Abstract
Pentatricopeptide repeat domain proteins are a large family of RNA-binding proteins involved in mitochondrial RNA editing, stability, and translation. Mitochondrial translation machinery defects are an expanding group of genetic diseases in humans. We describe a patient who presented with low birth weight, mental retardation, and optic atrophy. Brain MRI showed abnormal bilateral signals at the basal ganglia and brainstem, and the patient was diagnosed as Leigh syndrome. Exome sequencing revealed two potentially loss-of-function variants [c.415-2A>G, and c.1747_1748insCT (p.Phe583Serfs*3)] in PTCD3 (also known as MRPS39). PTCD3, a member of the pentatricopeptide repeat domain protein family, is a component of the small mitoribosomal subunit. The patient had marked decreases in mitochondrial complex I and IV levels and activities, oxygen consumption and ATP biosynthesis, and generalized mitochondrial translation defects in fibroblasts. Quantitative proteomic analysis revealed decreased levels of the small mitoribosomal subunits. Complementation experiments rescued oxidative phosphorylation complex I and IV levels and activities, ATP biosynthesis, and MT-RNR1 rRNA transcript level, providing functional validation of the pathogenicity of identified variants. This is the first report of an association of PTCD3 mutations with Leigh syndrome along with combined oxidative phosphorylation deficiencies caused by defects in the mitochondrial translation machinery.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The biogenesis of the mitochondrial oxidative phosphorylation (OXPHOS) system is complex owing to its dual control by the nuclear and mitochondrial genomes, and the proper function of mitochondria relies on the coordinated expression of both genomes [1]. Mitochondrial translation defects generally result in combined deficiency of multiple OXPHOS complexes, and decreased ATP production and cellular energy metabolism [2]. Mitochondrial gene expression is predominantly regulated by nuclear-encoded mitochondrial RNA-binding proteins (RBPs) that regulate RNA from transcription to degradation [3, 4].
The pentatricopeptide repeat (PPR) domain protein family is a helical repeat motif family of RBPs that are required for the regulation of mitochondrial gene expression at the post-transcriptional level [5, 6]. To date, seven PPR proteins have been identified in mammals, all of which are localized within the mitochondrial matrix [7]. These PPR proteins carry out essential roles in the regulation of transcription, mitochondrial RNA processing, stability, editing, protein synthesis, and cellular respiration [3].
Pentatricopeptide repeat domain-containing protein-3 (PTCD3), also known as MRPS39, is one of the mammalian-specific mitochondrial ribosomal supernumerary proteins [8]. The PTCD3 protein belongs to the PPR family, it has 15 PPR domains with an N-terminal mitochondrial targeting sequence and localizes in the mitochondria [9]. Cryo-electron microscopy studies of the bovine and human mitoribosome structures confirmed that PTCD3 is the largest protein component of the small mitoribosomal subunit (mt-SSU) [10,11,12]. PTCD3 resides at the head of mt-SSU and is adjacent to the entrance of the mRNA channel, which may enable it to guide mitochondrial mRNAs into the ribosomal mRNA channel at the entry site, playing a role in regulating translation initiation [10, 11, 13]. A PTCD3 knockdown study showed no effect on RNA metabolism, but protein synthesis was severely disrupted and caused an overall OXPHOS deficiency [9].
Leigh syndrome (OMIM 256000) is a well-recognized neurodegenerative metabolic disorder that usually has an infantile-onset [14]. Patients with Leigh syndrome present highly variable clinical features, involving the central nervous system, heart, muscle, liver, gastrointestinal, and renal tubular functions [15]. This disease was shown to be caused by mutations in genes encoding the structural components, assembly and regulatory factors of OXPHOS complexes, or electron carrier, protein transporters, and proteins involving cellular energy metabolism [16]. To date, mutations in more than 75 genes have been associated with this disease [17].
To date, PTCD3 has not been identified as a causative gene of mitochondrial disease. However, in this study, we demonstrate biallelic PTCD3 mutations in a Japanese patient with Leigh syndrome resulted in frameshift changes that generated premature stop codons causing loss-of-function of the PTCD3 protein, translational defects in mitochondrial DNA-encoded (mtDNA) protein, combined OXPHOS deficiency, and destabilization of the mt-SSU. Our findings show that PTCD3 mutations are associated with mitochondrial ribosomal protein defects, causing neurodegenerative disease and premature death.
Materials and methods
Whole-exome sequencing and Sanger sequence validation
Whole-exome library preparation and sequencing were performed using methods and a bioinformatic filtering pipeline ensuring the analysis of mtDNA as previously published [18]. Briefly, sequencing was performed using 100-bp paired-end reads on a HiSeq2500 (Illumina). The NCBI human genome reference (GRCh37/hg19) was used to align the sequencing reads. For validation of prioritized variants and haplotype phasing, Sanger sequencing was performed on genomic DNA (gDNA) prepared from the fibroblasts of patient and blood of family members using ABI3130XL and BigDye v3.1 Terminators (Applied Biosystems) system as per the manufacturer’s protocols. Sequencing primers are listed in Supplementary Table S1.
Cell culture
All cells were cultured at 37 °C and 5% CO2 in Dulbecco’s modified Eagle’s medium (DMEM; Nacalai Tesque Inc.) supplemented with 10% fetal bovine serum (Sigma-Aldrich) and 1% penicillin-streptomycin (Nacalai Tesque Inc.). Fetal human dermal fibroblasts (C1; Toyobo) and neonatal human dermal fibroblasts (C2; Toyobo) from healthy individuals and HEK293FT (C3; Invitrogen) were used as controls.
RNA extraction and cDNA synthesis
Total RNA was isolated from fibroblasts using TRIzol™ reagent (Invitrogen) as per manufacturer’s instructions. DNA was removed from RNA samples by RNase-free DNase I (Thermo Fisher Scientific). cDNA was synthesized from total RNA using ReverTra Ace reverse transcriptase (Toyobo). PCR was performed to amplify PTCD3 exons 1–10 from control and patient cDNA to investigate the pathogenicity of c.415-2A>G on mRNA splicing. Gel-purification of PCR products was done using Wizard SV gel and PCR clean-up system (Promega). The purified PCR products were sequenced using ABI3130XL and BigDye v3.1 Terminators (Applied Biosystems) system.
Quantitative reverse transcription PCR
qRT-PCR was performed to quantify the levels of PTCD3, MT-RNR1 (12S rRNA), and MT-RNR2 (16S rRNA) transcripts using Power SYBR Green PCR Master Mix (Life Technologies) and Mx3000P (Agilent Technologies). The mRNA expression levels were calculated relative to the mean expression levels of ACTB or GAPDH. Primers for qRT-PCR are listed in Supplementary Table S1.
Complementation assay
Patient and control fibroblasts were transduced with a lentiviral mammalian expression vector system expressing mitochondria-targeted red fluorescent protein TurboRFP (RFP‚ Evrogen) or wild-type PTCD3 (PTCD3wt) cDNA (NM_017952.5). The lentiviral vector CS-CA-MCS and In-Fusion HD Cloning Kit (Clontech Laboratories) were used to construct plasmids with gene of interest. The constructed plasmids [pCS-CA-MCS (candidate gene)-blast] contained a CAG promoter for mammalian cell expression, with or without a C-terminal V5 tag sequence, and blasticidin resistance gene as a selective marker. 2 × 106 HEK293FT cells were seeded in each 6-cm plates and co-transfected with ViraPower Packaging vectors (pLP1, pLP2, pLP/VSVG; Invitrogen) and a pCS-CA-ORF (candidate gene)-blast vector. Transfection was performed using Lipofectamine 2000 (Invitrogen). Primer sequences used to clone PTCD3 are listed in Supplementary Table S1.
