Abstract
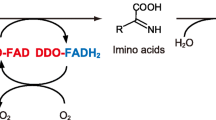
This chapter describes a method to assay the activity of reactive intermediate deaminases (Rid), a large family of conserved soluble enzymes, which have been proposed to prevent damages from metabolic intermediates such as the highly reactive and unstable compounds enamines/imines. In this method, the flavin adenine dinucleotide-dependent l- or d-amino acid oxidases generate an imino acid starting from a l- or d- amino acid, respectively. This reaction is coupled to the hydrolysis of the imino acid to the corresponding α-keto acid and ammonium ion catalyzed by a Rid enzyme. The spectrophotometric assay consists of measuring the decrease of the initial rate of formation of the semicarbazone, derived from the spontaneous reaction of the imino acid and semicarbazide, caused by the presence of the Rid enzyme. The set-up and testing of this method imply a preliminary characterization of the ability of the amino acid oxidase to release the imino acid required for the subsequent reactions. To this purpose, the activity of the l- or d-amino acid oxidases with different amino acids can be measured as production of hydrogen peroxide or formation of semicarbazone in parallel assays. The advantages and limitations of this assay of Rid activity are discussed.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
de Crecy-Lagard V, Haas D, Hanson AD (2018) Newly-discovered enzymes that function in metabolite damage-control. Curr Opin Chem Biol 47:101–108
Frelin O, Huang L, Hasnain G et al (2015) A directed-overflow and damage-control N-glycosidase in riboflavin biosynthesis. Biochem J 466(1):137–145
Linster CL, Van Schaftingen E, Hanson AD (2013) Metabolite damage and its repair or pre-emption. Nat Chem Biol 9(2):72–80
Peracchi A, Veiga-da-Cunha M, Kuhara T et al (2017) Nit1 is a metabolite repair enzyme that hydrolyzes deaminated glutathione. Proc Natl Acad Sci U S A 114(16):E3233–E3242
Borchert AJ, Ernst DC, Downs DM (2019) Reactive enamines and imines in vivo: lessons from the RidA paradigm. Trends Biochem Sci. https://doi.org/10.1016/j.tibs.2019.04.011
Lambrecht JA, Flynn JM, Downs DM (2012) Conserved YjgF protein family deaminates reactive enamine/imine intermediates of pyridoxal 5′-phosphate (PLP)-dependent enzyme reactions. J Biol Chem 287(5):3454–3461
Lambrecht JA, Schmitz GE, Downs DM (2013) RidA proteins prevent metabolic damage inflicted by PLP-dependent dehydratases in all domains of life. MBio 4(1):e00033–e00013
Degani G, Barbiroli A, Regazzoni L et al (2018) Imine deaminase activity and conformational stability of UK114, the mammalian member of the Rid protein family active in amino acid metabolism. Int J Mol Sci 19(4):945–963
Borchert AJ, Downs DM (2017) The response to 2-aminoacrylate differs in Escherichia coli and Salmonella enterica, despite shared metabolic components. J Bacteriol 199(14):e00140–e00117
Borchert AJ, Downs DM (2017) Endogenously generated 2-aminoacrylate inhibits motility in Salmonella enterica. Sci Rep 7(1):12971
Flynn JM, Downs DM (2013) In the absence of RidA, endogenous 2-aminoacrylate inactivates alanine racemases by modifying the pyridoxal 5′-phosphate cofactor. J Bacteriol 195(16):3603–3609
Flynn JM, Christopherson MR, Downs DM (2013) Decreased coenzyme A levels in RidA mutant strains of Salmonella enterica result from inactivated serine hydroxymethyltransferase. Mol Microbiol 89(4):751–759
Liu X, Zeng J, Chen X et al (2016) Crystal structures of RidA, an important enzyme for the prevention of toxic side products. Sci Rep 6:30494
Niehaus TD, Gerdes S, Hodge-Hanson K et al (2015) Genomic and experimental evidence for multiple metabolic functions in the RidA/YjgF/YER057c/UK114 (Rid) protein family. BMC Genomics 16:382
Hodge-Hanson KM, Downs DM (2017) Members of the Rid protein family have broad imine deaminase activity and can accelerate the Pseudomonas aeruginosa D-arginine dehydrogenase (DauA) reaction in vitro. PLoS One 12(9):e0185544
Burman JD, Stevenson CE, Sawers RG et al (2007) The crystal structure of Escherichia coli TdcF, a member of the highly conserved YjgF/YER057c/UK114 family. BMC Struct Biol 7:30
Bartorelli A, Bussolati B, Millesimo M et al (1996) Antibody-dependent cytotoxic activity on human cancer cells expressing UK 114 tumor membrane antigen. Int J Oncol 8(3):543–548
Bartorelli A, Biancardi C, Cavalca V et al (1996) Purification and partial characterization of proteins present in a perchloric acid extract of goat liver (UK101). J Tumor Marker Oncol 11(1):57–61
Bussolati G, Geuna M, Bussolati B et al (1997) Cytolytic and tumor inhibitory antibodies against UK114 protein in the sera of cancer patients. Int J Oncol 10:779–785
