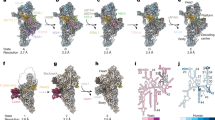
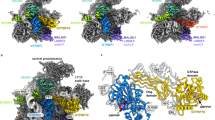
Abstract
Mitoribosome biogenesis is a complex and energetically costly process that involves RNA elements encoded in the mitochondrial genome and mitoribosomal proteins most frequently encoded in the nuclear genome. The process is catalyzed by extra-ribosomal proteins, nucleus-encoded assembly factors that act in all stages of the assembly process to coordinate the processing and maturation of ribosomal RNAs with the hierarchical association of ribosomal proteins. Biochemical studies and recent cryo-EM structures of mammalian mitoribosomes have provided hints regarding their assembly. In this general concept chapter, we will briefly describe the current knowledge, mainly regarding the mammalian mitoribosome biogenesis pathway and factors involved, and will emphasize the biological sources and approaches that have been applied to advance the field.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Küntzel H, Noll H (1967) Mitochondrial and cytoplasmic polysomes from Neurospora crassa. Nature 215(5108):1340–1345. https://doi.org/10.1038/2151340a0
O’Brien TW, Kalf GF (1967) Ribosomes from rat liver mitochondria. I. Isolation procedure and contamination studies. J Biol Chem 242(9):2172–2179
O’Brien TW, Kalf GF (1967) Ribosomes from rat liver mitochondira. II. Partial characterization. J Biol Chem 242(9):2180–2185
Zeng R, Smith E, Barrientos A (2018) Yeast mitoribosome large subunit assembly proceeds by hierarchical incorporation of protein clusters and modules on the inner membrane. Cell Metab 27(3):645–656
Bogenhagen DF, Ostermeyer-Fay AG, Haley JD et al (2018) Kinetics and mechanism of mammalian mitochondrial ribosome assembly. Cell Rep 22(7):1935–1944
Amunts A, Brown A, Bai X et al (2014) Structure of the yeast mitochondrial large ribosomal subunit. Science 343:1485–1489
Desai N, Brown A, Amunts A et al (2017) The structure of the yeast mitochondrial ribosome. Science 355(6324):528–531
Lenarčič T, Niemann M, Ramrath DJF et al (2022) Mitoribosomal small subunit maturation involves formation of initiation-like complexes. Proc Natl Acad Sci U S A 119(3). https://doi.org/10.1073/pnas.2114710118
Tobiasson V, Gahura O, Aibara S et al (2021) Interconnected assembly factors regulate the biogenesis of mitoribosomal large subunit. EMBO J 40(6):e106292. https://doi.org/10.15252/embj.2020106292
Soufari H, Waltz F, Parrot C et al (2020) Structure of the mature kinetoplastids mitoribosome and insights into its large subunit biogenesis. Proc Natl Acad Sci U S A 117(47):29851–29861. https://doi.org/10.1073/pnas.2011301117
Saurer M, Ramrath DJF, Niemann M et al (2019) Mitoribosomal small subunit biogenesis in trypanosomes involves an extensive assembly machinery. Science 365(6458):1144–1149
Greber BJ, Bieri P, Leibundgut M et al (2015) Ribosome. The complete structure of the 55S mammalian mitochondrial ribosome. Science 348(6232):303–308
Greber BJ, Boehringer D, Leibundgut M et al (2014) The complete structure of the large subunit of the mammalian mitochondrial ribosome. Nature 515(7526):283–286
Amunts A, Brown A, Toots J et al (2015) The structure of the human mitochondrial ribosome. Science 348(6230):95–98
Brown A, Amunts A, Bai XC et al (2014) Structure of the large ribosomal subunit from human mitochondria. Science 346(6210):718–722
Aibara S, Singh V, Modelska A et al (2020) Structural basis of mitochondrial translation. Elife 9. https://doi.org/10.7554/eLife.58362
Brown A, Rathore S, Kimanius D et al (2017) Structures of the human mitochondrial ribosome in native states of assembly. Nat Struct Mol Biol 24(10):866–869
Itoh Y, Khawaja A, Laptev I et al (2022) Mechanism of mitoribosomal small subunit biogenesis and preinitiation. Nature 606(7914):603–608. https://doi.org/10.1038/s41586-022-04795-x