OXPHOS enzyme activity assays
Spectrophotometric enzyme activity assays using mitochondria of fibroblasts and lentiviral-mediated transduced samples were performed as previously described [19]. OXPHOS enzyme activities were measured using Cary300 (Agilent Technologies) as per manufacturer’s instructions and were expressed as percentages of citrate synthase activity. Protein concentration was determined by the bicinchoninic acid assay (Pierce™ BCA Protein Assay Kit, Thermo Fisher Scientific).
SDS-PAGE, BN-PAGE, and immunoblotting
Mitochondrial extracts were prepared from fibroblasts as described previously [18]. Total cell lysates (TCL) were prepared using M-PER™ Mammalian Protein Extraction Reagent (Thermo Fisher Scientific). Ten to twenty micrograms mitochondria or TCL proteins were separated by electrophoresis on 7 or 10% SDS-PAGE gels, depending on the size of the protein of interest.
BN-PAGE was performed to separate individual OXPHOS complexes, supercomplexes, and mitoribosomal complexes. Mitochondrial fractions were solubilized in 1% Triton X-100 or 1% digitonin and separated on 4–16% NativePAGE Novex Bis-Tris Gel System (Life Technologies). Proteins were transferred onto PVDF membrane (GE Healthcare) using a semi-dry method. Following membrane blocking and antibody incubations, proteins of interest were detected using ECL reagents (GE Healthcare). Details of antibodies are provided in the Supplementary Methods section.
Microscale oxygraphy analysis
The oxygen consumption rates were analyzed by microscale oxygraphy with the Seahorse XF96 extracellular flux analyzer (Agilent technologies). Fibroblasts from patient or control were seeded in a 96-well plate at 2 × 104 cells/well with 80 μl of growth medium containing 25 mM glucose (Glu), and incubated for 24 h (37 °C, 5%CO2). After replacing the medium with 160 μL of unbuffered DMEM containing 1 mM sodium pyruvate, 2 mM glutamine, and 25 mM glucose or 10 mM galactose (Gal), the assay plates were incubated at 37 °C without CO2 for 1 h. Following the calibration of the sensor cartridge loaded with compounds including oligomycin (2 μM final concentration), carbonyl cyanide phenylhydrazone (FCCP, 0.4 μM final concentration), and rotenone (1 μM final concentration), experiments were started. The data obtained were normalized to the cell numbers determined using CyQUANT Cell Proliferation kit (Invitrogen).
In vitro metabolic labeling for mitochondrial translation products
Pulse labeling of mitochondrial translation products in control and patient fibroblasts was performed as previously described [20] with some modifications. When the cells were 90% confluent they were incubated in methionine-free DMEM with 100 μg/ml emetine (Enzo life science) to inhibit cytoplasmic protein translation for 15 min at 37 °C. Radiolabeling was performed with [35S]-methionine/[35S]-cysteine (11 mCi/ml‚ EXPRE35S35S[35S]-Protein Labeling Mix; PerkinElmer) at a concentration of 1 μl per ml methionine-free DMEM for 1 h at 37 °C. Then, 0.1 mM unlabeled methionine (Sigma-Aldrich) was added to the cells for 15 min incubation at 37 °C. Cells were then harvested and TCL were prepared using M-PER reagent. Seventy micrograms of protein was separated using a 12–18% polyacrylamide Tris-tricine gradient gel [20]. After separation, the protein-containing gel was dried using a gel dryer (Bio-Rad Laboratories) and exposed to a phosphor storage screen. Radiolabeled proteins were detected using a Typhoon Laser Scanner (GE Healthcare).
Proteomic analysis
Preparation of peptide samples for mass spectrometry analysis
Peptide samples for mass spectrometry were prepared similarly to that described previously [21]. Cells and mitochondrial extracts were suspended in modified RIPA buffer and disrupted using sonication. Extracted proteins (50 μg) in 100 μL of RIPA buffer were precipitated with 6 volumes of acetone at − 20 °C overnight. The precipitated proteins were pelleted by centrifugation at 17,400×g for 20 min and the proteins were suspended in 100 μL of 8 M urea, and 0.1 M NH4HCO3 buffer. Proteins were reduced with 10 mM DTT for 30 min at 37 °C and then alkylated with 40 mM iodoacetamide at 25 °C for 45 min, in the dark. Mass spectrometry-grade lysyl endopeptidase (Wako) was added at a 1:40 (w/w) enzyme/substrate concentration and the proteins were digested at 37 °C for 4 h. Subsequently, the samples were diluted with 0.1 M NH4HCO3 buffer to a final concentration of 1.5 M urea. Sequencing-grade trypsin (Promega) was added at a 1:50 (w/w) enzyme/substrate concentration and the samples were digested at 37 °C for 16 h. The digested protein samples were acidified to pH 2–3 with 10% (v/v) trifluoroacetic acid. The samples were desalted using MonoSpin C18 columns (GL Sciences) and the peptides were eluted from C18 in 0.1% (v/v) trifluoroacetic acid, and 50% (v/v) acetonitrile. The peptides were dried using a Genevac miVac DUO concentrator (SP Scientific). The dried peptides were suspended in 50 μL of 0.1% (v/v) formic acid, and 3% (v/v) acetonitrile for mass spectrometry analysis.
Mass spectrometry acquisition and data analysis
Every sample was measured in both data-dependent and data-independent modes performed on the Q-Exactive Plus mass spectrometer (Thermo Fisher Scientific), similarly to the approach previously described [22]. Alternatively, the peptides were separated by a 2-h linear gradient from 2 to 34% buffer B (0.1% formic acid, 80% acetonitrile) at 300 nl/min, followed by a linear increase to 95% buffer B in 2 min and then maintenance at 95% for 10 min. For data-dependent mass spectrometry (DDA-MS), profile mode raw files from the DIA-MS were searched against the canonical Uniprot complete database for mouse using Proteome Discoverer 2.2. Cysteine carbamidomethylation was set as a static modification. Methionine oxidation and protein N-terminal acetylation were set as variable modifications. A peptide spectral library was generated using the results from DDA-MS with the spectral library generation function in Spectronaut, similar to SpectraST [23]. For data-independent mass spectrometry (DIA-MS), the DIA method consisted of a survey scan from 400 to 1200 m/z at 70,000 resolution (AGC target 5e6, maximum injection time 120 ms) and 32 DIA windows at 35,000 resolution (AGC target 3e6 and auto for injection time). Stepped collision energy was 27%. The DIA-MS targeted data extraction was performed using Spectronaut, which applies a target-decoy model to estimate the false discovery rate (FDR) using the mProphet algorithm [24]. Peptide features were retained to reach the 1% FDR threshold. Protein abundances were estimated using the transitions from the first to third most intensive peptides and normalized with the total protein abundance in each sample. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE [25] partner repository (https://www.ebi.ac.uk/pride/) with the dataset identifier PXD010903.
Statistics
Results are presented as mean ± SEM for the number of experiments indicated in the figure legends. Statistical analysis was performed using two-tailed Student’s t test or one sample t test, as appropriate. *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001 were considered statistically significant.
