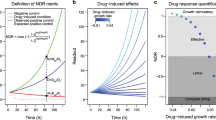
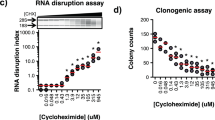
Abstract
During the preclinical stages of the drug discovery process, cell viability assays are fundamental tools for studying the phenotypic properties and overall health of cells following in vitro drug sensitivity screens. Therefore, it is important to optimize your viability assay of choice to obtain reproducible and replicable results, as well as use relevant drug response metrics (e.g., IC50, AUC, GR50, and GRmax) to identify candidate drugs for further evaluation in vivo. Herein, we used the resazurin reduction assay which is a quick, cost-effective, simple-to-use, and sensitive method for examining the phenotypic properties of cells. Using the MCF7 breast cancer cell line, we provide a detailed step-by-step protocol for optimizing drug sensitivity screens using the resazurin assay.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Riss TL, Moravec RA, Niles AL, Duellman S, Benink HA, Worzella TJ, Minor L (2004) Cell viability assays. In: Sittampalam GS, Grossman A, Brimacombe K et al (eds) Assay guidance manual. Eli Lilly & Company and the National Center for Advancing Translational Sciences, Bethesda, pp 353–377
Stoddart MJ (2011) Cell viability assays: introduction. Methods Mol Biol 740:1–6. https://doi.org/10.1007/978-1-61779-108-6_1
Adan A, Kiraz Y, Baran Y (2016) Cell proliferation and cytotoxicity assays. Curr Pharm Biotechnol 17(14):1213–1221. https://doi.org/10.2174/1389201017666160808160513
Kamiloglu S, Sari G, Ozdal T, Capanoglu E (2020) Guidelines for cell viability assays. Food Front 1(3):332–349. https://doi.org/10.1002/fft2.44
Aslantürk ÖS (2018) In vitro cytotoxicity and cell viability assays: principles, advantages, and disadvantages. In: Genotoxicity – a predictable risk to our actual world. IntechOpen, London, pp 13–30. https://doi.org/10.5772/intechopen.71923
Präbst K, Engelhardt H, Ringgeler S, Hübner H (2017) Basic colorimetric proliferation assays: MTT, WST, and Resazurin. Methods Mol Biol 1601:1–17. https://doi.org/10.1007/978-1-4939-6960-9_1
Shokrzadeh M, Modanloo M (2017) An overview of the most common methods for assessing cell viability. J Res Med Dent Sci 5(2):33. https://doi.org/10.5455/jrmds.2017526
Larsson P, Engqvist H, Biermann J, Werner Rönnerman E, Forssell-Aronsson E, Kovács A, Karlsson P, Helou K, Parris TZ (2020) Optimization of cell viability assays to improve replicability and reproducibility of cancer drug sensitivity screens. Sci Rep 10(1):5798. https://doi.org/10.1038/s41598-020-62848-5
Ivanov DP, Parker TL, Walker DA, Alexander C, Ashford MB, Gellert PR, Garnett MC (2014) Multiplexing spheroid volume, resazurin and acid phosphatase viability assays for high-throughput screening of tumour spheroids and stem cell neurospheres. PLoS One 9(8):e103817. https://doi.org/10.1371/journal.pone.0103817
Ryu AH, Eckalbar WL, Kreimer A, Yosef N, Ahituv N (2017) Use antibiotics in cell culture with caution: genome-wide identification of antibiotic-induced changes in gene expression and regulation. Sci Rep 7(1):7533. https://doi.org/10.1038/s41598-017-07757-w
Quast J-P (2022) Dose-response data analysis workflow. https://cran.r-project.org/web/packages/protti/vignettes/data_analysis_dose_response_workflow.html
Hafner M, Niepel M, Subramanian K, Sorger PK (2017) Designing drug-response experiments and quantifying their results. Curr Protoc Chem Biol 9(2):96–116. https://doi.org/10.1002/cpch.19
Hafner M, Niepel M, Chung M, Sorger PK (2016) Growth rate inhibition metrics correct for confounders in measuring sensitivity to cancer drugs. Nat Methods 13(6):521–527. https://doi.org/10.1038/nmeth.3853
Niepel M, Hafner M, Chung M, Sorger PK (2017) Measuring cancer drug sensitivity and resistance in cultured cells. Curr Protoc Chem Biol 9(2):55–74. https://doi.org/10.1002/cpch.21
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Science+Business Media, LLC, part of Springer Nature
About this protocol
Cite this protocol
Larsson, P., Parris, T.Z. (2023). Optimization of Cell Viability Assays for Drug Sensitivity Screens. In: Friedrich, O., Gilbert, D.F. (eds) Cell Viability Assays. Methods in Molecular Biology, vol 2644. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-3052-5_18
Download citation
DOI: https://doi.org/10.1007/978-1-0716-3052-5_18
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-3051-8
Online ISBN: 978-1-0716-3052-5
eBook Packages: Springer Protocols