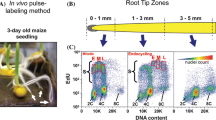
Abstract
DNA replication during S phase in eukaryotes is a highly regulated process that ensures the accurate transmission of genetic material to daughter cells during cell division. Replication follows a well-defined temporal program, which has been studied extensively in humans, Drosophila, and yeast, where it is clear that the replication process is both temporally and spatially ordered. The replication timing (RT) program is increasingly considered to be a functional readout of genomic features and chromatin organization. Although there is increasing evidence that plants display important differences in their DNA replication process compared to animals, RT programs in plants have not been extensively studied. To address this deficiency, we developed an improved protocol for the genome-wide RT analysis by sequencing newly replicated DNA (“Repli-seq”) and applied it to the characterization of RT in maize root tips. Our protocol uses 5-ethynyl-2′-deoxyuridine (EdU) to label replicating DNA in vivo in intact roots. Our protocol also eliminates the need for synchronization and frequently associated chemical perturbations as well as the need for cell cultures, which can accumulate genetic and epigenetic differences over time. EdU can be fluorescently labeled under mild conditions and does not degrade subnuclear structure, allowing for the differentiation of labeled and unlabeled nuclei by flow sorting, effectively eliminating contamination issues that can result from sorting on DNA content alone. We also developed an analysis pipeline for analyzing and classifying regions of replication and present it in a point-and-click application called Repliscan that eliminates the need for command line programming.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Klein KN, Gilbert DM (2016) Epigenetic vs. sequence-dependent control of eukaryotic replication timing. In: Kaplan D (ed) The initiation of DNA replication in eukaryotes. Springer International Publishing, pp 39–63
Marchal C, Sima J, Gilbert DM (2019) Control of DNA replication timing in the 3D genome. Nat Rev Mol Cell Biol 20(12):721–737
Gilbert DM, Takebayashi S-I, Ryba T, Lu J, Pope BD, Wilson KA, Hiratani I (2010) Space and time in the nucleus: developmental control of replication timing and chromosome architecture. Cold Spring Harb Symp Quant Biol 75:143–153
Schubeler D, Scalzo D, Kooperberg C, van Steensel B, Delrow J, Groudine M (2002) Genome-wide DNA replication profile for Drosophila melanogaster: a link between transcription and replication timing. Nat Genet 32(3):438–442
Woodfine K, Fiegler H, Beare DM, Collins JE, McCann OT, Young BD, Debernardi S, Mott R, Dunham I, Carter NP (2004) Replication timing of the human genome. Hum Mol Genet 13(2):191–202
Hiratani I, Gilbert DM (2009) Replication timing as an epigenetic mark. Epigenetics 4(2):93–97
Schwaiger M, Stadler MB, Bell O, Kohler H, Oakeley EJ, Schubeler D (2009) Chromatin state marks cell-type- and gender-specific replication of the Drosophila genome. Genes Dev 23(5):589–601
Hansen RS, Thomas S, Sandstrom R, Canfield TK, Thurman RE, Weaver M, Dorschner MO, Gartler SM, Stamatoyannopoulos JA (2010) Sequencing newly replicated DNA reveals widespread plasticity in human replication timing. Proc Natl Acad Sci U S A 107(1):139–144