Results
Clinical course and neuroimaging features
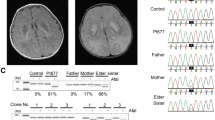
The patient was the second child of nonconsanguineous Japanese parents. Her growth retardation was identified at 24 weeks of the intrauterine stage. She was unable to gain weight and was delivered by an emergency caesarian section at 30 weeks of the gestational period. Her weight at birth was 632 g (− 4.26SD), height 28 cm (− 4.7SD), head circumference 23.2 cm (− 2.3SD), and Apgar score 5–7 (1′−5′). Soon after birth, she was under ventilatory support, and gradually developed limb rigidity, myoclonus, nystagmus, and psychomotor regression. She was hospitalized due to respiratory syncytial virus infection at 7 months of age and developed bronchitis at 13 months, for which she needed assisted ventilation, nasogastric tube feeding, and was treated with per oral clonazepam. At 14 months of age, she developed aspiration pneumonia; at that time, her height was 58.0 cm (− 7.1SD) and weight 5.05 kg (− 4.4SD). Analysis of OXPHOS complexes in cultured fibroblasts showed severe reduction in complex I (CI) activity, and decrease in CI, complex III (CIII), and complex IV (CIV) protein levels. Brain MRI showed bilateral symmetrical punctate lesions involving ventral side of the medulla, cerebral peduncle (Fig. 1a, b), and symmetrical hyperintense lesions involving the thalamus, caudate nucleus, and putamen of the basal ganglia (Fig. 1c, d), along with mild cerebral atrophy. The patient also developed bilateral optic atrophy and severe bilateral hearing loss. She had a progressive clinical course with typical brain MRI findings that were diagnostic of Leigh syndrome; she died of respiratory failure at 1 year 4 months old. No autopsy was performed.
Brain MRI of the patient. a, b Axial T2-weighted images of the brainstem showing symmetrical punctate lesions involving the ventral side of the medulla (arrows in a) and cerebral peduncle (arrows in b). c, d Axial T2-weighted images of the basal ganglia showing symmetrical hyperintense lesions involving the thalamus (arrows in c), caudate nucleus, and putamen (arrows in d)
Identification of PTCD3 frameshift variants in Leigh syndrome patient
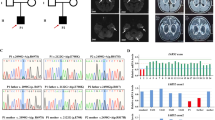
Whole-exome sequencing analysis was performed using patient DNA to identify the disease-causing gene. To narrow down the list of candidate genes from the exome sequencing data, variant filtering and prioritization were performed based on the strategy published previously where variants common in public DNA databases were removed and genes listed in MitoCarta2.0 were prioritized [18, 26]. We identified a single candidate gene PTCD3 (NM_017952.5), with heterozygous variants c.415-2A>G and c.1747_1748insCT. Sanger sequencing confirmed the autosomal-recessive PTCD3 variants in the patient’s parents (Fig. 2a), and the wild-type alleles in two siblings (Fig. 2b).
Identification of PTCD3 variants and gene structure. a Pedigree of the patient’s family with variants in the PTCD3 gene. b Sanger sequencing of gDNA from the affected individual and unaffected family members confirming the PTCD3 variants. The NCBI human genome reference (GRCh38/hg38) was used to align the sequencing data. c A graphical representation of the PTCD3 gene structure illustrating the mutations in gDNA, mRNA, and PTCD3 protein (not to scale). The illustration also shows the truncated protein (predicted using SmartBLAST) due to the c.1747_1748insCT variant. The following abbreviations are used: WT, wild-type; MLS, mitochondrial localization signal; UTR, untranslated region
The PTCD3 variant c.415-2A>G is reported in the Genome Aggregation Database (gnomAD) (in 1 of 243,016 alleles examined) with a very low minor allele frequency (< 0.0001) among the East Asian population [27]. This variant is not reported in the Exome Aggregation Consortium (ExAC) [27] and the Japanese population reference panel (3.5KJPN) [28]. PCR and sequencing of PTCD3 cDNA from exons 1–10 confirmed the presence of an alternative splice variant lacking exon 7 (Fig. 3a). This exon 7-skipped transcript was predicted to be caused by the abolishment of the consensus 3′ acceptor splice site in intron 6 and the use of acceptor site in intron 7, resulting in a shorter protein (Fig. 3b). This splice variant is reported in the Ensembl genome browser (ENST00000409277.3, GRCh37/hg19) [29] but expression level is low in all tissues analyzed in the Genotype-Tissue Expression (GTEx Analysis Release V7) database [30].
Characterization of PTCD3 variants in patient’s fibroblasts. a PCR products from the cDNA of C1 and patient’s (Pt) fibroblasts were generated by amplifying PTCD3 exons 1–10 to analyze splice site (c.415-2A>G) variant. Gel electrophoresis of amplified products showed two bands of different molecular weights in the patient. Sanger sequencing of gel-purified PCR products showed the top band corresponds to the amplicon observed in C1 and lower band corresponds to a splice variant lacking exon 7. M-5 represents molecular marker 5. b Schematic illustration showing the abnormally spliced transcript generated from the c.415-2A>G allele. The red letter indicates the mutated nucleotide and solid ash bars represent exons. c SDS-PAGE immunoblot analysis of PTCD3 in TCL and mitochondrial (MT) extracts from of C1, C2, and Pt fibroblasts showed the lower abundance of PTCD3 protein in the patient. ACTB (β-actin) and HSP60 antibodies were used as loading controls. d qRT-PCR revealed lower expression of the PTCD3 transcript in the patient. ACTB was used as an endogenous control. All values are reported as mean ± SEM (n = 3). ***p < 0.001, calculated by Student’s t test. e The expression of mature mitochondrial rRNAs was measured by qRT-PCR and expressed relatively to GAPDH. MT-RNR1 expression was reduced in the patient compared with that in the controls. All values are reported as mean ± SEM (n = 3). *p < 0.05, **p < 0.01, and ***p < 0.001 are considered as statistically significant
The patient was also heterozygous for the c.1747_1748insCT variant in exon 21, predicted to cause a premature stop codon (p.Phe583Serfs*3), resulting in a shorter mutant protein (Fig. 2c). There are no reports of this variant in the ExAC, 3.5KJPN, or gnomAD databases.
The PTCD3 variants are associated with reduced expression of PTCD3 protein, and PTCD3 and MT-RNR1 transcripts
SDS-PAGE and immunoblotting showed a reduced level of PTCD3 protein in patient’s fibroblasts (Fig. 3c). qRT-PCR revealed a reduced steady-state level of PTCD3 mRNA (Fig. 3d). Reduced expression of MT-RNR1 and increased expression of MT-RNR2 which is a component of the large mitoribosomal subunit (mt-LSU) (Fig. 3e) were also observed, suggesting PTCD3 is necessary for MT-RNR1 transcript stability and its loss leads to upregulation of the MT-RNR2 transcript.
PTCD3 is required for the normal levels of OXPHOS CI and CIV, and CI/CIII2/CIV supercomplexes
We performed SDS-PAGE immunoblotting of patient’s mitochondria with antibodies specific for OXPHOS subunits which revealed a loss of mtDNA-encoded CIV subunit COXII and nDNA-encoded CI subunit NDUFB8 in the patient, while the levels of other OXPHOS complex subunits were comparable with those of the controls (Fig. 4a).
Loss of PTCD3 impairs the abundance of OXPHOS complexes, mt-SSU stability, and mitochondrial translation. a SDS-PAGE analysis of the OXPHOS complex subunits in controls’ and patient’s mitochondria showed significant loss of CIV, as well as CI subunits. A positive control (PC) of mitochondria from an MRPS23 patient [18] was also shown. b BN-PAGE analysis revealed reduced amounts of fully assembled CI and IV in patient fibroblasts. Antibodies specific for mitochondrial complexes CI, CII, CIII, and CIV were used. CII was used as a loading control. c BN-PAGE immunoblotting revealed the destabilization of supercomplexes CI/CIII2/CIV1–4 and CI/CIII2, and a reduced level of CIV in mitochondrial proteins. Antibodies were used as mentioned above (Fig. 4b). TOM40 was used as a loading control. d BN-PAGE immunoblot analysis of mt-SSU and mt-LSU complexes showed reduced abundance of the mt-SSU proteins, but the abundance of mt-LSU was unaffected in the patient. Proteins were solubilized in 1% digitonin and probed by antibodies specific for the mt-SSU (PTCD3, MRPS29, and MRPS23) and the mt-LSU (MRPL11 and MRPL37). e Spectrophotometric analysis of OXPHOS enzyme activities showed significantly reduced CI and CIV activities in the patient. All values are reported as mean ± SEM (n = 3), ***p < 0.001. f De novo synthesis of mtDNA-encoded proteins analyzed by [35S] methionine/cysteine labeling demonstrated significant translational inhibition of proteins in the patient compared to the control individuals. Coomassie Brilliant Blue (CBB) staining was used to confirm equal loading of the samples
We also observed a reduced abundance of fully assembled CI and CIV, whereas the levels of CIII and the nuclear-encoded complex II (CII) remain unchanged in patient (Fig. 4b). Increased efficiency of the OXPHOS complexes is dependent on supercomplex formation [31]; therefore, we further analyzed the stability of OXPHOS supercomplexes using milder detergent digitonin in BN-PAGE. The supercomplexes are formed by the association of one CI, a CIII dimer (CIII2), and one to four copies of CIV (CIV1–4). This analysis revealed that the stability of the CI/CIII2, and CI/CIII2/CIV1–4 supercomplexes was compromised in the patient (Fig. 4c). However, there was an increased level of CIII2/CIV supercomplex and CIII2 in the patient (Fig. 4c).