Cheng J, Berninghausen O, Beckmann R (2021) A distinct assembly pathway of the human 39S late pre-mitoribosome. Nat Commun 12(1):4544. https://doi.org/10.1038/s41467-021-24818-x
Cipullo M, Gesé GV, Khawaja A et al (2021) Structural basis for late maturation steps of the human mitoribosomal large subunit. Nat Commun 12(1):3673. https://doi.org/10.1038/s41467-021-23617-8
Hillen HS, Lavdovskaia E, Nadler F et al (2021) Structural basis of GTPase-mediated mitochondrial ribosome biogenesis and recycling. Nat Commun 12(1):3672. https://doi.org/10.1038/s41467-021-23702-y
Lenarčič T, Jaskolowski M, Leibundgut M et al (2021) Stepwise maturation of the peptidyl transferase region of human mitoribosomes. Nat Commun 12(1):3671. https://doi.org/10.1038/s41467-021-23811-8
Chandrasekaran V, Desai N, Burton NO et al (2021) Visualizing formation of the active site in the mitochondrial ribosome. Elife 10. https://doi.org/10.7554/eLife.68806
Smits P, Smeitink JA, van den Heuvel LP et al (2007) Reconstructing the evolution of the mitochondrial ribosomal proteome. Nucleic Acids Res 35(14):4686–4703. https://doi.org/10.1093/nar/gkm441
Petrov AS, Wood EC, Bernier CR et al (2019) Structural patching fosters divergence of mitochondrial ribosomes. Mol Biol Evol 36(2):207–219
van der Sluis EO, Bauerschmitt H, Becker T et al (2015) Parallel structural evolution of mitochondrial ribosomes and OXPHOS complexes. Genome Biol Evol 7(5):1235–1251. https://doi.org/10.1093/gbe/evv061
De Silva D, Fontanesi F, Barrientos A (2013) The DEAD-Box protein Mrh4 functions in the assembly of the mitochondrial large ribosomal subunit. Cell Metab 18:712–725
Maiti P, Kim HJ, Tu YT et al (2018) Human GTPBP10 is required for mitoribosome maturation. Nucleic Acids Res 13. https://doi.org/10.1093/nar/gky938
Ferrari A, Del’Olio S, Barrientos A (2021) The diseased mitoribosome. FEBS Lett 595(8):1025–1061. https://doi.org/10.1002/1873-3468.14024
Maiti P, Lavdovskaia E, Barrientos A et al (2020) Role of GTPases in driving mitoribosome assembly. Trends Cell Biol 3(4):284–297
Lopez Sanchez MIG, Krüger A, Shiriaev DI et al (2021) Human mitoribosome biogenesis and its emerging links to disease. Int J Mol Sci 22(8). https://doi.org/10.3390/ijms22083827
Kummer E, Ban N (2021) Mechanisms and regulation of protein synthesis in mitochondria. Nat Rev Mol Cell Biol 22(5):307–325. https://doi.org/10.1038/s41580-021-00332-2
Lavdovskaia E, Hillen HS, Richter-Dennerlein R (2022) Hierarchical folding of the catalytic core during mitochondrial ribosome biogenesis. Trends Cell Biol 32(3):182–185. https://doi.org/10.1016/j.tcb.2021.09.004
Kummer E, Leibundgut M, Rackham O et al (2018) Unique features of mammalian mitochondrial translation initiation revealed by cryo-EM. Nature 560(7717):263–267
Singh V, Itoh Y, Huynen MA et al (2022) Activation mechanism of mitochondrial translation by LRPPRC-SLIRP. bioRxiv:2022.2006.2020.496763. https://doi.org/10.1101/2022.06.20.496763
Chen Y, Hagopian K, Bibus D et al (2013) The influence of dietary lipid composition on liver mitochondria from mice following 1 month of calorie restriction. Biosci Rep 33(1):83–95. https://doi.org/10.1042/BSR20120060
Singh V, Itoh I, Del’Olio S et al (2023) The complete structure of human mitoribosome, roles of mito-specific protein elements, cofactors and rRNA modifications. Nat Commun In press
Greber BJ, Ban N (2016) Structure and function of the mitochondrial ribosome. Annu Rev Biochem 85:103–132
Itoh Y, Singh V, Khawaja A, Naschberger A, Nguyen MD, Rorbach J, Amunts A (2022) Structure of the mitoribosomal small subunit with streptomycin reveals Fe-S clusters and physiological molecules. eLife 11:e77460