Eaton ML, Prinz JA, MacAlpine HK, Tretyakov G, Kharchenko PV, MacAlpine DM (2011) Chromatin signatures of the Drosophila replication program. Genome Res 21(2):164–174
Lubelsky Y, Prinz JA, DeNapoli L, Li Y, Belsky JA, MacAlpine DM (2014) DNA replication and transcription programs respond to the same chromatin cues. Genome Res 24(7):1102–1114
Gilbert DM (2002) Replication timing and transcriptional control: beyond cause and effect. Curr Opin Cell Biol 14:377–383
Ryba T, Hiratani I, Lu J, Itoh M, Kulik M, Zhang J, Schulz TC, Robins AJ, Dalton S, Gilbert DM (2010) Evolutionarily conserved replication timing profiles predict long-range chromatin interactions and distinguish closely related cell types. Genome Res 20(6):761–770
Yaffe E, Farkash-Amar S, Polten A, Yakhini Z, Tanay A, Simon I (2010) Comparative analysis of DNA replication timing reveals conserved large-scale chromosomal architecture. PLoS Genet 6(7):e1001011
Pope BD, Ryba T, Dileep V, Yue F, Wu W, Denas O, Vera DL, Wang Y, Hansen RS, Canfield TK, Thurman RE, Cheng Y, Gulsoy G, Dennis JH, Snyder MP, Stamatoyannopoulos JA, Taylor J, Hardison RC, Kahveci T, Ren B, Gilbert DM (2014) Topologically associating domains are stable units of replication-timing regulation. Nature 515(7527):402–405
Rivera-Mulia JC, Buckley Q, Sasaki T, Zimmerman J, Didier RA, Nazor K, Loring JF, Lian Z, Weissman S, Robins AJ, Schulz TC, Menendez L, Kulik MJ, Dalton S, Gabr H, Kahveci T, Gilbert DM (2015) Dynamic changes in replication timing and gene expression during lineage specification of human pluripotent stem cells. Genome Res 25(8):1091–1103
Agier N, Delmas S, Zhang Q, Fleiss A, Jaszczyszyn Y, van Dijk E, Thermes C, Weigt M, Cosentino-Lagomarsino M, Fischer G (2018) The evolution of the temporal program of genome replication. Nat Commun 9(1):2199
Thorpe SD, Charpentier M (2017) Highlight on the dynamic organization of the nucleus. Nucleus 8(1):2–10
Shultz RW, Tatineni VM, Hanley-Bowdoin L, Thompson WF (2007) Genome-wide analysis of the core DNA replication machinery in the higher plants Arabidopsis and rice. Plant Physiol 144(4):1697–1714
Gnan S, Flyamer IM, Klein KN, Castelli E, Rapp A, Maiser A, Chen N, Weber P, Enervald E, Cardoso MC, Bickmore WA, Gilbert DM, Buonomo SCB (2020) Nuclear organisation and replication timing are coupled through RIF1-PP1 interaction. bioRxiv. https://doi.org/10.1101/812156
Sreesankar E, Senthilkumar R, Barathi V, Mishra R, Mishra K (2012) Functional diversification of yeast telomere associated protein, Rif1, in higher eukaryotes. BMC Genomics 13:255
Wheeler E, Brooks AM, Concia L, Vera DL, Wear EE, LeBlanc C, Ramu U, Vaughn MW, Bass HW, Martienssen RA, Thompson WF, Hanley-Bowdoin L (2020) Arabidopsis DNA replication initiates in intergenic, AT-rich open chromatin. Plant Physiol 183(1):206–220
Bass HW, Hoffman GG, Lee TJ, Wear EE, Joseph SR, Allen GC, Hanley-Bowdoin L, Thompson WF (2015) Defining multiple, distinct, and shared spatiotemporal patterns of DNA replication and endoreduplication from 3D image analysis of developing maize (Zea mays L.) root tip nuclei. Plant Mol Biol 89(4–5):339–351
Savadel SD, Bass HW (2017) Take a look at plant DNA replication: recent insights and new questions. Plant Signal Behav 12(4):e1311437
Mickelson-Young L, Wear E, Mulvaney P, Lee TJ, Szymanski ES, Allen G, Hanley-Bowdoin L, Thompson W (2016) A flow cytometric method for estimating S-phase duration in plants. J Exp Bot 67(21):6077–6087