The enzyme activities of CI and CIV were also significantly reduced in the patient’s fibroblasts (Fig. 4e), consistent with the reduced levels of CI and CIV proteins relative to that in controls (Fig. 4b, c). From these findings, we can conclude that OXPHOS complexes were impaired in the patient due to the loss of PTCD3.
PTCD3 is required for mt-SSU stability and mitochondrial translation
To evaluate whether the decreased level of PTCD3 protein affects the mitoribosome, we performed BN-PAGE analysis to detect mt-SSU and mt-LSU complexes in the controls’ and patient’s fibroblasts. A recent study showed the apparent molecular masses of mt-SSU and mt-LSU are ~ 3 MDa [32]. We found a decreased abundance of the mt-SSU in the patient, whereas the levels of mt-LSU were unaffected (Fig. 4d). The mt-SSU and mt-LSU were detected by using antibodies against PTCD3, MRPS29 and MRPS23, and MRPL11 and MRPL37, respectively (Fig. 4d).
To test whether the reduced level of mt-SSU complex impairs de novo mitochondrial protein synthesis, we performed in vitro pulse labeling of mitochondrial translation products with [35S]-methionine/cysteine in the presence of emetine, an inhibitor of cytosolic protein translation. We observed a generalized defect in the translation of mtDNA-encoded polypeptides in the patient’s fibroblasts compared to controls (Fig. 4f), suggesting a vital role of PTCD3 in mitochondrial translation. Overall, these results validate that loss of PTCD3 decreases the level of mt-SSU, and impairs mitochondrial protein synthesis, leading to combined OXPHOS defects.
The PTCD3 variants lead to reduced mitochondrial respiration
In Gal-based medium, cells depend mostly on OXPHOS to produce their ATP for survival and are more susceptible to mitochondrial toxins [33, 34]. Patient-derived fibroblasts with mitochondrial dysfunction cannot survive when cultured in Gal medium [35]. Thus, to study mitochondrial dysfunction, culturing of cells in Gal appears to be a good alternative to Glu medium. A microrespirometry assay on intact cells using flux analyzer was performed to evaluate the functional bioenergetic capacity of the patient’s fibroblasts under Glu or Gal medium, and reduced mitochondrial respiration was observed in the patient fibroblasts under both conditions (Fig. 5a).
PTCD3 variants cause dysfunctional mitochondrial respiration and ATP production in patient’s fibroblasts. a Microscale oxygraphy analysis of fibroblasts cultured in Glu and Gal medium was performed with sequential injections of oligomycin, FCCP, and rotenone; measurement was performed for ≥ 14 technical replicates in each conditions. b Mitochondrial ATP-linked OCR in control and patient fibroblasts is expressed as per 2 × 104 cells. Data are presented as mean ± SEM; **p < 0.01, ***p < 0.001, compared with C1, calculated by two-tailed Student’s t test. Galactose/glucose ratio represents the ratio of ATP-linked OCR in Gal and Glu medium. c Maximal respiration was significantly reduced (p < 0.001) in the patient’s fibroblasts in both Glu and Gal culture conditions. Data are presented as mean ± SEM; ***p < 0.001, compared with C1, calculated by two-tailed Student’s t test. d ECAR in fibroblasts from the patient was significantly increased compared with that in the controls. ***p < 0.001 compared with C1, calculated by Student’s t test. e The coupling efficiency of respiration was considerably reduced in patient’s fibroblasts compared to controls. ***p < 0.001, compared with C1, calculation was done by two-tailed Student’s t test
Oligomycin is an inhibitor of ATP synthase, and ATP-linked oxygen consumption rate (OCR) can be calculated by OCRbasal-OCRoligomycin. The ATP-linked OCR was significantly reduced in the patient’s cells compared with that in the C1 in Glu and Gal medium (Fig. 5b), and the ratio of ATP-linked OCR in Gal versus Glu medium was also reduced significantly in patient’s cells (Fig. 5b). We carried out additional measurements of ATP concentration in patient fibroblasts cultured in Glu and Gal medium, using a luciferase/luminscence assay with luciferin as the substrate. This assay confirmed the significant reductions in ATP production in patient’s fibroblasts (Supplementary Fig. S1a).
The addition of FCCP, an ionophore that directly transports protons across the inner mitochondrial membrane, enabled an estimation of the maximal OCR (OCRFCCP-OCRrotenone), which is indicative of the functional capacity of OXPHOS. The patient-derived cells exhibited a marked reduction in maximal respiration or OCR compared with that of C1 in both conditions (Fig. 5c).
The extracellular acidification rate (ECAR) was measured to determine glycolytic stress level during the conversion of glucose into lactic acid. The patient’s fibroblasts showed a significant increase in ECAR activity (Fig. 5d). An increase in ECAR is indicative of increased glycolysis to compensate for the loss of ATP generation from OXPHOS. The coupling efficiency of respiration (percentage of ATP-linked OCR relative to basal OCR) was significantly reduced in the patient fibroblasts relative to that in C1 (Fig. 5e). This indicated an increased proton leakage across the mitochondrial membrane, impairing ATP production efficiency in patient’s fibroblasts. These findings strongly suggest that PTCD3 mutations interfere with mitochondrial metabolism.
Proteomic analysis showed the PTCD3 variants cause reduction of small mitoribosomal and OXPHOS subunit proteins
We performed quantitative mass spectrometry proteomic analysis of fibroblasts to detect changes in cellular proteins due to the presence of PTCD3 variants. In this analysis, 3991 proteins were quantified; among them, 807 are mitochondrial proteins within the MitoCarta2.0 database. Among those mitochondrial proteins, 30 are from the mt-SSU and 48 are from the mt-LSU. The protein-protein interaction networks were generated using the STRING 10.5 database [36] for all MitoCarta2.0-reported proteins with a threshold of > 2-fold downregulation in the patient (Fig. 6a, Supplementary Table S2a). The interaction networks contained 98 nodes in clusters established according to Gene Ontology (GO) molecular function. Proteins contributing to oxidoreductase activity (GO:0016491), structural constituent of ribosome (GO:0003735), RNA-binding (GO:0044822), and cofactor binding (GO:0048037) pathways are highly enriched in the interaction networks (Fig. 6a). The abundance of mt-SSU, CI, CIII, and CIV proteins was significantly reduced in the patient compared with those in C1, while the proteins from the mt-LSU, CII, and CV were mostly unchanged (Fig. 6b, Supplementary Table S2b). The profound decreases in the mt-SSU proteins without a reduction in the mt-LSU protein levels in the patient support the findings from BN-PAGE analysis that PTCD3 variants cause destabilization of the mt-SSU complex but not mt-LSU (Fig. 4d). Heatmap clustering of OXPHOS CI-V and mitoribosomal proteins highlighted that most of the CI, CIV, and mt-SSU proteins were downregulated in the patient than in C1. Furthermore, we observed a pronounced upregulation of the mt-LSU protein MRPL57 in the patient (Fig. 6c, Supplementary Table S2c). Taken together, the quantitative proteomic data revealed a severe and generalized reduction of CI, CIV, and mt-SSU proteins which was caused by the loss of PTCD3 protein.
