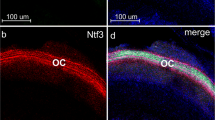
Abstract
Biological processes are largely governed by the RNA molecules and resulting peptides that are encoded by an organism’s DNA. For decades, our understanding of biology has been vastly enhanced through study of the distribution and abundance of RNA molecules. Studies of the inner ear are no exception, and approaches like qPCR, RNA-seq, and in situ hybridization (ISH) have contributed greatly to our understanding of inner ear development and function. While qPCR and RNA-seq provide sensitive and broad measures of RNA quantity, they can be limited in their ability to resolve RNA localization. Thus, ISH remains a vital technique for inner ear studies. However, traditional ISH approaches can be technically challenging, time-consuming, suffer from high background, and are generally limited to the investigation of only a single RNA of interest. Recent advances in ISH approaches have overcome many of these limitations allowing for speed, high signal-to-noise, and the ability to perform multiplexed ISH where several transcripts of interest can be visualized in the same tissue or section. One such approach is RNAscope which is a commercially available option that allows for ease of use and, for many transcripts, the ability to achieve absolute quantification of RNA molecules per cell. Here we outline RNAscope methods that have been optimized for inner ear (and related) tissues and allow for relatively rapid labeling of RNA transcripts of interest in fixed tissues. Furthermore, these methods elucidate how RNAscope labeling can be imaged with brightfield or fluorescence microscopy, how it allows for quantification as well as localization, how it can be multiplexed to visualize multiple transcripts simultaneously, and how it can be combined with immunocytochemistry so that RNA and proteins may be visualized in the same sample.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Crick F (1970) Central dogma of molecular biology. Nature 227(5258):561–563. https://doi.org/10.1038/227561a0
Xie F, Timme KA, Wood JR (2018) Using single molecule mRNA fluorescent in situ hybridization (RNA-FISH) to quantify mRNAs in individual murine oocytes and embryos. Sci Rep:1–11. https://doi.org/10.1038/s41598-018-26345-0
Soares RJ, Maglieri G, Gutschner T et al (2018) Evaluation of fluorescence in situ hybridization techniques to study long non-coding RNA expression in cultured cells. Nucleic Acids Res 46(1):e4. https://doi.org/10.1093/nar/gkx946
Wang F, Flanagan J, Su N et al (2012) RNAscope : a novel in situ. RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J Mol Diagn 4(1):22–29. https://doi.org/10.1016/j.jmoldx.2011.08.002
Yao RW, Wang Y, Chen LL (2019) Cellular functions of long noncoding RNAs. Nat Cell Biol 21(5):542–551. https://doi.org/10.1038/s41556-019-0311-8
Gebert LFR, MacRae IJ (2019) Regulation of microRNA function in animals. Nat Rev Mol Cell Biol 20(1):21–37. https://doi.org/10.1038/s41580-018-0045-7
Wang Z, Gerstein M, Snyder M (2009) RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 10(1):57–63. https://doi.org/10.1038/nrg2484
Lee JH, Daugharthy ER, Scheiman J et al (2015) Fluorescent in situ sequencing (FISSEQ) of RNA for gene expression profiling in intact cells and tissues. Nat Protoc 10(3):442–458. https://doi.org/10.1038/nprot.2014.191
Maniatis S, Äijö T, Vickovic S, Braine C et al (2019) Spatiotemporal dynamics of molecular pathology in amyotrophic lateral sclerosis. Science 364(6435):89–93. https://doi.org/10.1126/science.aav9776
Thrane K, Eriksson H, Maaskola J et al (2018) Spatially resolved transcriptomics enables dissection of genetic heterogeneity in stage III cutaneous malignant melanoma. Cancer Res 78(20):5970–5979. https://doi.org/10.1158/0008-5472.CAN-18-0747
Judice TN, Nelson NC, Beisel CL et al (2002) Cochlear whole mount in situ hybridization: identification of longitudinal and radial gradients. Brain Res Brain Res Protoc 9(1):65–76. https://doi.org/10.1016/s1385-299x(01)00138-6
Kersigo J, Pan N, Lederman JD et al (2018) A RNAscope whole mount approach that can be combined with immunofluorescence to quantify differential distribution of mRNA. Cell Tissue Res 374(2):251–262. https://doi.org/10.1007/s00441-018-2864-4