Lee TJ, Pascuzzi PE, Settlage SB, Shultz RW, Tanurdzic M, Rabinowicz PD, Menges M, Zheng P, Main D, Murray JA, Sosinski B, Allen GC, Martienssen RA, Hanley-Bowdoin L, Vaughn MW, Thompson WF (2010) Arabidopsis thaliana chromosome 4 replicates in two phases that correlate with chromatin state. PLoS Genet 6(6):e1000982
Wear EE, Song J, Zynda GJ, LeBlanc C, Lee TJ, Mickelson-Young L, Concia L, Mulvaney P, Szymanski ES, Allen GC, Martienssen RA, Vaughn MW, Hanley-Bowdoin L, Thompson WF (2017) Genomic analysis of the DNA replication timing program during mitotic S phase in maize (Zea mays) root tips. Plant Cell 29(9):2126–2149
Concia L, Brooks AM, Wheeler E, Zynda GJ, Wear EE, LeBlanc C, Song J, Lee TJ, Pascuzzi PE, Martienssen RA, Vaughn MW, Thompson WF, Hanley-Bowdoin L (2018) Genome-wide analysis of the Arabidopsis replication timing program. Plant Physiol 176(3):2166–2185
Wear EE, Song J, Zynda GJ, Mickelson-Young L, LeBlanc C, Lee TJ, Deppong DO, Allen GC, Martienssen RA, Vaughn MW, Hanley-Bowdoin L, Thompson WF (2020) Comparing DNA replication programs reveals large timing shifts at centromeres of endocycling cells in maize roots. PLoS Genet 16(10):e1008623
Zynda GJ, Song JW, Concia L, Wear EE, Hanley-Bowdoin L, Thompson WF, Vaughn MW (2017) Repliscan: a tool for classifying replication timing regions. BMC Bioinform 18:1–14
Bass HW, Wear EE, Lee TJ, Hoffman GG, Gumber HK, Allen GC, Thompson WF, Hanley-Bowdoin L (2014) A maize root tip system to study DNA replication programmes in somatic and endocycling nuclei during plant development. J Exp Bot 65(10):2747–2756
Galbraith DW, Harkins KR, Maddox JM, Ayres NM, Sharma DP, Firoozabady E (1983) Rapid flow cytometric analysis of the cell cycle in intact plant tissues. Science 220(4601):1049–1051
Wear EE, Concia L, Brooks AM, Markham EA, Lee TJ, Allen GC, Thompson WF, Hanley-Bowdoin L (2016) Isolation of plant nuclei at defined cell cycle stages using EdU labeling and flow cytometry. Methods Mol Biol 1370:69–86
Salic A, Mitchison TJ (2008) A chemical method for fast and sensitive detection of DNA synthesis in vivo. Proc Natl Acad Sci U S A 105(7):2415–2420
Kotogany E, Dudits D, Horvath GV, Ayaydin F (2010) A rapid and robust assay for detection of S-phase cell cycle progression in plant cells and tissues by using ethynyl deoxyuridine. Plant Methods 6(5)
Dolezel J, Cihalikova J, Weiserova J, Lucretti S (1999) Cell cycle synchronization in plant root meristems. Methods Cell Sci 21:95–107
Planchais S, Glab N, Inze D, Bergounioux C (2000) Chemical inhibitors: a tool for plant cell cycle studies. FEBS Lett 476:78–83
Menges M, Murray JA (2002) Synchronous Arabidopsis suspension cultures for analysis of cell-cycle gene activity. Plant J 30(2):203–212
Lee M, Phillips RL (1988) The chromosomal basis of somaclonal variation. Annu Rev Plant Physiol Plant Mol Biol 39:413–437
Phillips RL, Kaeppler SM, Olhoft P (1994) Genetic instability of plant tissue cultures: breakdown of normal controls. Proc Natl Acad Sci U S A 91(12):5222–5226
Tanurdzic M, Vaughn MW, Jiang H, Lee TJ, Slotkin RK, Sosinski B, Thompson WF, Doerge RW, Martienssen RA (2008) Epigenomic consequences of immortalized plant cell suspension culture. PLoS Biol 6(12):2880–2895
Dolezel J, Greilhuber J, Suda J (2007) Estimation of nuclear DNA content in plants using flow cytometry. Nat Protoc 2(9):2233–2244