Proteomic analysis and physical interaction of proteins in patient’s fibroblasts. a Protein association networks for the proteins quantified by mass spectrometry were analyzed using the STRING 10.5 platform with a confidence level of 0.4. Individual nodes represent individual proteins. Colored lines connecting the nodes indicate different evidence types for protein interaction. Red nodes indicate oxidoreductase proteins, blue nodes are proteins involved in the formation of ribosomal structures, green nodes represent RNA-binding proteins, while white nodes indicate proteins involved in other pathways. b Boxplots of the log2 fold change of the OXPHOS and mitoribosomal proteins in the Pt represented as the expression ratio relative to that of C1. One sample t-test was performed to determine the level of significance, ***p < 0.001, and ****p < 0.0001. The thick middle lines represent the median values, while the lower and upper limits of the boxes represent the 25th and the 75th percentiles of relative expression values. Each dot represents a single protein. c Heatmap and hierarchical clustering of quantified proteins in the Pt compared to C1 (> 2-fold change) identified from the proteomic analysis. Data analysis was based on spectral count data after exporting them into R computing environment. Data were converted to log2 scale. Columns represent samples; rows represent proteins. Blue represents downregulation and red represents upregulation
Lentiviral-mediated expression of wild-type PTCD3 rescues mitochondrial dysfunction
We performed a cellular complementation experiment to determine whether PTCD3wt can rescue the mitochondrial defects in the patient. We generated control and patient cell lines stably expressing either RFP as a negative control or PTCD3wt. SDS-PAGE immunoblotting confirmed that the expression of the PTCD3 protein, and OXPHOS complex CI and CIV subunits were rescued in the patient fibroblast following PTCD3wt transduction (Fig. 7a). BN-PAGE analysis revealed that levels of assembled CI and CIV were rescued in patient’s fibroblasts (Fig. 7b). The levels of OXPHOS CI/CIII2/CIV1–4 supercomplexes were higher in the patient’s fibroblasts complemented with PTCD3wt than in the patient’s RFP cells (Fig. 7c), indicating the restoration of OXPHOS supercomplexes by PTCD3wt. The mt-SSU was restored in PTCD3wt-expressing patient’s fibroblasts, whereas the mt-LSU was unaltered (Fig. 7d). qRT-PCR revealed complete restoration of MT-RNR1 transcript level, while PTCD3 expression had no effect in the MT-RNR1 transcript level in the controls (Fig. 7e).
Lentiviral-mediated transfection of wild-type PTCD3 restored mitochondrial functions in the patient’s fibroblasts. a Lentiviral-mediated transfection was performed with PTCD3wt with or without C-terminally tagged V5 (PTCD3-V5 and PTCD3, respectively). SDS-PAGE immunoblot analysis showing the restoration of PTCD3 protein level, and CI and CIV subunits in patient’s fibroblasts. HSP60 was used as a loading control. b BN-PAGE immunoblot analysis showed increases in CI and CIV levels in both Pt PTCD3-V5 and Pt PTCD3 fibroblasts. Mitochondria were solubilized by 1% Triton X-100. c BN-PAGE analysis showed the stabilization of CI/CIII2/CIV1–4 and CI/CIII2 supercomplexes in PTCD3wt-transfected patient’s fibroblasts. Tom40 was used as a loading control. d BN-PAGE analysis of 1% digitonin-permeabilized mitochondria from fibroblasts of PTCD3wt-transfected control individuals and the patient demonstrated no change in the mt-LSU level, whereas the mt-SSU level was restored in the patient’s fibroblasts. e qRT-PCR data showing the restoration of MT-RNR1 transcript level in the patient’s fibroblasts. Data are presented as an expression level relative to that of ACTB. Results are the mean ± SEM of three experiments. **p < 0.01, compared to RFP-transfected patient’s fibroblasts. f OXPHOS complex activities revealed significantly higher CI and CIV activities in the patient’s cells. Data are presented as mean ± SEM of four individual experiments. ***p < 0.001, calculated by Student’s t test. g Measurement of ATP-linked OCR showing the restoration of ATP-linked OCR under both Glu and Gal. Data are presented as mean ± SEM; *p < 0.05, **p < 0.01, by two-tailed Student’s t test. h ECAR was significantly reduced in patient’s fibroblasts growing in Glu. Data are presented as mean ± SEM; **p < 0.01, by two-tailed Student’s t test
OXPHOS enzymatic activities revealed that PTCD3 expression significantly increased the CI and CIV activities in the patient (Fig. 7f). The decreased ATP-linked OCR and ATP production level in patient were also significantly improved in both Glu and Gal by expression of PTCD3wt (Fig. 7g, Supplementary Fig. S1b). Maximal respiration in Glu was partially restored in PTCD3wt-transfected patient’s cells (Fig. 7h), however, the production of lactic acid was significantly reduced (Fig. 7h). These data suggested that the overall cellular respiration in patient’s fibroblasts was restored by expression of PTCD3wt. Therefore, the complementation assay data confirmed the pathogenicity of PTCD3 variants in the patient.
Discussion
PTCD3 is an RBP of the PPR protein family, plays an important role in mitochondrial protein translation, whereas other PPR proteins such as POLRMT, LRPPRC, MRPP3, PTCD1, and PTCD2 play roles in mitochondrial RNA metabolism [3, 9, 37, 38]. In this study, we identified a Japanese family with autosomal-recessive PTCD3 mutations and investigated the importance of PTCD3 for mitochondrial translation, mt-SSU stability, and OXPHOS functions.
We showed that PTCD3 mutations cause impaired translation of mtDNA-encoded proteins, and resulting in combined deficiency of OXPHOS CI, and CIV, as well as reduced ATP production and cellular respiration in the patient. Marked reduction in PTCD3 mRNA and protein levels in the patient highlighted the deleterious effect of those mutations. Moreover, the complementation assay restored OXPHOS CI and CIV assembly, and enzymatic activity, MT-RNR1 transcript level, as well as ATP production in the patient cells, establishing PTCD3 as a novel causative gene of OXPHOS disease and Leigh syndrome.
The transcription of human mitochondrial RNAs is driven by promoters in the mtDNA noncoding region to form polycistronic transcription units [1]. The subsequent RNAs are translated by the mitoribosome through several steps, such as initiation, elongation, termination, and recycling [39, 40]. Mitochondrial translation is initiated by the recruitment of the mitochondrial mRNA to the mt-SSU. The recruitment is aided by PTCD3, which is located in proximity of the entrance of mRNA channel [10, 12, 13]. Therefore, translation of mitochondrial mRNA is affected by the loss of PTCD3 protein (Fig. 4f). BN-PAGE data showed the level of assembled mt-SSU complex was decreased in the patient. The proteomic data showed the significantly decreased steady-state levels of most protein components of mt-SSU in the patient. Together with qRT-PCR data where the level of MT-RNR1, the RNA component of mt-SSU was significantly depleted in the patient, demonstrated a crucial role of PTCD3 in the assembly and stability of mt-SSU. Proteomic analysis demonstrated the importance of a more comprehensive understanding of the molecular mechanisms involved in human diseases. Nevertheless, PTCD3 knockdown in 143 osteosarcoma cells showed reduced PTCD3 did not destabilize the mt-SSU [9]. Previous studies showed mutations in the mt-SSU proteins MRPS16, MRPS22 [41] and MRPS34 [42] did not affect mt-LSU; however, in Tfb1m knockout mice, the loss of TFB1M which is associated with mt-SSU resulted in an increase of mt-LSU [43]. Furthermore, the loss of mt-LSU in the Ptcd1 knockout mice resulted in an accumulation of mt-SSU [7]. Mt-LSU was present at normal levels in the PTCD3 patient, indicating that mt-LSU was still fully assembled despite the significant loss of the mt-SSU. The MT-RNR2 level was increased in the patient fibroblasts, suggesting a compensatory mechanism for the impaired coordination of mitoribosomal assembly due to the compromised mt-SSU level.