Yusupova GZ, Yusupov MM, Cate JH et al (2001) The path of messenger RNA through the ribosome. Cell 106(2):233–241. https://doi.org/10.1016/s0092-8674(01)00435-4
Takyar S, Hickerson RP, Noller HF (2005) mRNA helicase activity of the ribosome. Cell 120:49–58
Helm M, Brulé H, Friede D et al (2000) Search for characteristic structural features of mammalian mitochondrial tRNAs. RNA 6(10):1356–1379. https://doi.org/10.1017/s1355838200001047
Rorbach J, Gao F, Powell CA et al (2016) Human mitochondrial ribosomes can switch their structural RNA composition. Proc Natl Acad Sci U S A 113(43):12198–12201. https://doi.org/10.1073/pnas.1609338113
Greber BJ, Boehringer D, Leitner A et al (2014) Architecture of the large subunit of the mammalian mitochondrial ribosome. Nature 505(7484):515–519. https://doi.org/10.1038/nature12890
Antonicka H, Choquet K, Lin ZY et al (2017) A pseudouridine synthase module is essential for mitochondrial protein synthesis and cell viability. EMBO Rep 18(1):28–38
Rackham O, Busch JD, Matic S et al (2016) Hierarchical RNA processing is required for mitochondrial ribosome assembly. Cell Rep 16(7):1874–1890
Lee KW, Bogenhagen DF (2014) Assignment of 2′-O-methyltransferases to modification sites on the mammalian mitochondrial large subunit 16 S ribosomal RNA (rRNA). J Biol Chem 289(36):24936–24942. https://doi.org/10.21074/jbc.C24114.581868. Epub 582014 Jul 581829
Rorbach J, Boesch P, Gammage PA et al (2014) MRM2 and MRM3 are involved in biogenesis of the large subunit of the mitochondrial ribosome. Mol Biol Cell 9:01–0014
Jaskolowski M, Ramrath DJF, Bieri P et al (2020) Structural insights into the mechanism of mitoribosomal large subunit biogenesis. Mol Cell 79(4):629–644
Chen SS, Williamson JR (2013) Characterization of the ribosome biogenesis landscape in E. coli using quantitative mass spectrometry. J Mol Biol 425(4):767–779
Davis JH, Tan YZ, Carragher B et al (2016) Modular assembly of the bacterial large ribosomal subunit. Cell 167(6):1610–1622
Snider J, Wang D, Bogenhagen DF et al (2019) Pulse SILAC approaches to the measurement of cellular dynamics. Adv Exp Med Biol 1140:575–583. https://doi.org/10.1007/978-3-030-15950-4_34
Lindahl L (1975) Intermediates and time kinetics of the in vivo assembly of Escherichia coli ribosomes. J Mol Biol 92(1):15–37
Davis JH, Williamson JR (2017) Structure and dynamics of bacterial ribosome biogenesis. Philos Trans R Soc Lond Ser B Biol Sci 372(1716). https://doi.org/10.1098/rstb.2016.0181
Antonicka H, Sasarman F, Nishimura T et al (2013) The mitochondrial RNA-binding protein GRSF1 localizes to RNA granules and is required for posttranscriptional mitochondrial gene expression. Cell Metab 17(3):386–398
Jourdain AA, Koppen M, Wydro M et al (2013) GRSF1 regulates RNA processing in mitochondrial RNA granules. Cell Metab 17(3):399–410
Antonicka H, Shoubridge EA (2015) Mitochondrial RNA granules are centers for posttranscriptional RNA processing and ribosome biogenesis. Cell Rep 10(6):920–932
Tu YT, Barrientos A (2015) The human mitochondrial DEAD-box protein DDX28 resides in RNA granules and functions in mitoribosome assembly. Cell Rep 10(6):854–864
Shajani Z, Sykes MT, Williamson JR (2011) Assembly of bacterial ribosomes. Annu Rev Biochem 80:501–526
Britton RA (2009) Role of GTPases in bacterial ribosome assembly. Annu Rev Microbiol 63:155–176
Barrientos A, Korr D, Barwell KJ et al (2003) MTG1 codes for a conserved protein required for mitochondrial translation. Mol Biol Cell 14(6):2292–2302
Paul MF, Alushin GM, Barros MH et al (2012) The putative GTPase encoded by MTG3 functions in a novel pathway for regulating assembly of the small subunit of yeast mitochondrial ribosomes. J Biol Chem 287(29):24346–24355