Gendrel AV, Lippman Z, Martienssen R, Colot V (2005) Profiling histone modification patterns in plants using genomic tiling microarrays. Nat Methods 2(3):213–218
Galbraith DW, Anderson MT, Herzenberg LA (1999) Flow cytometric analysis and FACS sorting of cells based on GFP accumulation. Methods Cell Biol 58:315–341
Galbraith DW, Bartos J, Dolezel J (2005) Flow cytometry and cell sorting in plant biotechnology. In: Skylar LA (ed) Flow cytometry for biotechnology. Oxford University Press, New York, pp 291–322
McCoy J (2002) Basic principles of flow cytometry. Hematol Oncol Clin North Am 16(2):229–243
Shapiro H (2003) Practical flow cytometry, 4th edn. Wiley-Liss, Hoboken, NJ
Picot J, Guerin CL, Le Van Kim C, Boulanger CM (2012) Flow cytometry: retrospective, fundamentals and recent instrumentation. Cytotechnology 64(2):109–130
Darzynkiewicz Z, Zhao H (2014) Cell cycle analysis by flow cytometry. In: Encyclopedia of life sciences. John Wiley & Sons, Ltd, Chichester
Wersto RP, Chrest FJ, Leary JF, Morris C, Stetler-Stevenson M, Gabrielson E (2001) Doublet discrimination in DNA cell-cycle analysis. Cytometry 46(5):296–306
Illumina (2017) Best practices for manually normalizing library concentrations. https://support.illumina.com/bulletins/2017/03/best-practices-for-manually-normalizing-library-concentrations.html. Accessed 21 Oct 2020
Li H (2013) Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. ArXiv 1303
Li H, Ruan J, Durbin R (2008) Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res 18(11):1851–1858
Broad-Institute (2016) Picard: a set of command line tools (in Java) for manipulating high-throughput sequencing (HTS) data and formats such as SAM/BAM/CRAM and VCF. https://broadinstitute.github.io/picard/. Accessed 20 Oct 2020
Krueger F (2012) Trim Galore!: a wrapper tool around Cutadapt and FastQC to consistently apply quality and adapter trimming to FastQ files. https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/. Accessed 20 Oct 2020
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, Genome Project Data Processing S (2009) The sequence alignment/map format and SAMtools. Bioinformatics 25(16):2078–2079
Ramirez F, Dundar F, Diehl S, Gruning BA, Manke T (2014) deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res 42(Web Server issue):W187–W191
Merchant N, Lyons E, Goff S, Vaughn M, Ware D, Micklos D, Antin P (2016) The iPlant collaborative: cyberinfrastructure for enabling data to discovery for the life sciences. PLoS Biol 14(1):e1002342
Diaz A, Park K, Lim DA, Song JS (2012) Normalization, bias correction, and peak calling for ChIP-seq. Stat Appl Genet Mol Biol 11(3):Article 9
Percival DB, Walden AT (2000) Wavelet methods for time series analysis. Cambridge University Press, Cambridge
Diermeier-Daucher S, Clarke ST, Hill D, Vollmann-Zwerenz A, Bradford JA, Brockhoff G (2009) Cell type specific applicability of 5-ethynyl-2′-deoxyuridine (EdU) for dynamic proliferation assessment in flow cytometry. Cytometry A 75(6):535–546
Kohlmeier F, Maya-Mendoza A, Jackson DA (2013) EdU induces DNA damage response and cell death in mESC in culture. Chromosome Res 21(1):87–100
Ross HH, Rahman M, Levkoff LH, Millette S, Martin-Carreras T, Dunbar EM, Reynolds BA, Laywell ED (2011) Ethynyldeoxyuridine (EdU) suppresses in vitro population expansion and in vivo tumor progression of human glioblastoma cells. J Neuro-Oncol 105(3):485–498
