matovina S, Wang J, Hauri-Hohl M, Martin R, Kapolou K, Walz JS, Velz J, Moch H, Regli L, Silginer M, Weller M, Löffler MW, Erhard F, Schlosser A, Kohlbacher O, Stevanović S, Rammensee H-G, Neidert MC (2021) HLA Ligand Atlas: a benign reference of HLA-presented peptides to improve T-cell-based cancer immunotherapy. J Immunother Cancer 9:e002071. https://doi.org/10.1136/jitc-2020-002071
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Science+Business Media, LLC, part of Springer Nature
About this protocol
Cite this protocol
Farriol-Duran, R., Vallejo-Vallés, M., Amengual-Rigo, P., Floor, M., Guallar, V. (2023). NetCleave: An Open-Source Algorithm for Predicting C-Terminal Antigen Processing for MHC-I and MHC-II. In: Reche, P.A. (eds) Computational Vaccine Design. Methods in Molecular Biology, vol 2673. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-3239-0_15
Download citation
DOI: https://doi.org/10.1007/978-1-0716-3239-0_15
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-3238-3
Online ISBN: 978-1-0716-3239-0
eBook Packages: Springer Protocols