Proteins localized in the same organelle or function in common biological processes tend to be coregulated [44]. Furthermore, it is crucial to know about the specific interaction partners of a protein to understand its function, and protein-protein interaction networks from coexpression data is a powerful approach to achieve this. The protein networks revealed that the severely downregulated proteins were mostly associated with OXPHOS, ribosomal structural integrity, and RNA-binding pathways. Downregulation of several mitochondrial ribosomal structural proteins (MRPS5, MRPS11, MRPS14, MRPS23, MRPS24, MRPS31, MRPS33, MRPS35 and NDUFA7) indicates that the structural integrity of mt-SSU was disrupted owing to the PTCD3 variants. Since PTCD3 is an RBP, downregulation of other RBPs (CHCHD1, MRPS29, LRPPRC, ACO1, FDPS, PTRH1, and MRPS28) suggests functionally related proteins are coregulated at the protein level. Mitochondria are a major source of reactive oxygen species (ROS) and generate free radicals as a byproduct of metabolism. An imbalance between the generation of ROS and free radical scavenger systems results in oxidative damage to cells, and are associated with neurodegeneration [45]. Mutations in mitochondrial OXPHOS CI-CIV induce ROS production and neuronal cell damage [46]. Within the protein networks, downregulated proteins with oxidoreductases activity are mostly mitochondrial CI and CIV subunits (Supplementary Table S2a), which can contribute to an increased generation of free radicals. In mammalian mitochondria, there is a multi-leveled ROS defense network of antioxidants such as glutathione peroxidase, glutathione reductase, superoxide dismutase, Cytochrome c, and catalases [45]. Oxidoreductases such as SOD2, and GSTZ1 were downregulated in the patient and they are antioxidants that scavenge ROS from mitochondria. Mutations of oxidoreductases MAOA, ALDH3A2, and GLDC are associated with mental retardation and severe neurological symptoms [47,48,49]. In the patient, downregulation of proteins involving multiple biological processes such as cellular respiration, mitochondrial ATP synthesis and translation, OXPHOS complex and mitochondrial metabolism shows the extensive impact and severity of the disease caused by PTCD3 mutations.
OXPHOS CI, CIII, CIV, and CV contain mtDNA-encoded subunits, and CII only contains nDNA-encoded subunits. A disrupted mitochondrial protein translation is likely to impact on all OXPHOS complexes that contain mtDNA-encoded subunits. BN-PAGE showed protein levels of CI and CIV, and CI/III2/IV supercomplexes had significant reductions in the patient as a consequence of PTCD3 mutations; however, there was no significant decrease in CIII level but an increased CIII2 dimer. Furthermore, we found no deficiency in CIII activity in the patient’s fibroblasts. Proteomic data showed CI, CIII and CIV proteins were decreased in the patient. This small but statistically significant decrease in CIII level in the patient was not detectable by immunoblotting. The relatively stable CIII2 dimer contributed to the normal CIII activity. A similar increase in CIII2 dimer with decreased CI/III/IV supercomplexes was described in human 143B cells with no ND1 protein [50]. Proteomic analysis could not identify any changes in CV protein levels suggesting that this complex is relatively stable. A similar finding of no significant changes in CV proteins was observed in the patient with MRPS34 mutations [42].
Mutations in the mitochondrial translation machinery are associated with diverse range of clinical presentations and prognoses. The current patient presented with limb rigidity and myoclonus, nystagmus, psychomotor regression, optic atrophy, hearing loss, and combined OXPHOS complex deficiency. The most characteristic neuroradiological findings in Leigh syndrome are bilateral, symmetric focal hyperintensities in the basal ganglia, thalamus, substantia nigra, and brainstem nuclei [51]. The involvement of the brainstem and cerebral white matter is an indication of disease progression and involvement of lower brainstem causes respiratory failure and sudden death [52, 53]. Therefore, the brainstem lesions and early death of the PTCD3 patient are correlated with the progressive nature of Leigh syndrome.
To our best knowledge, this is the first report of PTCD3 variants as a novel cause of mitochondrial disease. Our findings emphasize that mutations in genes encoding members of the PPR protein family contribute to defects in mitochondrial translation associated with Leigh syndrome. These findings may enable carrier testing and provide improved options for prenatal diagnosis, thereby reducing the disease-related burden on society.
Data availability
The data generated during the current study are available in the supplemental online content and from the corresponding author on request.
References
Gustafsson CM, Falkenberg M, Larsson NG (2016) Maintenance and expression of mammalian mitochondrial DNA. Annu Rev Biochem 85:133–160. https://doi.org/10.1146/annurev-biochem-060815-014402
Boczonadi V, Horvath R (2014) Mitochondria: impaired mitochondrial translation in human disease. Int J Biochem Cell Biol 48:77–84. https://doi.org/10.1016/j.biocel.2013.12.011
Rackham O, Mercer TR, Filipovska A (2012) The human mitochondrial transcriptome and the RNA-binding proteins that regulate its expression. Wiley Interdiscip Rev RNA 3(5):675–695. https://doi.org/10.1002/wrna.1128
Hallberg BM, Larsson NG (2014) Making proteins in the powerhouse. Cell Metab 20(2):226–240. https://doi.org/10.1016/j.cmet.2014.07.001
Aubourg S, Boudet N, Kreis M, Lecharny A (2000) In Arabidopsis thaliana, 1% of the genome codes for a novel protein family unique to plants. Plant Mol Biol 42(4):603–613. https://doi.org/10.1023/a:1006352315928
Small ID, Peeters N (2000) The PPR motif—a TPR-related motif prevalent in plant organellar proteins. Trends Biochem Sci 25(2):45–47
Perks KL, Rossetti G, Kuznetsova I, Hughes LA, Ermer JA, Ferreira N, Busch JD, Rudler DL, Spahr H, Schondorf T, Shearwood AJ, Viola HM, Siira SJ, Hool LC, Milenkovic D, Larsson NG, Rackham O, Filipovska A (2018) PTCD1 is required for 16S rRNA maturation complex stability and mitochondrial ribosome assembly. Cell Rep 23(1):127–142. https://doi.org/10.1016/j.celrep.2018.03.033
Filipovska A, Rackham O (2013) Pentatricopeptide repeats: modular blocks for building RNA-binding proteins. RNA Biol 10(9):1426–1432. https://doi.org/10.4161/rna.24769
Davies SM, Rackham O, Shearwood AM, Hamilton KL, Narsai R, Whelan J, Filipovska A (2009) Pentatricopeptide repeat domain protein 3 associates with the mitochondrial small ribosomal subunit and regulates translation. FEBS Lett 583(12):1853–1858. https://doi.org/10.1016/j.febslet.2009.04.048