Zhao H, Halicka HD, Li JW, Biela E, Berniak K, Dobrucki J, Darzynkiewicz Z (2013) DNA damage signaling, impairment of cell cycle progression, and apoptosis triggered by 5-Ethynyl-2'-deoxyuridine incorporated into DNA. Cytometry A 83(11):979–988
Baluska F (1990) Nuclear size, DNA content, and chromatin condensation are different in individual tissues of the maize root apex. Protoplasma 158(1–2):45–52
Baluska F, Mancuso S (2013) Root apex transition zone as oscillatory zone. Front Plant Sci 4:Article 354
Rostovtsev VV, Green LG, Fokin VV, Sharpless KB (2002) A stepwise huisgen cycloaddition process: copper(I)-catalyzed regioselective “ligation” of azides and terminal alkynes. Angew Chem Int Ed 41(14):2596–2599
Endaya B, Cavanagh B, Alowaidi F, Walker T, de Pennington N, Ng JM, Lam PY, Mackay-Sim A, Neuzil J, Meedeniya AC (2016) Isolating dividing neural and brain tumour cells for gene expression profiling. J Neurosci Methods 257:121–133
Prutz W, Butler J, Land E (1990) Interaction of copper(I) with nucleic acids. Int J Radiat Biol 58(2)
Soares E, Hebbelinck K, Soares H (2003) Toxic effects caused by heavy metals in the yeast Saccharomyces cerevisiae: a comparative study. Can J Microbiol 49:336–343
Galbraith DW (1989) Analysis of higher plants by flow cytometry and cell sorting. Int Rev Cytol 116:165–228
Bauer K (1993) Quality control issues in DNA content flow cytometry. Ann N Y Acad Sci 677:59–77
Houtz B, Trotter J, Sasaki D (2004) BD FACService technotes: customer focused solutions. Customer Focused Solutions. https://static.bdbiosciences.com/documents/BD_Research_Sorting_TechBulletin.pdf. Accessed 22 Oct 2020
Schnable PS, Ware D, Fulton RS, Stein JC, Wei F, Pasternak S, Liang C, Zhang J, Fulton L, Graves TA, Minx P, Reily AD, Courtney L, Kruchowski SS, Tomlinson C, Strong C, Delehaunty K, Fronick C, Courtney B, Rock SM, Belter E, Du F, Kim K, Abbott RM, Cotton M, Levy A, Marchetto P, Ochoa K, Jackson SM, Gillam B, Chen W, Yan L, Higginbotham J, Cardenas M, Waligorski J, Applebaum E, Phelps L, Falcone J, Kanchi K, Thane T, Scimone A, Thane N, Henke J, Wang T, Ruppert J, Shah N, Rotter K, Hodges J, Ingenthron E, Cordes M, Kohlberg S, Sgro J, Delgado B, Mead K, Chinwalla A, Leonard S, Crouse K, Collura K, Kudrna D, Currie J, He R, Angelova A, Rajasekar S, Mueller T, Lomeli R, Scara G, Ko A, Delaney K, Wissotski M, Lopez G, Campos D, Braidotti M, Ashley E, Golser W, Kim H, Lee S, Lin J, Dujmic Z, Kim W, Talag J, Zuccolo A, Fan C, Sebastian A, Kramer M, Spiegel L, Nascimento L, Zutavern T, Miller B, Ambroise C, Muller S, Spooner W, Narechania A, Ren L, Wei S, Kumari S, Faga B, Levy MJ, McMahan L, Van Buren P, Vaughn MW, Ying K, Yeh CT, Emrich SJ, Jia Y, Kalyanaraman A, Hsia AP, Barbazuk WB, Baucom RS, Brutnell TP, Carpita NC, Chaparro C, Chia JM, Deragon JM, Estill JC, Fu Y, Jeddeloh JA, Han Y, Lee H, Li P, Lisch DR, Liu S, Liu Z, Nagel DH, McCann MC, SanMiguel P, Myers AM, Nettleton D, Nguyen J, Penning BW, Ponnala L, Schneider KL, Schwartz DC, Sharma A, Soderlund C, Springer NM, Sun Q, Wang H, Waterman M, Westerman R, Wolfgruber TK, Yang L, Yu Y, Zhang L, Zhou S, Zhu Q, Bennetzen JL, Dawe RK, Jiang J, Jiang N, Presting GG, Wessler SR, Aluru S, Martienssen RA, Clifton SW, McCombie WR, Wing RA, Wilson RK (2009) The B73 maize genome: complexity, diversity, and dynamics. Science 326(5956):1112–1115