Lambrecht JA, Browne BA, Downs DM (2010) Members of the YjgF/YER057c/UK114 family of proteins inhibit phosphoribosylamine synthesis in vitro. J Biol Chem 285(45):34401–34407
Ernst DC, Lambrecht JA, Schomer RA et al (2014) Endogenous synthesis of 2-aminoacrylate contributes to cysteine sensitivity in Salmonella enterica. J Bacteriol 196(18):3335–3342
Niehaus TD, Nguyen TN, Gidda SK et al (2014) Arabidopsis and maize RidA proteins preempt reactive enamine/imine damage to branched-chain amino acid biosynthesis in plastids. Plant Cell 26(7):3010–3022
Hafner EW, Wellner D (1979) Reactivity of the imino acids formed in the amino acid oxidase reaction. Biochemistry 18(3):411–417
Campillo-Brocal JC, Lucas-Elio P, Sanchez-Amat A (2015) Distribution in different organisms of amino acid oxidases with FAD or a quinone as cofactor and their role as antimicrobial proteins in marine bacteria. Mar Drugs 13(12):7403–7418
Geueke B, Hummel W (2003) Heterologous expression of Rhodococcus opacus L-amino acid oxidase in Streptomyces lividans. Protein Expr Purif 28(2):303–309
Amano M, Mizuguchi H, Sano T et al (2015) Recombinant expression, molecular characterization and crystal structure of antitumor enzyme, L-lysine α-oxidase from Trichoderma viride. J Biochem 157(6):549–559
Arima J, Tamura T, Kusakabe H et al (2003) Recombinant expression, biochemical characterization and stabilization through proteolysis of an L-glutamate oxidase from Streptomyces sp. X-119-6. J Biochem 134(6):805–812
Hossain GS, Li J, Shin HD et al (2014) L-Amino acid oxidases from microbial sources: types, properties, functions, and applications. Appl Microbiol Biotechnol 98(4):1507–1515
Pollegioni L, Molla G (2011) New biotech applications from evolved D-amino acid oxidases. Trends Biotechnol 29(6):276–283
Subramanian K, Gora A, Spruijt R et al (2018) Modulating D-amino acid oxidase (DAAO) substrate specificity through facilitated solvent access. PLoS One 13(6):e0198990
Xu XL, Grant GA (2016) Mutagenic and chemical analyses provide new insight into enzyme activation and mechanism of the type 2 iron-sulfur L-serine dehydratase from Legionella pneumophila. Arch Biochem Biophys 596:108–117
Takahashi S, Abe K, Kera Y (2015) Bacterial D-amino acid oxidases: recent findings and future perspectives. Bioengineered 6(4):237–241
Curti B, Ronchi S, Branzoli U et al (1973) Improved purification, amino acid analysis and molecular weight of homogenous D-amino acid oxidase from pig kidney. Biochim Biophys Acta 327(2):266–273
Hillebrand GG, Dye JL, Suelter CH (1979) Formation of an intermediate and its rate of conversion to pyruvate during the tryptophanase-catalyzed degradation of S-o-nitrophenyl-L-cysteine. Biochemistry 18(9):1751–1755
Vanoni MA, Curti B (2007) D-amino acid oxidase activity assays. In: Konno R (ed) D-amino acids: a new frontier in amino acid and protein research. Hauppauge, Nova Science Publishers, pp 467–476
Rosini E, Caldinelli L, Piubelli L (2017) Assays of D-amino acid oxidase activity. Front Mol Biosci 4:102
Bayse GS, Michaels AW, Morrison M (1972) The peroxidase-catalyzed oxidation of tyrosine. Biochim Biophys Acta 284(1):34–42
Cook PF, Cleland WW (2007) Enzyme kinetics and mechanism. Chapter 3 Enzyme assays. Garland Science, New York
Acknowledgments
The authors are grateful to Prof. Alberto Bartorelli Cusani and Dr. Francesco Baggi Sisini for the generous financial support to this research. We wish to thank Mr. Alessandro Lucini Paioni for the assistance in the enzymatic assays. S.D. is a recipient of a fellowship financed by Alalia S.r.l (Turin, Italy). G.D. is a recipient of a Post-Doctoral fellowship from the University of Milan (Italy). Stefania Digiovanni and Genny Degani contributed equally to this work.
Author information
Authors and Affiliations
Corresponding authors
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 Springer Science+Business Media, LLC, part of Springer Nature
About this protocol
Cite this protocol
Digiovanni, S., Degani, G., Popolo, L., Vanoni, M.A. (2021). Using d- and l-Amino Acid Oxidases to Generate the Imino Acid Substrate to Measure the Activity of the Novel Rid (Enamine/Imine Deaminase) Class of Enzymes. In: Barile, M. (eds) Flavins and Flavoproteins. Methods in Molecular Biology, vol 2280. Springer, New York, NY. https://doi.org/10.1007/978-1-0716-1286-6_13
Download citation
DOI: https://doi.org/10.1007/978-1-0716-1286-6_13
Published:
Publisher Name: Springer, New York, NY
Print ISBN: 978-1-0716-1285-9
Online ISBN: 978-1-0716-1286-6
eBook Packages: Springer Protocols