Amunts A, Brown A, Toots J, Scheres SHW, Ramakrishnan V (2015) The structure of the human mitochondrial ribosome. Science 348(6230):95–98. https://doi.org/10.1126/science.aaa1193
Kaushal PS, Sharma MR, Booth TM, Haque EM, Tung CS, Sanbonmatsu KY, Spremulli LL, Agrawal RK (2014) Cryo-EM structure of the small subunit of the mammalian mitochondrial ribosome. Proc Natl Acad Sci U S A 111(20):7284–7289. https://doi.org/10.1073/pnas.1401657111
Greber BJ, Bieri P, Leibundgut M, Leitner A, Aebersold R, Boehringer D, Ban N (2015) Ribosome. The complete structure of the 55S mammalian mitochondrial ribosome. Science 348(6232):303–308. https://doi.org/10.1126/science.aaa3872
Kummer E, Leibundgut M, Rackham O, Lee RG, Boehringer D, Filipovska A, Ban N (2018) Unique features of mammalian mitochondrial translation initiation revealed by cryo-EM. Nature 560(7717):263–267. https://doi.org/10.1038/s41586-018-0373-y
Leigh D (1951) Subacute necrotizing encephalomyelopathy in an infant. J Neurol Neurosurg Psychiatry 14(3):216–221. https://doi.org/10.1136/jnnp.14.3.216
Lopez LC, Schuelke M, Quinzii CM, Kanki T, Rodenburg RJ, Naini A, Dimauro S, Hirano M (2006) Leigh syndrome with nephropathy and CoQ10 deficiency due to decaprenyl diphosphate synthase subunit 2 (PDSS2) mutations. Am J Hum Genet 79(6):1125–1129. https://doi.org/10.1086/510023
Ruhoy IS, Saneto RP (2014) The genetics of Leigh syndrome and its implications for clinical practice and risk management. Appl Clin Genet 7:221–234. https://doi.org/10.2147/TACG.S46176
Lake NJ, Compton AG, Rahman S, Thorburn DR (2016) Leigh syndrome: one disorder, more than 75 monogenic causes. Ann Neurol 79(2):190–203. https://doi.org/10.1002/ana.24551
Kohda M, Tokuzawa Y, Kishita Y, Nyuzuki H, Moriyama Y, Mizuno Y, Hirata T, Yatsuka Y, Yamashita-Sugahara Y, Nakachi Y, Kato H, Okuda A, Tamaru S, Borna NN, Banshoya K, Aigaki T, Sato-Miyata Y, Ohnuma K, Suzuki T, Nagao A, Maehata H, Matsuda F, Higasa K, Nagasaki M, Yasuda J, Yamamoto M, Fushimi T, Shimura M, Kaiho-Ichimoto K, Harashima H, Yamazaki T, Mori M, Murayama K, Ohtake A, Okazaki Y (2016) A comprehensive genomic analysis reveals the genetic landscape of mitochondrial respiratory chain complex deficiencies. PLoS Genet 12(1):e1005679. https://doi.org/10.1371/journal.pgen.1005679
Kirby DM, Thorburn DR, Turnbull DM, Taylor RW (2007) Biochemical assays of respiratory chain complex activity. In: Pon L, Schon E (eds) Methods in cell biology, vol 80. Academic Press, pp 93–119. https://doi.org/10.1016/S0091-679X(06)80004-X
McKenzie M, Lazarou M, Ryan MT (2009) Chapter 18 analysis of respiratory chain complex assembly with radiolabeled nuclear- and mitochondrial-encoded subunits. In: Allison W, Scheffler I (eds) Mitochondrial function, part a: mitochondrial electron transport complexes and reactive oxygen species. Methods in enzymology. Elsevier, pp 321–339. https://doi.org/10.1016/s0076-6879(08)04418-2
Wu Y, Williams EG, Dubuis S, Mottis A, Jovaisaite V, Houten SM, Argmann CA, Faridi P, Wolski W, Kutalik Z, Zamboni N, Auwerx J, Aebersold R (2014) Multilayered genetic and omics dissection of mitochondrial activity in a mouse reference population. Cell 158(6):1415–1430. https://doi.org/10.1016/j.cell.2014.07.039
Bruderer R, Bernhardt OM, Gandhi T, Miladinovic SM, Cheng LY, Messner S, Ehrenberger T, Zanotelli V, Butscheid Y, Escher C, Vitek O, Rinner O, Reiter L (2015) Extending the limits of quantitative proteome profiling with data-independent acquisition and application to acetaminophen-treated three-dimensional liver microtissues. Mol Cell Proteomics 14(5):1400–1410. https://doi.org/10.1074/mcp.M114.044305
Lam H, Deutsch EW, Eddes JS, Eng JK, King N, Stein SE, Aebersold R (2007) Development and validation of a spectral library searching method for peptide identification from MS/MS. Proteomics 7(5):655–667. https://doi.org/10.1002/pmic.200600625
Reiter L, Rinner O, Picotti P, Huttenhain R, Beck M, Brusniak MY, Hengartner MO, Aebersold R (2011) mProphet: automated data processing and statistical validation for large-scale SRM experiments. Nat Methods 8(5):430–435. https://doi.org/10.1038/nmeth.1584
Vizcaino JA, Csordas A, del-Toro N, Dianes JA, Griss J, Lavidas I, Mayer G, Perez-Riverol Y, Reisinger F, Ternent T, Xu QW, Wang R, Hermjakob H (2016) 2016 update of the PRIDE database and its related tools. Nucleic Acids Res 44(D1):D447–D456. https://doi.org/10.1093/nar/gkv1145
Calvo SE, Clauser KR, Mootha VK (2016) MitoCarta2.0: an updated inventory of mammalian mitochondrial proteins. Nucleic Acids Res 44(D1):D1251–D1257. https://doi.org/10.1093/nar/gkv1003
Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O'Donnell-Luria AH, Ware JS, Hill AJ, Cummings BB, Tukiainen T, Birnbaum DP, Kosmicki JA, Duncan LE, Estrada K, Zhao F, Zou J, Pierce-Hoffman E, Berghout J, Cooper DN et al (2016) Analysis of protein-coding genetic variation in 60,706 humans. Nature 536(7616):285–291. https://doi.org/10.1038/nature19057
Nagasaki M, Yasuda J, Katsuoka F, Nariai N, Kojima K, Kawai Y, Yamaguchi-Kabata Y, Yokozawa J, Danjoh I, Saito S, Sato Y, Mimori T, Tsuda K, Saito R, Pan X, Nishikawa S, Ito S, Kuroki Y, Tanabe O, Fuse N et al (2015) Rare variant discovery by deep whole-genome sequencing of 1,070 Japanese individuals. Nat Commun 6:8018. https://doi.org/10.1038/ncomms9018
Zerbino DR, Achuthan P, Akanni W, Amode MR, Barrell D, Bhai J, Billis K, Cummins C, Gall A, Giron CG, Gil L, Gordon L, Haggerty L, Haskell E, Hourlier T, Izuogu OG, Janacek SH, Juettemann T, To JK, Laird MR et al (2018) Ensembl 2018. Nucleic Acids Res 46(D1):D754–D761. https://doi.org/10.1093/nar/gkx1098
Gamazon ER, Segre AV, van de Bunt M, Wen X, Xi HS, Hormozdiari F, Ongen H, Konkashbaev A, Derks EM, Aguet F, Quan J, Consortium GT, Nicolae DL, Eskin E, Kellis M, Getz G, McCarthy MI, Dermitzakis ET, Cox NJ, Ardlie KG (2018) Using an atlas of gene regulation across 44 human tissues to inform complex disease- and trait-associated variation. Nat Genet 50(7):956–967. https://doi.org/10.1038/s41588-018-0154-4
Acin-Perez R, Fernandez-Silva P, Peleato ML, Perez-Martos A, Enriquez JA (2008) Respiratory active mitochondrial supercomplexes. Mol Cell 32(4):529–539. https://doi.org/10.1016/j.molcel.2008.10.021
Gardeitchik T, Mohamed M, Ruzzenente B, Karall D, Guerrero-Castillo S, Dalloyaux D, van den Brand M, van Kraaij S, van Asbeck E, Assouline Z, Rio M, de Lonlay P, Scholl-Buergi S, Wolthuis D, Hoischen A, Rodenburg RJ, Sperl W, Urban Z, Brandt U, Mayr JA et al (2018) Bi-allelic mutations in the mitochondrial ribosomal protein MRPS2 cause sensorineural hearing loss, hypoglycemia, and multiple OXPHOS complex deficiencies. Am J Hum Genet 102(4):685–695. https://doi.org/10.1016/j.ajhg.2018.02.012