Head SR, Komori HK, LaMere SA, Whisenant T, Van Nieuwerburgh F, Salomon DR, Ordoukhanian P (2014) Library construction for next-generation sequencing: overviews and challenges. Biotechniques 56(2):61–64
10x Genomics (2016) SPRIselect: DNA ratios affect the size range of library fragments. https://support.10xgenomics.com/genome-exome/index/doc/technical-note-spriselectdna-ratios-affect-the-size-range-of-library-fragments. Accessed 21 Oct 2020
Farouni R, Djambazian H, Ferri LE, Ragoussis J, Najafabadi HS (2020) Model-based analysis of sample index hopping reveals its widespread artifacts in multiplexed single-cell RNA-sequencing. Nat Commun 11(1):2704
Illumina (2017) Effects of index misassignment on multiplexing and downstream analysis. https://www.illumina.com/content/dam/illumina-marketing/documents/products/whitepapers/index-hopping-white-paper-770-2017-004.pdf. Accessed 23 Jul 2021
MacConaill LE, Burns RT, Nag A, Coleman HA, Slevin MK, Giorda K, Light M, Lai K, Jarosz M, McNeill MS, Ducar MD, Meyerson M, Thorner AR (2018) Unique, dual-indexed sequencing adapters with UMIs effectively eliminate index cross-talk and significantly improve sensitivity of massively parallel sequencing. BMC Genomics 19(1):30
Illumina (2018) Understanding unique dual indexes (UDI) and associated library prep kits. https://support.illumina.com/bulletins/2018/08/understanding-unique-dual-indexes%2D%2Dudi%2D%2Dand-associated-library-p.html. Accessed 21 Oct 2020
Illumina (2016) Converting ng/ul to nM when calculating dsDNA library concentration. https://support.illumina.com/bulletins/2016/11/converting-ngl-to-nm-when-calculating-dsdna-library-concentration-.html. Accessed 21 Oct 2020
Bronner IF, Quail MA, Swerdlow H, Turner DJ (2009) Improved protocols for the Illumina genome analyzer sequencing system. Curr Protoc Hum Genet 62:18.2.1–18.2.27. https://doi.org/10.1002/0471142905.hg1802s62
Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408(6814):796–815
Lee H, Lee KW, Lee T, Park D, Chung J, Lee C, Park WY, Son DS (2018) Performance evaluation method for read mapping tool in clinical panel sequencing. Genes Genomics 40(2):189–197
Illumina (2020) Illumina adapter sequences. https://support.illumina.com/downloads/illumina-adapter-sequences-document-1000000002694.html. Accessed 21 Oct 2020
Krueger F (2019) Taking appropriate QC measure for RRBS-type or other -Seq application with Trim Galore! https://github.com/FelixKrueger/TrimGalore/blob/master/Docs/Trim_Galore_User_Guide.md. Accessed 21 Oct 2020
Andrews S (2017) FastQC a quality control application for high throughput sequence data. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/. Accessed 20 Oct 2020
Van’t Hof J (1996) DNA replication in plants. In: DNA replication in eukaryotic cells. Cold Spring Harbor Press, Cold Spring Harbor, NY
Acknowledgments
This work was supported by grants from the NSF Plant Genome Research Program (NSF IOS-1025830 and IOS-2025811 to L.H.B.). Leigh Mickelson-Young and Emily E. Wear contributed equally to this work.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
1 Electronic Supplementary Material
Fig. S1
Trim Galore! Parameter window. The default parameters are entered, as well the fastqc checkbox selected to generate the FASTQC report. Any known Illumina adapters and index sequences should be entered in the Adapter sequence to be trimmed textbox. Trim Galore! will auto-detect adapters if this parameter is left blank (PDF 409 kb)