Salehi P, Nelson CN, Chen Y et al (2018) Detection of single mRNAs in individual cells of the auditory system. Hear Res 367:88–96. https://doi.org/10.1016/j.heares.2018.07.008
Gold EM, Vasilevko V, Hasselmann J et al (2018) Repeated mild closed head injuries induce long-term white matter pathology and neuronal loss that are correlated with behavioral deficits. ASN Neuro 10:1759091418781921
John HA, Birnstiel ML, Jones KW (1969) RNA-DNA hybrids at the cytological level. Nature 223(5206):582–587. https://doi.org/10.1038/223582a0
Gall JG, Pardue ML (1969) Formation and detection of RNA-DNA hybrid molecules in cytological preparations. Proc Natl Acad Sci 63(2):378–383. https://doi.org/10.1073/pnas.63.2.378
Xia L, Yin S, Wang J (2012) Inner ear gene transfection in neonatal mice using adeno-associated viral vector: a comparison of two approaches. PLoS One 7(8):e43218. https://doi.org/10.1371/journal.pone.0043218
Jolly S, Lang V, Koelzer VH et al (2019) Single-cell quantification of mRNA expression in the human brain. Sci Rep 9(1):12353. https://doi.org/10.1038/s41598-019-48787-w
Chan S, Filézac de L’Etang A et al (2018) A method for manual and automated multiplex RNAscope in situ hybridization and immunocytochemistry on cytospin samples. PLoS One 13(11):e0207619. https://doi.org/10.1371/journal.pone.0207619
Yin VP (2018) In Situ detection of MicroRNA expression with RNAscope probes. Methods Mol Biol 1649:197–208. https://doi.org/10.1007/978-1-4939-7213-5_13
Grandi FC, De Tomassi L, Mustapha M (2020) Single-cell RNA analysis of type I spiral ganglion neurons reveals a Lmx1a population in the cochlea. Front Mol Neurosci 13:83. https://doi.org/10.3389/fnmol.2020.00083
Roccio M, Perny M, Ealy M et al (2018) Molecular characterization and prospective isolation of human fetal cochlear hair cell progenitors. Nat Commun 9(1):1–14. https://doi.org/10.1038/s41467-018-06334-7
Lingle CJ, Martinez-Espinosa PL, Yang-Hood A et al (2019) LRRC52 regulates BK channel function and localization in mouse cochlear inner hair cells. Proc Natl Acad Sci U S A 116(37):18397–18403. https://doi.org/10.1073/pnas.1907065116
Yu KS, Frumm SM, Park JS et al (2019) Development of the mouse and human cochlea at single cell resolution. Cell Commun Signal 11:78. https://doi.org/10.1186/1478-811X-11-78
Sherrill HE, Jean P, Driver EC et al (2019) Pou4f1 defines a subgroup of Type I spiral ganglion neurons and is necessary for normal inner hair cell presynaptic Ca2+ signaling. J Neurosci 39(27):5284–5298. https://doi.org/10.1523/JNEUROSCI.2728-18.2019
Wiwatpanit T, Lorenzen SM, Cantú JA et al (2018) Trans-differentiation of outer hair cells into inner hair cells in the absence of INSM1. Nature 563(7733):691–695. https://doi.org/10.1038/s41586-018-0570-8
Shrestha BR, Chia C, Wu L et al (2018) Sensory neuron diversity in the inner ear is shaped by activity. Cell 174(5):1229–1246. https://doi.org/10.1016/j.cell.2018.07.007
Oivanen M, Kuusela S, Lönnberg H (1998) Kinetics and mechanisms for the cleavage and isomerization of the phosphodiester bonds of RNA by bronsted acids and bases. Chem Rev 98(3):961–990. https://doi.org/10.1021/cr960425x
Järvinen P, Oivanen M, Lönnberg H (1991) Interconversion and phosphoester hydrolysis of 2′,5′- and 3′,5′-dinucleoside monophosphates: kinetics and mechanisms. J Org Chem 56(18):5396–5401. https://doi.org/10.1021/jo00018a037
Taylor CR, Shi SR, Cote RJ (1996) Antigen retrieval for immunohistochemistry status and need for greater standardization. Applied Immunohistochemistry and Molecular Morphology. Appl Immunohistochem 4(3):144–166
Urieli-Shoval S, Meek RL, Hanson RH et al (1992) Preservation of RNA for in situ hybridization: Carnoy’s versus formaldehyde fixation. J Histochem Cytochem 40(12):1879–1885. https://doi.org/10.1177/40.12.1280665
Yan F, Wu X, Crawford M et al (1992) The search for an optimal DNA, RNA, and protein detection by in situ hybridization, immunohistochemistry, and solution-based methods. Methods 52(4):281–286. https://doi.org/10.1016/j.ymeth.2010.09.005
Ryan AF, Watts AG, Simmons DM (1991) Preservation of mRNA during in situ hybridization in the cochlea. Hear Res 56(1-2):148–152. https://doi.org/10.1016/0378-5955(91)90164-5