Marroquin LD, Hynes J, Dykens JA, Jamieson JD, Will Y (2007) Circumventing the Crabtree effect: replacing media glucose with galactose increases susceptibility of HepG2 cells to mitochondrial toxicants. Toxicol Sci 97(2):539–547. https://doi.org/10.1093/toxsci/kfm052
Rossignol R, Gilkerson R, Aggeler R, Yamagata K, Remington SJ, Capaldi RA (2004) Energy substrate modulates mitochondrial structure and oxidative capacity in cancer cells. Cancer Res 64(3):985–993. https://doi.org/10.1158/0008-5472.Can-03-1101
Robinson BH, Petrova-Benedict R, Buncic JR, Wallace DC (1992) Nonviability of cells with oxidative defects in galactose medium: a screening test for affected patient fibroblasts. Biochem Med Metab Biol 48(2):122–126. https://doi.org/10.1016/0885-4505(92)90056-5
Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, Jensen LJ, von Mering C (2017) The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res 45(D1):D362–D368. https://doi.org/10.1093/nar/gkw937
Rackham O, Shearwood A-MJ, Mercer TR, Davies SMK, Mattick JS, Filipovska A (2011) Long noncoding RNAs are generated from the mitochondrial genome and regulated by nuclear-encoded proteins. RNA 17(12):2085–2093. https://doi.org/10.1261/rna.029405.111
Davies SM, Lopez Sanchez MI, Narsai R, Shearwood AM, Razif MF, Small ID, Whelan J, Rackham O, Filipovska A (2012) MRPS27 is a pentatricopeptide repeat domain protein required for the translation of mitochondrially encoded proteins. FEBS Lett 586(20):3555–3561. https://doi.org/10.1016/j.febslet.2012.07.043
Christian BE, Spremulli LL (2012) Mechanism of protein biosynthesis in mammalian mitochondria. Biochim Biophys Acta 1819(9–10):1035–1054. https://doi.org/10.1016/j.bbagrm.2011.11.009
Ott M, Amunts A, Brown A (2016) Organization and regulation of mitochondrial protein synthesis. Annu Rev Biochem 85:77–101. https://doi.org/10.1146/annurev-biochem-060815-014334
Emdadul Haque M, Grasso D, Miller C, Spremulli LL, Saada A (2008) The effect of mutated mitochondrial ribosomal proteins S16 and S22 on the assembly of the small and large ribosomal subunits in human mitochondria. Mitochondrion 8(3):254–261. https://doi.org/10.1016/j.mito.2008.04.004
Lake NJ, Webb BD, Stroud DA, Richman TR, Ruzzenente B, Compton AG, Mountford HS, Pulman J, Zangarelli C, Rio M, Boddaert N, Assouline Z, Sherpa MD, Schadt EE, Houten SM, Byrnes J, McCormick EM, Zolkipli-Cunningham Z, Haude K, Zhang Z et al (2017) Biallelic mutations in MRPS34 Lead to instability of the small mitoribosomal subunit and Leigh syndrome. Am J Hum Genet 101(2):239–254. https://doi.org/10.1016/j.ajhg.2017.07.005
Metodiev MD, Lesko N, Park CB, Camara Y, Shi Y, Wibom R, Hultenby K, Gustafsson CM, Larsson NG (2009) Methylation of 12S rRNA is necessary for in vivo stability of the small subunit of the mammalian mitochondrial ribosome. Cell Metab 9(4):386–397. https://doi.org/10.1016/j.cmet.2009.03.001
Foster LJ, de Hoog CL, Zhang Y, Zhang Y, Xie X, Mootha VK, Mann M (2006) A mammalian organelle map by protein correlation profiling. Cell 125(1):187–199. https://doi.org/10.1016/j.cell.2006.03.022
Andreyev AY, Kushnareva YE, Starkov AA (2005) Mitochondrial metabolism of reactive oxygen species. Biochemistry (Mosc) 70(2):200–214
Gandhi S, Abramov AY (2012) Mechanism of oxidative stress in neurodegeneration. Oxidative Med Cell Longev 2012:428010–428011. https://doi.org/10.1155/2012/428010
Brunner HG, Nelen M, Breakefield XO, Ropers HH, van Oost BA (1993) Abnormal behavior associated with a point mutation in the structural gene for monoamine oxidase A. Science 262(5133):578–580
Ganemo A, Jagell S, Vahlquist A (2009) Sjogren-larsson syndrome: a study of clinical symptoms and dermatological treatment in 34 Swedish patients. Acta Derm Venereol 89(1):68–73. https://doi.org/10.2340/00015555-0561
Liu S, Wang Z, Liang J, Chen N, OuYang H, Zeng W, Chen L, Xie X, Jiang J (2017) Two novel mutations in the glycine decarboxylase gene in a boy with classic nonketotic hyperglycinemia: case report. Arch Argent Pediatr 115(4):e225–e229. https://doi.org/10.5546/aap.2017.eng.e225
Lim SC, Hroudova J, Van Bergen NJ, Lopez Sanchez MI, Trounce IA, McKenzie M (2016) Loss of mitochondrial DNA-encoded protein ND1 results in disruption of complex I biogenesis during early stages of assembly. FASEB J 30(6):2236–2248. https://doi.org/10.1096/fj.201500137R
Savoiardo M, Zeviani M, Uziel G, Farina L (2002) MRI in Leigh syndrome with SURF1 gene mutation. Ann Neurol 51(1):138–139. https://doi.org/10.1002/ana.10031
Finsterer J (2008) Leigh and Leigh-like syndrome in children and adults. Pediatr Neurol 39(4):223–235. https://doi.org/10.1016/j.pediatrneurol.2008.07.013
Saneto RP, Friedman SD, Shaw DW (2008) Neuroimaging of mitochondrial disease. Mitochondrion 8(5–6):396–413. https://doi.org/10.1016/j.mito.2008.05.003
Acknowledgements
We thank Dr. Hiroyuki Miyoshi of Keio University and RIKEN BioResource Center for the CS-CA-MCS plasmid. The authors are thankful to the Biomedical Research Center, Saitama Medical University for supporting to do the experiment. The authors thank Edanz Group (www.edanzediting.com/ac) for editing a draft of this manuscript.
Funding
This work was supported in part by a grant of the “Practical Research Project for Rare/Intractable Diseases” (Fund ID: 18ek0109273s0102 and 18ek0109177s0103) and “Program for an Integrated Database of Clinical and Genomic Information” (Fund ID: 18kk0205002s9903) from Japan Agency for Medical Research and Development (AMED) (http://www.amed.go.jp/en/), and also MEXT-Supported Program for the Private University Research Branding Project. NNB is a recipient of the Uehara Memorial Foundation Research Fellowship. SCL is a JSPS International Research Fellow.
Author information
Authors and Affiliations
Contributions
NNB, YK, and YO designed the study. Drafting of the manuscript was performed by NNB. NNB, SCL, MS, YW, YY, and HH acquired data. NNB, YK, MK, and KM analyzed data. TF and KI provided the patient’s clinical information. YH is the attending physician of the patient. YO, AO, and KM gave critical comments. YO led the project.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Study approval
The study was approved by the ethics committee of Juntendo University and was performed after receiving written informed consent from the parents of the patient.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Borna, N.N., Kishita, Y., Kohda, M. et al. Mitochondrial ribosomal protein PTCD3 mutations cause oxidative phosphorylation defects with Leigh syndrome. Neurogenetics 20, 9–25 (2019). https://doi.org/10.1007/s10048-018-0561-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10048-018-0561-9