Dennerlein S, Rozanska A, Wydro M et al (2010) Human ERAL1 is a mitochondrial RNA chaperone involved in the assembly of the 28S small mitochondrial ribosomal subunit. Biochem J 430(3):551–558
Metodiev MD, Spahr H, Loguercio Polosa P et al (2014) NSUN4 is a dual function mitochondrial protein required for both methylation of 12S rRNA and coordination of mitoribosomal assembly. PLoS Genet 10(2):e1004110
De Silva D, Poliquin S, Zeng R et al (2017) The DEAD-box helicase Mss116 plays distinct roles in mitochondrial ribogenesis and mRNA-specific translation. Nucleic Acids Res 45(11):6628–6643
Dalla Rosa I, Durigon R, Pearce SF et al (2014) MPV17L2 is required for ribosome assembly in mitochondria. Nucleic Acids Res 42(13):8500–8515
De Silva D, Tu YT, Amunts A et al (2015) Mitochondrial ribosome assembly in health and disease. Cell Cycle 14(14):2226–2250
Boczonadi V, Horvath R (2014) Mitochondria: impaired mitochondrial translation in human disease. Int J Biochem Cell Biol 48(100):77–84
Gopisetty G, Thangarajan R (2016) Mammalian mitochondrial ribosomal small subunit (MRPS) genes: a putative role in human disease. Gene 589(1):27–35
Pulman J, Ruzzenente B, Bianchi L et al (2019) Mutations in the MRPS28 gene encoding the small mitoribosomal subunit protein bS1m in a patient with intrauterine growth retardation, craniofacial dysmorphism and multisystemic involvement. Hum Mol Genet 28(9):1445–1462
Bugiardini E, Mitchell AL, Rosa ID et al (2019) MRPS25 mutations impair mitochondrial translation and cause encephalomyopathy. Hum Mol Genet 28(16):2711–2719
Borna NN, Kishita Y, Kohda M et al (2019) Mitochondrial ribosomal protein PTCD3 mutations cause oxidative phosphorylation defects with Leigh syndrome. Neurogenetics 20(1):9–25
Di Nottia M, Marchese M, Verrigni D et al (2020) A homozygous MRPL24 mutation causes a complex movement disorder and affects the mitoribosome assembly. Neurobiol Dis 141:104880
Gardeitchik T, Mohamed M, Ruzzenente B et al (2018) Bi-allelic mutations in the mitochondrial ribosomal protein MRPS2 cause sensorineural hearing loss, hypoglycemia, and multiple OXPHOS complex deficiencies. Am J Hum Genet 102(4):685–695. https://doi.org/10.1016/j.ajhg.2018.1002.1012. Epub 2018 Mar 1022
O’Brien TW, O’Brien BJ, Norman RA (2005) Nuclear MRP genes and mitochondrial disease. Gene 354:147–151
Saada A, Shaag A, Arnon S et al (2007) Antenatal mitochondrial disease caused by mitochondrial ribosomal protein (MRPS22) mutation. J Med Genet 44(12):784–786
Emdadul Haque M, Grasso D, Miller C et al (2008) The effect of mutated mitochondrial ribosomal proteins S16 and S22 on the assembly of the small and large ribosomal subunits in human mitochondria. Mitochondrion 8(3):254–261
Rebelo-Guiomar P, Pellegrino S, Dent KC et al (2022) A late-stage assembly checkpoint of the human mitochondrial ribosome large subunit. Nat Commun 13(1):929. https://doi.org/10.1038/s41467-022-28503-5
Klinge S, Woolford JL Jr (2019) Ribosome assembly coming into focus. Nat Rev Mol Cell Biol 20(2):116–131. https://doi.org/10.1038/s41580-018-0078-y
Srivastava AK, Schlessinger D (1990) Mechanism and regulation of bacterial ribosomal RNA processing. Annu Rev Microbiol 44:105–129. https://doi.org/10.1146/annurev.mi.44.100190.000541
Montoya J, Christianson T, Levens D et al (1982) Identification of initiation sites for heavy-strand and light-strand transcription in human mitochondrial DNA. Proc Natl Acad Sci U S A 79(23):7195–7199
Terzioglu M, Ruzzenente B, Harmel J et al (2013) MTERF1 binds mtDNA to prevent transcriptional interference at the light-strand promoter but is dispensable for rRNA gene transcription regulation. Cell Metab 17(4):618–626. https://doi.org/10.1016/j.cmet.2013.03.006
Litonin D, Sologub M, Shi Y et al (2010) Human mitochondrial transcription revisited: only TFAM and TFB2M are required for transcription of the mitochondrial genes in vitro. J Biol Chem 285(24):18129–18133. https://doi.org/10.1074/jbc.C110.128918