Cunningham LL (2006) The adult mouse utricle as an in vitro preparation for studies of ototoxic-drug-induced sensory hair cell death. Brain Res 1091(1):277–281. https://doi.org/10.1016/j.brainres.2006.01.128
Montgomery SC, Cox BC (2016) Whole mount dissection and immunofluorescence of the adult mouse cochlea. J Vis Exp 107:53561. https://doi.org/10.3791/53561
Driver EC, Kelley MW (2010) Transfection of mouse cochlear explants by electroporation. Curr Protoc Neuroscipr Chapter 4:Unit 4.34.1–Unit 4.34.10. https://doi.org/10.1002/0471142301.ns0434s51
Ogier JM, Burt RA, Drury HR et al (2019) Organotypic culture of neonatal murine inner ear explants. Front Cell Neurosci 13:170. https://doi.org/10.3389/fncel.2019.00170
Bode J, Kruwel T, Tews B (2017) Light sheet fluorescence microscopy combined with optical cleaning methods as a novel imaging tool in biomedical research. EMJ Innov 1(1):67–74
Carraro M, Paroutis P, Woodside M, Harrison R V (2015) Improved imaging of cleared samples with ZEISS Lightsheet Z.1: refractive index on demand. Zeiss Appl Note:1–7
Sun S, Babola T, Pregernig G et al (2018) Hair cell mechanotransduction regulates spontaneous activity and spiral ganglion subtype specification in the auditory system. Cell 174(5):1247–1263.e15. https://doi.org/10.1016/j.cell.2018.07.008
Hoa M, Olszewski R, Li X et al (2020) Characterizing adult cochlear supporting cell transcriptional diversity using single-cell RNA-Seq: validation in the adult mouse and translational implications for the adult human cochlea. Front Mol Neurosci 13:13. https://doi.org/10.3389/fnmol.2020.00013
Gross-Thebing T, Paksa A, Raz E (2014) Simultaneous high-resolution detection of multiple transcripts combined with localization of proteins in whole-mount embryos. BMC Biol 12:55. https://doi.org/10.1186/s12915-014-0055-7
Maynard KR, Tippani M, Takahashi Y et al (2020) dotdotdot: an automated approach to quantify multiplex single molecule fluorescent in situ hybridization (smFISH) images in complex tissues. Nucleic Acid Res 48(11):e66. https://doi.org/10.1093/nar/gkaa312
Carine Stapel L, Lombardot B, Broaddus C et al (2016) Automated detection and quantification of single RNAs at cellular resolution in zebrafish embryos. Development 143(3):540–546. https://doi.org/10.1242/dev.128918
Trcek T, Lionnet T, Shroff H, Lehmann R (2017) mRNA quantification using single-molecule FISH in Drosophila embryos. Nat Protoc 12(7):1326–1348. https://doi.org/10.1038/nprot.2017.030
Femino AM, Fay FS, Fogarty K, Singer RH (1998) Visualization of single RNA transcripts in situ. Science 280(5363):585–590. https://doi.org/10.1126/science.280.5363.585
Raj A, van den Bogaard P, Rifkin SA et al (2008) Imaging individual mRNA molecules using multiple singly labeled probes. Nat Methods 5(10):877–879. https://doi.org/10.1038/nmeth.1253
Tsanov N, Samacoits A, Chouaib R et al (2016) SmiFISH and FISH-quant—a flexible single RNA detection approach with super-resolution capability. Nucleic Acids Res 44(22):e165. https://doi.org/10.1093/nar/gkw784
Shah S, Lubeck E, Schwarzkopf M et al (2016) Single-molecule RNA detection at depth by hybridization chain reaction and tissue hydrogel embedding and clearing. Development 143(15):2862–2867. https://doi.org/10.1242/dev.138560
Gaspar I, Wippich F, Ephrussi A (2017) Enzymatic production of single-molecule FISH and RNA capture probes. RNA 23(10):1582–1591. https://doi.org/10.1261/rna.061184.117
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Science+Business Media, LLC, part of Springer Nature
About this protocol
Cite this protocol
Ghosh, S., Casey, G., Stansak, K.L., Thapa, P., Walters, B.J. (2022). An Efficient Method to Detect Messenger RNA (mRNA) in the Inner Ear by RNAscope In Situ Hybridization. In: Groves, A.K. (eds) Developmental, Physiological, and Functional Neurobiology of the Inner Ear. Neuromethods, vol 176. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-2022-9_6
Download citation
DOI: https://doi.org/10.1007/978-1-0716-2022-9_6
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-2021-2
Online ISBN: 978-1-0716-2022-9
eBook Packages: Springer Protocols