Fig. S2
BWA-MEM parameters and output options windows. The BWA-MEM default parameters (a) and default output options (b) are auto-populated (PDF 365 kb)
Fig. S3
SAMtools view parameters window. (a) The SAMtools view app recommended parameters are selected. (b) The output from Plot distribution of MAPQ is a plot of the mapping quality (MAPQ) distribution found in the alignment file before filtering (PDF 292 kb)
Fig. S4
Picard MarkDuplicates parameter window. The recommended parameters for marking and removing duplicate read alignments (PDF 186 kb)
Fig. S5
SAMtools Flagstat output window. SAMtools Flagstat generates mapping statistics outputted as a nameable .txt file (PDF 136 kb)
Fig. S6
DeepTools multiBamSummary parameters window. (a) The recommended Correlation type is selected. A desired static bin size (bins in base pairs) should also be selected. (b) The Pearson correlation coefficients between individual biological replicates for early, middle, and late S samples represented as a heatmap with a hierarchical clustering dendrogram (PDF 226 kb)
Fig. S8
Repliscan recommended parameters for maize B73 test dataset. For the maize test dataset, which has been filtered to remove alignments with MAPQ <10 and randomly downsampled, we recommend adjusting the Analysis bin size in base pairs as well as the remove parameter with the dependent parameter percentile cutoff. These setting adjustments are shown in the parameter window. See Notes 21 and 22 for a detailed description (PDF 220 kb)
Fig. S9
Repliscan input configuration example. (a) An input.txt configuration file is needed to assign individual files to analysis name labels. Individual files should be delimited by a tab. The name labels are used in output file naming and RT class segmentation naming. (b) Repliscan Inputs window requiring reference genome in Fasta format, a configuration file with a list of .bam files (input.txt), and a directory containing the .bam files (PDF 340 kb)
Fig. S10
Repliscan optional output example. Selecting the plot checkbox generates additional output files. (a) Included in these output files is a plot of the distribution of natural log transformed reads per bin in the G1 data and the selected cutoff (gray shaded area) from the remove and percentile cutoff parameters. For maize data that has been filtered to remove alignments with MAPQ <10, the distribution is negatively skewed (see Note 22). (b) An example of the .out file, which includes the parameter settings used and the auto-tuned RT class segmentation thresholds for individual chromosomes (see Subheading 3.11, step 8). The plots shown represent the files from the maize B73 test dataset with recommended parameters (Fig. S8) (PDF 520 kb)
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Science+Business Media, LLC, part of Springer Nature
About this protocol
Cite this protocol
Mickelson-Young, L., Wear, E.E., Song, J., Zynda, G.J., Hanley-Bowdoin, L., Thompson, W.F. (2022). A Protocol for Genome-Wide Analysis of DNA Replication Timing in Intact Root Tips. In: Caillaud, MC. (eds) Plant Cell Division. Methods in Molecular Biology, vol 2382. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-1744-1_3
Download citation
DOI: https://doi.org/10.1007/978-1-0716-1744-1_3
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-1743-4
Online ISBN: 978-1-0716-1744-1
eBook Packages: Springer Protocols