Siira SJ, Rossetti G, Richman TR et al (2018) Concerted regulation of mitochondrial and nuclear non-coding RNAs by a dual-targeted RNase Z. EMBO Rep 19(10). https://doi.org/10.15252/embr.201846198
Jedynak-Slyvka M, Jabczynska A, Szczesny RJ (2021) Human mitochondrial RNA processing and modifications: overview. Int J Mol Sci 22(15). https://doi.org/10.3390/ijms22157999
Uchiumi T, Ohgaki K, Yagi M et al (2010) ERAL1 is associated with mitochondrial ribosome and elimination of ERAL1 leads to mitochondrial dysfunction and growth retardation. Nucleic Acids Res 38(16):5554–5568
Seidel-Rogol BL, McCulloch V, Shadel GS (2003) Human mitochondrial transcription factor B1 methylates ribosomal RNA at a conserved stem-loop. Nat Genet 33(1):23–24
Szczepanowska K, Maiti P, Kukat A et al (2016) CLPP coordinates mitoribosomal assembly through the regulation of ERAL1 levels. EMBO J 35(23):2566–2583
Summer S, Smirnova A, Gabriele A et al (2020) YBEY is an essential biogenesis factor for mitochondrial ribosomes. Nucleic Acids Res 48(17):9762–9786. https://doi.org/10.1093/nar/gkaa148
Kolanczyk M, Pech M, Zemojtel T et al (2011) NOA1 is an essential GTPase required for mitochondrial protein synthesis. Mol Biol Cell 22(1):1–11
He J, Cooper HM, Reyes A et al (2012) Human C4orf14 interacts with the mitochondrial nucleoid and is involved in the biogenesis of the small mitochondrial ribosomal subunit. Nucleic Acids Res 40:6097–6108
Reyes A, Favia P, Vidoni S et al (2020) RCC1L (WBSCR16) isoforms coordinate mitochondrial ribosome assembly through their interaction with GTPases. PLoS Genet 16(7):e1008923
Van Haute L, Hendrick AG, D’Souza AR et al (2019) METTL15 introduces N4-methylcytidine into human mitochondrial 12S rRNA and is required for mitoribosome biogenesis. Nucleic Acids Res 47(19):10267–10281. https://doi.org/10.1093/nar/gkz735
Fung S, Nishimura T, Sasarman F et al (2013) The conserved interaction of C7orf30 with MRPL14 promotes biogenesis of the mitochondrial large ribosomal subunit and mitochondrial translation. Mol Biol Cell 24(3):184–193
Rorbach J, Gammage PA, Minczuk M (2012) C7orf30 is necessary for biogenesis of the large subunit of the mitochondrial ribosome. Nucleic Acids Res 40:4097–4109
Popow J, Alleaume AM, Curk T et al (2015) FASTKD2 is an RNA-binding protein required for mitochondrial RNA processing and translation. RNA 21(11):1873–1884. https://doi.org/10.1261/rna.052365.115
Kim H-J, Barrientos A (2018) MTG1 couples mitoribosome large subunit assembly and intersubunit bridge formation. Nucleic Acid Res 46:8435
Maiti P, Antonicka H, Gingras A-C et al (2020) Human GTPBP5 (MTG2) fuels mitoribosome large subunit maturation by facilitating 16S rRNA methylation. Nucleic Acids Res 48(14):7924–7943
Lee KW, Okot-Kotber C, Lacomb JF et al (2013) Mitochondrial rRNA methyltransferase family members are positioned to modify nascent rRNA in foci near the mtDNA nucleoid. J Biol Chem 288(43):31386–31399
Nikolay R, Hilal T, Schmidt S et al (2021) Snapshots of native pre-50S ribosomes reveal a biogenesis factor network and evolutionary specialization. Mol Cell 81(6):1200–1215.e1209. https://doi.org/10.1016/j.molcel.2021.02.006
Zaganelli S, Rebelo-Guiomar P, Maundrell K et al (2017) The pseudouridine synthase RPUSD4 is an essential component of mitochondrial RNA granules. J Biol Chem 292(11):4519–4532
Lavdovskaia E, Denks K, Nadler F et al (2020) Dual function of GTPBP6 in biogenesis and recycling of human mitochondrial ribosomes. Nucleic Acids Res. https://doi.org/10.1093/nar/gkaa1132
Kim HJ, Maiti P, Barrientos A (2017) Mitochondrial ribosomes in cancer. Semin Cancer Biol 47:67–81
Ban N, Beckmann R, Cate JH et al (2014) A new system for naming ribosomal proteins. Curr Opin Struct Biol 24:165–169. https://doi.org/10.1016/j.sbi.2014.01.002
Metodiev MD, Lesko N, Park CB et al (2009) Methylation of 12S rRNA is necessary for in vivo stability of the small subunit of the mammalian mitochondrial ribosome. Cell Metab 9(4):386–397
Liu X, Shen S, Wu P et al (2019) Structural insights into dimethylation of 12S rRNA by TFB1M: indispensable role in translation of mitochondrial genes and mitochondrial function. Nucleic Acids Res 47(14):7648–7665. https://doi.org/10.1093/nar/gkz505
Powell CA, Minczuk M (2020) TRMT2B is responsible for both tRNA and rRNA m(5)U-methylation in human mitochondria. RNA Biol 17(4):451–462. https://doi.org/10.1080/15476286.2020.1712544
Laptev I, Shvetsova E, Levitskii S et al (2020) Mouse Trmt2B protein is a dual specific mitochondrial metyltransferase responsible for m(5)U formation in both tRNA and rRNA. RNA Biol 17(4):441–450. https://doi.org/10.1080/15476286.2019.1694733
Shi Z, Xu S, Xing S et al (2019) Mettl17, a regulator of mitochondrial ribosomal RNA modifications, is required for the translation of mitochondrial coding genes. FASEB J 33(11):13040–13050. https://doi.org/10.1096/fj.201901331R
Tang T, Zheng B, Chen SH et al (2009) hNOA1 interacts with complex I and DAP3 and regulates mitochondrial respiration and apoptosis. J Biol Chem 284(8):5414–5424. https://doi.org/10.1074/jbc.M807797200
Bar-Yaacov D, Frumkin I, Yashiro Y et al (2016) Mitochondrial 16S rRNA is methylated by tRNA methyltransferase TRMT61B in all vertebrates. PLoS Biol 14(9):e1002557. https://doi.org/10.1371/journal.pbio.1002557
Arroyo JD, Jourdain AA, Calvo SE et al (2016) A genome-wide CRISPR death screen identifies genes essential for oxidative phosphorylation. Cell Metab 24(6):875–885. https://doi.org/10.1016/j.cmet.2016.08.017
Rathore A, Chu Q, Tan D et al (2018) MIEF1 microprotein regulates mitochondrial translation. Biochemistry 57(38):5564–5575
Dibley MG, Formosa LE, Lyu B et al (2020) The mitochondrial acyl-carrier protein interaction network highlights important roles for LYRM family members in complex i and mitoribosome assembly. Mol Cell Proteomics 19(1):65–77. https://doi.org/10.1074/mcp.RA119.001784
Wredenberg A, Lagouge M, Bratic A et al (2013) MTERF3 regulates mitochondrial ribosome biogenesis in invertebrates and mammals. PLoS Genet 9(1):e1003178
Camara Y, Asin-Cayuela J, Park CB et al (2011) MTERF4 regulates translation by targeting the methyltransferase NSUN4 to the mammalian mitochondrial ribosome. Cell Metab 13(5):527–539
Fogal V, Richardson AD, Karmali PP et al (2010) Mitochondrial p32 protein is a critical regulator of tumor metabolism via maintenance of oxidative phosphorylation. Mol Cell Biol 30(6):1303–1318. https://doi.org/10.1128/mcb.01101-09
Yagi M, Uchiumi T, Takazaki S et al (2012) p32/gC1qR is indispensable for fetal development and mitochondrial translation: importance of its RNA-binding ability. Nucleic Acids Res 40(19):9717–9737. https://doi.org/10.1093/nar/gks774
Acknowledgments
This research was supported by an NIGMS-MIRA award [R35GM118141 to A.B.] and an NICHD F30 predoctoral M.D./Ph.D. fellowship [F30HD107939 to J.C.M.].
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Science+Business Media, LLC, part of Springer Nature
About this protocol
Cite this protocol
Conor Moran, J., Del’Olio, S., Choi, A., Zhong, H., Barrientos, A. (2023). Mitoribosome Biogenesis. In: Barrientos, A., Fontanesi, F. (eds) The Mitoribosome. Methods in Molecular Biology, vol 2661. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-3171-3_3
Download citation
DOI: https://doi.org/10.1007/978-1-0716-3171-3_3
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-3170-6
Online ISBN: 978-1-0716-3171-3
eBook Packages: Springer Protocols